Abstract
The C-terminal region of Brh2 (Brh2CT), the BRCA2 homolog in Ustilago maydis, is highly conserved and aligns with the DSS1/DNA-binding domain (DBD) of mammalian BRCA2, while the N-terminal region (Brh2NT) is poorly conserved and has no obvious functional domain except for the single Rad51-interacting BRC element. Paradoxically, Brh2NT, but not Brh2CT, complements the DNA repair and recombination deficiency of the brh2 mutant. We show here that Brh2NT exhibits an unexpected DNA binding activity with properties similar to that of the full-length protein. Deletion mapping localized the region responsible for the DNA binding activity to a stretch of residues between the BRC element and the canonical DBD. A heterologous DNA-binding domain from the large subunit of replication protein A substituted for the endogenous binding region within Brh2NT in supporting DNA repair. Rad51-promoted strand invasion was stimulated by Brh2NT, but required the presence of the BRC element. The findings suggest a model in which Brh2NT serves as the principal site for association with DNA, while the Brh2CT provides a means for regulation.
The repair of damaged DNA by homologous recombination is a high fidelity and universal process for maintaining genome integrity. At the heart of it is the use of an undamaged homologous DNA sequence as a template to direct repair of damaged regions. In eukaryotes, Rad51 plays the key role of promoting the search for the homologous sequence (1). Rad51 assembles into a complex with single-stranded (ss)2 DNA that has been revealed after some processing of the damaged duplex to form a nucleoprotein filament, which is the active element for promoting homologous pairing and DNA strand exchange. BRCA2 is an important regulator of Rad51 activity (2, 3). Investigations using peptides or synthetic fusion proteins designed to model component elements of human BRCA2 suggest both positive and negative roles for BRCA2 in organizing Rad51 filaments (4–8).
Biochemical studies with Brh2, the BRCA2 homolog from the fungus Ustilago maydis, have demonstrated that it mediates Rad51 assembly on a protruding single-strand DNA tail (9). Interaction of Rad51 with Brh2 is through the single BRC element located in the N-terminal region of the protein (10). The BRC element comprises a sequence of about 30 amino acids (aa) that mimics a critical determinant of the Rad51 polymerization interface (11, 12). Rad51 also interacts with Brh2 through a second, poorly defined locus, the CRE, located at the extreme C terminus of Brh2 (13). These N-terminal and C-terminal elements communicate with each other in some as yet unknown way to support proper Rad51 filament assembly. Such communication requires an interplay that is coordinated by Dss1 (13), a small versatile acidic protein that interacts with Brh2 in a region corresponding to a segment of the DSS1/DNA-binding domain (DBD) in mammalian BRCA2 (14). The mammalian BRCA2 DBD domain consists of a tandem array of three oligonucleotide/oligosaccharide-binding (OB) folds and a helix-rich domain that is laced to the adjacent OB fold by intertwining with DSS1 (15). Sequence alignment with BRCA2 indicates that a region corresponding to part of the helix-rich domain and the adjacent OB1 and OB2 folds is conserved in Brh2, but that OB3 is absent. 36 of 40 residues in BRCA2 DBD that contact DSS1 are identical or conservatively substituted in Brh2 (14).
Mutants lacking Brh2 are extremely sensitive to DNA damage by UV, ionizing radiation, and other genotoxic agents, are deficient in recombination, and are defective in meiosis (16). Deletion of the gene encoding Dss1 results in a virtual phenocopy of the brh2 mutant (14). In view of the physical interaction between Brh2 and Dss1, the implication is that Dss1 serves as a positive activator for Brh2. Other studies also provide support for the notion that Dss1 serves as a regulator. Removal of a tract of 40 amino acids from the C terminus of Brh2 is enough to cause complete loss of function, as measured by lack of activity in complementing the UV sensitivity of the brh2 mutant. Yet paradoxically, by removing much longer tracts from the C terminus (with truncation points between residues 503 and 589) that delete the region corresponding to the Dss1/DNA-binding domain (the DBD), resistance to DNA damage is restored substantially when these severely truncated proteins are expressed not only in the brh2 mutant, but also in the dss1 mutant (17). This is surprising because the only obvious functional motif recognizable in the remnant N-terminal region of the protein is the BRC element and which is considered to serve in an essentially negative manner by blocking formation or destabilizing the Rad51 filament (4, 18, 19). These findings imply that the longer deletions remove a negative regulatory element from Brh2.
The large subunit of replication protein A (RPA70) has a modular arrangement of four domains including an N-terminal protein-protein interaction domain and three ssDNA-binding OB folds (20), but no residues in common with those in the BRCA2 DBD known from the crystal structure to contact Dss1. Replacing the DBD of Brh2 with the DNA-binding domain comprised of RPA70 OB folds A, B, and C creates a hybrid that is even more competent in restoring resistance to DNA damage and extremely elevated in recombination regardless of Dss1 status (17). These observations suggest that the N-terminal BRC-containing domain has an inherent ability to organize Rad51 and to support DNA repair independently of Dss1, but that the addition of a domain with strong single-strand DNA binding activity enhances the Rad51-organizing activity. Analogous findings were made in a mammalian system in which a synthetic protein consisting of BRCA2 BRC elements fused to RPA70 was found to suppress the DNA repair and recombination defects of BRCA2-deficient cells (6). Based on these observations it can be imagined that a Rad51-interaction element coupled with a single-strand-specific DNA binding activity constitutes a primitive mechanism to enable mediator function.
Biochemical testing with the mammalian BRCA2 DBD (15) and studies using a synthetic construct that consisted of the human BRCA2 DBD fused with a polypeptide spanning BRC3 and BRC4 confirmed the DNA binding ability and preference for single-stranded DNA of the BRCA2 DBD (7), as was predicted based on its co-crystal structure with oligothymidylate (oligo(dT9)). In experiments with a synthetic version of BRCA2 consisting of the region spanning BRC3 and BRC4 fused to the DBD, stimulation of Rad51 nucleation onto RPA-coated single-stranded DNA was shown to depend on both the Rad51 interaction ability of the BRC domain and DNA binding activity of the DBD (7).
A simple mediator model would suggest that Brh2 binds Rad51 by interaction with the BRC element and ferries it to single-stranded DNA through the DNA binding function in the DBD. Yet our observations that the N-terminal region of Brh2 can restore resistance to DNA damage and recombination proficiency in the absence of the canonical Dss1/OB fold-containing DNA-binding domain and without dependence on Dss1 (17) suggest that this part of the protein has some inherent ability to enhance Rad51 filament formation. A simple explanation is that the N-terminal region of Brh2 contains an unrecognized DNA-binding domain that, in concert with the BRC, mediates delivery of Rad51 to damaged sites even in the absence of the apparently canonical Dss1/DNA-binding domain. In the present investigation we tested the idea that the N-terminal region of Brh2 has an inherent DNA binding ability.
EXPERIMENTAL PROCEDURES
U. maydis Genetic Methods—Manipulations of U. maydis strains, culture methods, gene transfer procedures, and survival after UV and γ radiation have been described previously (13, 21). U. maydis strains included UCM350 (wild type for recombination function), UCM565 (brh2), and UCM54 (rec2). Brh2 derivatives used in complementation studies and protein expression were prepared by PCR amplification of regions as specified in figures using the cloned Brh2 gene as template. Similarly, a region from RPA70 spanning OB folds A and B (codons 144–436) was amplified from the cloned gene and fused in-frame with a fragment encoding Brh2 residues 1–373. Brh2 and derivatives were expressed under control of the moderate glyceraldehyde-3-phosphate dehydrogenase promoter on self-replicating plasmids.
Protein Purification—Brh2 and Brh2CT proteins tagged with maltose-binding protein (MBP) in complex with His-tagged Dss1 were purified after overexpression in Escherichia coli as described previously (13, 22). Brh2NT terminating at residue Met-551 and other truncated forms of the Brh2NT deleted of N- and C-terminal residues were purified as fusion proteins with an N-terminal MBP tag and C-terminal His tag. Purification followed the same protocol developed for the full-length protein involving sequential affinity chromatography on Ni2+-NTA (nitrilotriacetic acid-agarose), amylose resin, and monoQ beads, but included an additional step of salt gradient elution from HiTrap heparin-agarose (GE Healthcare). Brh2NT protein devoid of affinity tag was prepared from a fusion protein with an N-terminal MBP tag and C-terminal His tag, which was engineered to contain a tobacco etch virus (TEV) protease recognition site in the linker regions connecting the MBP and His tags to Brh2NT. After cleavage with TEV protease, Brh2CT was purified by salt gradient elution from monoQ resin using an AKTA FPLC chromatography system (GE Healthcare). Protein was stored in 50 mm Tris-HCl, pH 7.5, 200 mm KCl, 1 mm dithiothreitol, 1 mm EDTA, 10% glycerol. His-tagged Dss1 was purified after expression in E. coli. The procedure included affinity purification on Ni2+-NTA, gradient elution from monoQ beads, and molecular sieve chromatography on Superose 6 gel filtration column. TEV protease engineered to be highly stabilized was purified from an overproducing E. coli strain (23) transformed with pRK793 graciously provided by David S. Waugh (NCI, National Institutes of Health, Frederick, MD). Mouse BRCA2 DBD was generously provided by Jie Fan (Nikola Pavletich Laboratory, Memorial Sloan-Kettering Cancer Center, New York, NY).
DNA Dynamics—DNA binding and D-loop formation were assayed using labeled oligonucleotides by the methods described previously (22, 24). For DNA binding by electrophoretic mobility shift assay, reactions were performed with 3.3 nm DNA in 25 mm HEPES, pH 7.5, 55 mm KCl, 2 mm MgCl2, at 37 °C for 15 min. Glutaraldehyde was added to 0.2% and after an additional 10 min, 0.1 m Tris-HCl, pH 8.0, was added. Products were examined after electrophoresis at room temperature in 1% agarose gels cast in 40 mm Tris acetate, pH 7.6, 1 mm EDTA. Oligonucleotide substrates design was the same as in the previous studies. For analysis of conformational and structural specificity in Brh2, binding oligonucleotides were labeled at the 5′-end with 32P, and images of gels were determined by phosphorimaging. The fraction of DNA in complexes was determined using ImageQuaNT software (Molecular Dynamics). For standard binding comparisons an ss60mer oligonucleotide (derived from phage φX174 sequence 103–162) labeled at the 5′-end with the fluorescent dye IRD800 (MWG Biotech AG) was used as substrate. For D-loop reactions an ss80mer oligonucleotide (derived from pBluescript II sequence 1–80) was used as substrate. Gel images were collected with an Odyssey infrared detection platform (LI-COR Biosciences). All DNA concentrations are expressed in molecules, rather than nucleotide.
RESULTS
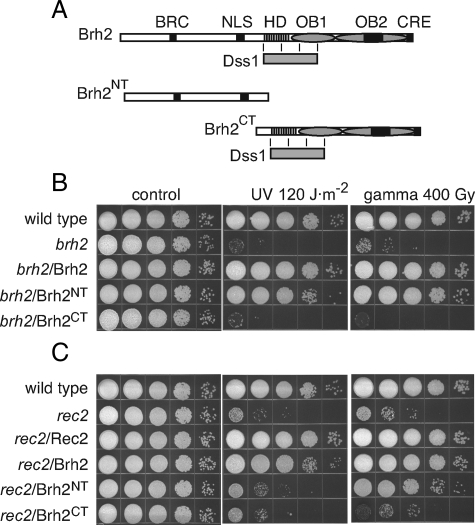
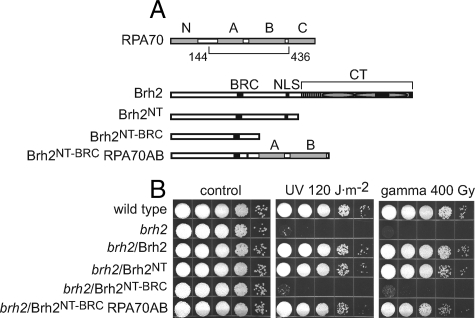
Biological Activity of Brh2 Lacking the DBD—To define in more detail the biological capabilities of Brh2 N-terminal domain we compared a representative truncation mutant with the DBD removed (Brh21–551 or Brh2NT hereafter) with a Brh2 truncation expressing only the DBD (Brh2505–1075 or Brh2CT hereafter) for ability to complement radiation sensitivity when expressed in a brh2-null mutant. Schematic representations are shown with approximate locations of the Rad51-interacting elements BRC and CRE, a putative nuclear localization signal, the helix-rich domain HD and the two OB folds corresponding to the canonical DBD of BRCA2, and Dss1 with its region of interaction in Brh2 (Fig. 1A). At doses of UV and γ radiation that resulted in about the same degree of killing of the brh2D mutant, Brh2NT exhibited a level of complementation approaching that of the full-length Brh2, while Brh2CT showed little activity (Fig. 1B). These results demonstrate that the Brh2NT is active in repair of DNA lesions introduced as a consequence of both UV and ionizing radiation.
FIGURE 1.
Domain mapping by rescue of radiation resistance. A, schematic illustration of the domain maps of Brh2 (1075 aa), Brh2NT (551 aa), and Brh2CT (570 aa). Dss1 (gray bar) is shown below Brh2 and Brh2CT with the approximate boundaries of the interacting region. Rad51-interacting elements BRC and CRE; putative nuclear localization signal NLS: black bars. HD: vertical hatch. OB1 and OB2 (ovals). Tower insert in OB2, black bar. B and C, survival of the indicated strains after treatment with UV or γ-rays. brh2 or rec2 mutant cells were transformed as indicated with self-replicating plasmids expressing Brh2, Brh2NT, Brh2CT, or Rec2. Cell suspensions were adjusted to ∼2 × 107 per ml, diluted in 10-fold serial dilutions, and aliquots (10 μl) of each were spotted from left to right as shown. Survival is indicated as growth of colonies 3 days after irradiation.
To test the actions of Brh2NT further, we examined its ability to suppress the radiation sensitivity of rec2. Rec2 is the single Rad51 paralog in U. maydis, and mutants depleted of it are also sensitive to UV and ionizing radiation. Unlike dss1, radiation resistance of the rec2 mutant can be substantially restored by expressing the full-length Brh2 (10) ectopically from a moderately strong, constitutive promoter. Here we found that Brh2NT was also effective in restoring resistance of rec2 to γ radiation (Fig. 1C). Nevertheless, Brh2NT was not comparable in activity in restoring resistance to UV, suggesting that the response to DNA lesions arising as a consequence of UV damage is not the same by full-length Brh2 and Brh2NT in the absence of Rec2 function. In contrast, Brh2CT showed little to no ability to suppress the sensitivity of rec2 to UV or γ radiation. The basis for how Brh2 restores radiation resistance to rec2 is not known, but one suggestion is that by increasing the level of Brh2, Rad51 and auxiliary factors can access DNA lesions in a manner ordinarily dependent on Rec2 function. That the response of Brh2NT to UV versus γ-ray-induced damage differs from full-length Brh2 most likely reflects differences in the nature of the lesions, and how they are processed. Single-stranded gaps arise as a consequence of DNA replication past UV photoproducts (25), while double-strand breaks result from ionizing radiation. When promoted by Brh2NT, repair of the former is dependent on Rec2, but apparently not the latter. Collectively, these results confirm and extend the conclusions that the N-terminal region of Brh2 indeed has an inherent biological activity comparable to a large degree with the full-length Brh2 protein and that the canonical DNA-binding domain is to some extent dispensable for function. A simple explanation to account for the biological function is that Brh2NT is still able to bind DNA through an unrecognized DNA-binding domain.
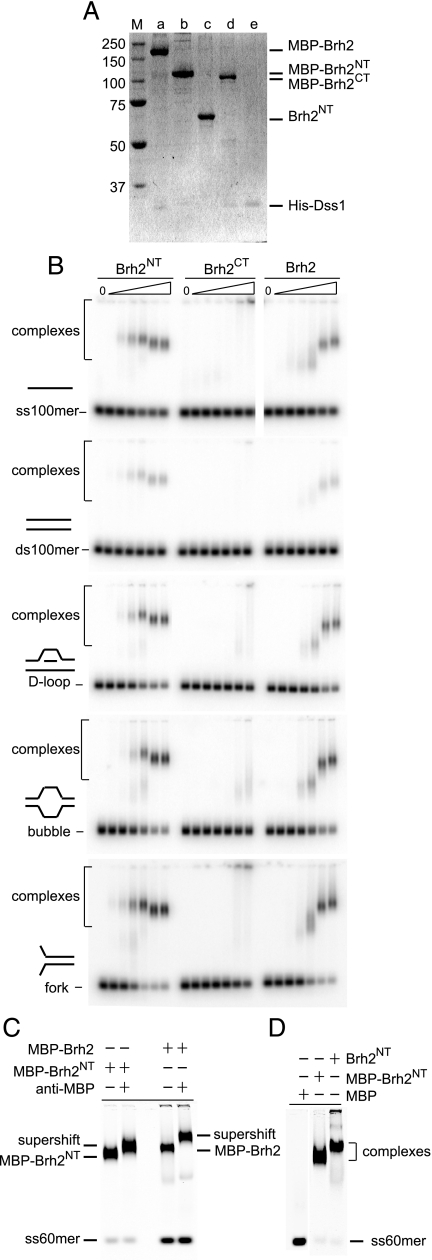
DNA Binding Activity within Brh2NT—We overproduced and purified Brh2NT and Brh2CT separately to compare them directly for DNA binding activity. As an aid to purification the proteins were fused to an MBP affinity tag. Full-length Brh2 and Brh2CT were necessarily produced as heterotypic complexes with Dss1 (His-tagged) (13), but are referred to below as Brh2 and Brh2CT, respectively, for simplicity unless stated otherwise (Fig. 2A). As previous analysis showed that biological activity of Brh2 was not compromised by the presence of the MBP tag, nor that of Dss1 by the His tag (13, 17), we used the tagged proteins in the experiments reported below, unless indicated differently.
FIGURE 2.
DNA binding activity associated with the N-terminal region of Brh2. A, proteins in the analysis. SDS-polyacrylamide gel (10%) with ∼3 pmol each of the indicated proteins run alongside a ladder of Precision Plus protein standards (BioRad) after staining with Simplyblue Safe stain (Invitrogen), lane a, MBP-Brh2/His-Dss1; lane b, MBP-Brh2NT; lane c, Brh2NT with MBP tag removed by TEV protease cleavage; lane d, MBP-Brh2CT/His-Dss1; lane e, His-Dss1. B, mobility shift determinations for DNA binding were determined using 32P-labeled oligonucleotides in the indicated conformations with Brh2 or derivative truncated proteins in the following final concentrations from left to right: 0, 2, 4, 10, 20, 100, and 200 nm. C, supershift determination was performed by addition of anti-MBP monoclonal antibody (New England Biolabs) 15 min after initiating a standard DNA binding reaction with IRD800-labeled ss60mer and 200 nm Brh2 or Brh2NT. D, mobility shift determinations for DNA binding were determined using IRD800-labeled ss60mer with MBP alone, MBP-Brh2NT, or Brh2NT without the MBP tag at 100 nm.
DNA binding was measured using an electrophoretic mobility shift assay with 5′ 32P-labeled or 5′ IRD800 fluorophore-labeled oligonucleotides as substrate. Full-length Brh2, as noted previously (22), exhibited a marked preference for single-stranded versus duplex DNA and formed complexes that migrated in almost ladder-like fashion with progressively slower mobility as protein concentration increased (Fig. 2B), probably reflecting increasing mass of complexes as more and more protein molecules associated with individual DNA molecules. Brh2CT, comprising the canonical DBD, bound DNA with lower affinity-forming complexes that were not as discrete and that included a fraction that failed to enter the gel. To learn whether the MBP tag interfered with DNA binding in the case of Brh2CT, we removed the tag by specific proteolytic cleavage through an engineered TEV recognition site in the linker region fusing the MBP to Brh2CT, but found that the untagged protein was no different in DNA binding properties (data not shown). The Brh2NT fragment bound DNA with capacity and preference for single-stranded conformation matching the full-length protein (Fig. 2B). The complexes migrated somewhat differently from those formed with full-length Brh2. Absent was the ladder of slower mobility complexes. Instead, with increasing protein concentration, the complexes ran with slightly faster mobility. The basis for this slightly increased mobility is not clear but could mean that the Brh2NT complexes assume a more compact form. Moreover, the mobility of the complexes formed with Brh2NT was about the same as with full-length Brh2 despite the substantial difference in the molecular mass between the two proteins. This suggests that the stoichiometry of protein bound per DNA molecule in the complexes is higher with Brh2NT compared with full-length Brh2 such that the total mass of the complexes formed in each case is about the same.
In previous studies on the binding specificity of Brh2, it was found that oligonucleotide model structures with D-loops, i.e. a duplex with an internal displaced ss loop, or branches were preferred substrates and were bound with higher affinity than ssDNA (22). A set of three of these substrates was used to test the specificity of the Brh2 truncations. These included a D-loop, a bubble, i.e. a duplex with an unpaired internal stretch, and a fork, i.e. a duplex with protruding unpaired 5′ and 3′-ss tails. Binding by Brh2CT was weak, and poorly distinguished complexes were formed; whereas in contrast the specificity and binding capacity of Brh2NT paralleled full-length Brh2 (Fig. 2B). As with ssDNA the Brh2NT complexes migrated slightly faster with increasing levels of protein. These studies suggest that the DNA substrate specificity exhibited by the full-length Brh2 protein is governed primarily by determinants within the N-terminal region of the protein.
To test whether the DNA binding activity observed with the Brh2NT was an inherent property of the Brh2NT protein and not due to a contaminant in the preparations, we used tag-specific antibodies to see if the complexes could be supershifted. Antibodies to the MBP affinity tag were added after formation of Brh2NT- or Brh2-ssDNA complexes, and the electrophoretic mobility was then analyzed. DNA-bound complexes formed with both full-length Brh2 and Brh2NT were shifted to a slower mobility (Fig. 2C) indicating that the protein present in the shifted DNA complexes carried the MBP tag.
To examine whether the DNA-binding activity was due to the MBP tag, we removed the tag by specific proteolytic cleavage through an engineered TEV recognition site in the linker region fusing the MBP to Brh2NT (Fig. 2A). This protein deleted of the MBP tag bound DNA with similar affinity as the tagged protein strongly supporting the notion that a DNA binding activity was inherent in the Brh2 N-terminal region (Fig. 2D). The complexes formed with the untagged Brh2NT ran with a slightly slower mobility compared with the tagged protein. The reason for this is not clear, but one possibility is that the MBP tag sterically hinders loading of adjacent Brh2NT molecules along the length of the oligonucleotide. Thus, without the MBP tag slightly more Brh2NT might be able to bind to the DNA to form a more slowly migrating complex. No mobility shift of the DNA substrate was observed in reactions containing MBP alone (data not shown). These controls show that a previously unrecognized DNA binding activity is associated with the N-terminal region of Brh2.
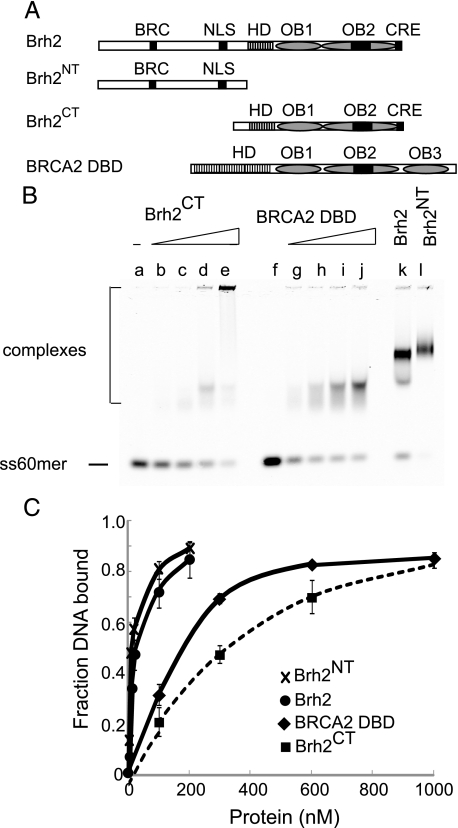
DNA Binding Activity of Brh2 Compared with BRCA2 DBD—In the seminal structural study describing the tandem arrangement of the helical domain and three OB folds of the mammalian BRCA2 DBD, the DNA binding activity and conformational preference for ssDNA were confirmed by direct biochemical experimentation (15). Protein-bound complexes with oligo(dT) in the range of 32–36 residues were reported to form avidly with 300 nm BRCA2 DBD. We compared murine BRCA2 DBD side-by-side with Brh2 and derivative proteins (Fig. 3A) using the ss60mer natural sequence DNA substrate. Under our reaction conditions with this longer DNA oligomer half of the DNA substrate was shifted by addition of 180 nm BRCA2 DBD to form complexes that were fairly uniform in mobility (Fig. 3B). By comparison 310 nm Brh2CT was required to shift half the input DNA. Complexes that formed were diffuse or else failed to enter the gel. The stronger binding by BRCA2 DBD could reflect a contribution from the third OB fold, which makes contacts in its binding groove with DNA according to the atomic structure (15) and which appears to be missing from Brh2. On the other hand a much lower concentration of Brh2 (20 nm) or Brh2NT (10 nm) was required to shift half the DNA, indicating that these proteins bound DNA tighter than BRCA2 DBD by more than an order of magnitude (Fig. 3C). These observations suggest that the primary site within Brh2 for associating with DNA resides in the N-terminal region and support the hypothesis that the biological activity manifested by the N-terminal region is due to the coupling of the BRC element to a DNA binding activity.
FIGURE 3.
Comparison of DNA binding activities. A, schematic representation of Brh2, Brh2NT, Brh2CT, and mouse BRCA2 DBD (736 amino acids) drawn approximately to scale. The mouse DBD has a more extensive HD and a third OB fold. B, mobility shift determinations shown for DNA binding were determined using IRD800-labeled ss60mer and increasing concentrations of Brh2CT and BRCA2 DBD as follows: lanes a, f, no protein; lanes b, g, 75 nm; lanes c, h, 300 nm; lanes d, i, 600 nm; lanes e, j, 1000 nm. Controls included lane k, 100 nm Brh2; lane l, 100 nm Brh2NT. C, binding results are presented graphically showing means of three independent determinations with standard deviations. Also shown for comparison are binding curves with Brh2 and Brh2NT at concentrations used in Fig. 2B.
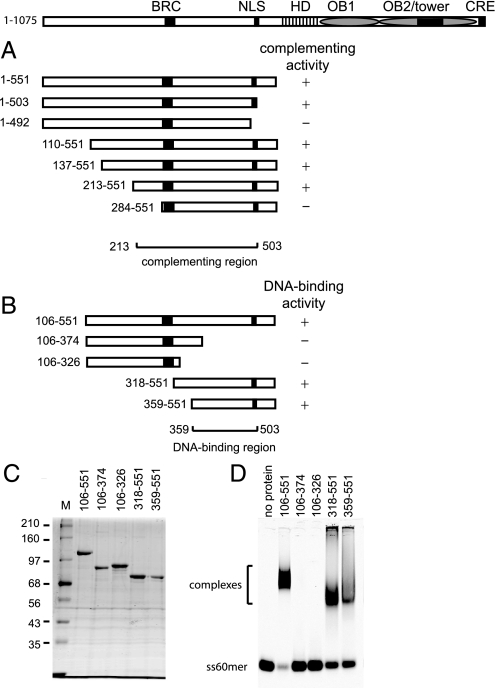
Mapping the Brh2NT DNA-binding Domain—In our previous genetic studies showing that biologically active variants of Brh2 independent of Dss1 could be obtained by truncating the entire DBD, we performed a deletion analysis to define the region within the N-terminal fragment responsible for complementing UV sensitivity of the brh2 mutant (17). Truncation from the N terminus was without effect upon removal of 213 residues, but when the deletion endpoint was within 10 residues of the BRC element there was >90% loss of activity (Fig. 4A). Starting at residue 551, truncation in the proximal direction was without effect up to residue 503, but truncation to residue 492, which eliminated the putative nuclear localization signal (KRPR 496–499) and cut into a motif (FVTPFK 488–493) shown to be critical for DMC1 interaction with human BRCA2 (26), caused >90% loss of activity.
FIGURE 4.
Mapping the DNA-binding domain within the Brh2 N-terminal region. A, activity of Brh2 gene fragments encoding the schematically depicted N-terminal polypeptides in complementing the UV sensitivity of brh2 (17). B, schematic illustration summarizing the results of Brh2 fragments tested for DNA binding. C, proteins in the analysis. SDS-polyacrylamide gel with ∼1.5 pmol each of the indicated Brh2 protein derivatives with coordinates as shown. D, mobility shift determinations for DNA binding were determined using IRD800-labeled ss60mer with Brh2 protein derivatives as shown in B at 100 nm.
Based on the findings that the complementing activity could be localized to a region between Brh2 residues 213–503, it seemed reasonable to assume that the DNA-binding domain would lie within the same sequence. To confirm this notion, we wished to test truncated Brh2 polypeptides directly for DNA binding activity. Therefore, we constructed a nested series of deletion alleles that spanned the sequence and expressed these in E. coli with the intention of purifying the protein product for biochemical analysis. Not all of the constructs overproduced protein and several that did produce protein products in forms that appeared badly degraded and were therefore of little use. This was particularly true of truncations that deleted residues from the distal end of Brh2NT (e.g. Brh2106–551 was overproduced as a soluble protein of uniform size while Brh2106–503 appeared as a polydisperse collection of truncated fragments). Nevertheless, four overlapping deletion products expressed well and were obtained in sufficient quantity to allow biochemical characterization (Fig. 4, B and C). When tested directly for DNA interaction, the N-terminal fragments Brh2318–551 and Brh2359–551 exhibited substantial binding activity. Interpolating from the complementation assay for biological activity, we surmise that the DNA-binding domain within the N-terminal region of Brh2 resides between residues 359–503.
Substituting the N-terminal DNA-binding Domain with a Heterologous Domain—When the 144-residue amino acid sequence harboring the N-terminal DNA-binding domain was used as query in a BLAST search no candidate with convincing similarity was identified. Inspection of the sequence, however, revealed it to be highly basic (pI 11.5) raising the possibility that interaction of DNA is promoted through a positively charged surface in this region of the protein. Nevertheless, from the BLAST search one weak hit (25% identity, 41% similarity, E: 8.1) was to a segment of a predicted prokaryotic endonuclease precursor. Using both of these sequences to jumpstart a PSI-BLAST search to provide a profile hidden Markov model (HMM), we performed a data base search with the HHpred server for HMM-HMM comparison (27). The top hits from this search consisted of OB fold domains (supplemental Fig. S1), which are characterized by five β-strands, but can be extremely divergent in amino acid sequence, often to the point where no domain is evident by sequence comparison. Given the possibility that the DNA-binding domain within the Brh2NT might be an OB fold, we decided to replace the sequence comprising it with a known OB fold domain to see if the N-terminal region could still perform biological function. We chose to use a sequence constituting the A and B OB folds from the 70-kDa subunit of the single-strand DNA-binding protein RPA (Fig. 5A), because the binding properties of the RPA70-AB polypeptide are well-established (20, 28). Brh2NT deleted of the DNA-binding region, but retaining the proximal sequence including the BRC element (residues 1–373, termed Brh2NT-BRC) was completely inactive in complementing the UV or γ-ray sensitivity of the brh2-null mutant (Fig. 5B). When this fragment, however, was fused to a region of RPA70 spanning the A and B OB folds (termed Brh2NT-BRC-RPA70AB), radiation resistance was restored to a level approaching that achieved by Brh2NT. These observations suggest that a threshold requirement for Brh2 function in DNA repair can be met by providing a BRC element coupled with a heterologous single-strand DNA-binding domain.
FIGURE 5.
Swapping the N-terminal DNA-binding domain. A, schematic illustration of the domain maps of RPA70 (623 aa), Brh2 (1075 aa), Brh2NT (551 aa), Brh2NT-BRC (373 aa), and Brh2NT-BRC-RPA70AB (654 aa). RPA70 OB folds N, A, B, and C were identified by alignment of the U. maydis RPA70 sequence (623 aa) with that of the human protein (616 aa) and are shown as gray rectangles. The DNA binding region spanning OB folds AB that was fused with the Brh2 N-terminal region is indicated. For Brh2 the Rad51-interacting element BRC and putative nuclear localization signal NLS are shown as black bars. The CT including helix-rich region, OB1, OB2 with tower, and CRE as in Fig. 1 are shown grouped. The junction where Brh2NT-BRC is fused with RPA70AB is shown as a narrow black bar. B, survival of the indicated strains after treatment with UV or γ-rays. brh2 mutant was transformed with self-replicating plasmids expressing proteins as indicated. Survival was determined as described in the legend to Fig. 1.
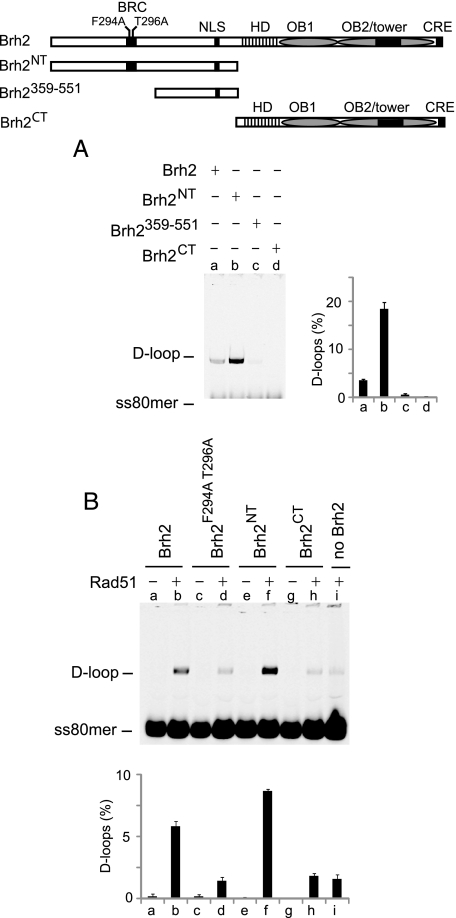
Rad51-independent and -dependent D-loop Activities Promoted by Brh2—Besides its role in mediating Rad51 nucleation on RPA-coated ssDNA, Brh2 was previously also found capable of promoting certain DNA-DNA interactions including annealing of complementary single strands, strand exchange over short lengths with oligonucleotide substrates, and a form of DNA strand invasion as measured by D-loop formation (22, 24). Unlike Rad51, the Brh2-promoted reactions have no requirement for ATP as a cofactor and no need for preincubation with ssDNA. We tested activity in D-loop formation by a standard assay using superhelical plasmid DNA (pBluescript II) and homologous ss80mer oligonucleotide as substrates, but without addition of the Rad51 cofactors ATP and divalent cation, which becomes inhibitory at levels of 4 mm and above. By this assay D-loops are detected by gel electrophoresis following deproteinization of reaction mixtures by a shift in mobility of the labeled 80mer as it is taken up by the plasmid DNA. It was surprising to observe that Brh2NT promoted D-loop formation to an extent about 4-fold greater than full-length Brh2 (Fig. 6A, lanes a and b). Only slight reaction was evident with Brh2359–551, and none was apparent with Brh2CT (Fig. 6A, lanes c and d). These findings show that the specificity for structure and conformation in DNA binding as well as the activity in promoting DNA-DNA transactions resides in and is directed by the N-terminal domain of Brh2, not the canonical DBD in the C-terminal region. Further, it appears that at least for D-loop formation the DNA binding activity within Brh2359–551 is missing some determinants necessary for efficient reaction.
FIGURE 6.
Promoting D-loop formation. A, D-loop formation by Brh2 or derivatives was measured in reactions using 18 nm pBluescript II plasmid DNA and 6 nm homologous IRD800-labeled ss80mer with addition of 200 nm Brh2 (lane a) or derivatives Brh2NT (lane b), Brh2359–551 (lane c), Brh2CT (lane d). D-loop formation was quantified and expressed as percent of ss60mer incorporated into product. Results are presented graphically showing means of three independent determinations with standard deviations. B, stimulation of Rad51-promoted D-loop formation. IRD800-labeled ss80mer (6 nm) was preincubated with Rad51 (600 nm), ATP (2 mm), and Ca2+ (4 mm). D-loop reactions were started by addition of pBluescript II plasmid DNA (18 nm) and Brh2 or derivative (200 nm) as indicated.
It was demonstrated previously that D-loop formation promoted by Rad51 could be stimulated by Brh2 even when Rad51 was added to saturating levels and that the stimulation was dependent on a functional BRC element (24). Optimal conditions required preincubating Rad51 with ssDNA to enable nucleoprotein filament formation and pre-incubating plasmid DNA with Brh2, presumably to enable formation of a protein-DNA complex. A mechanism envisioned was that Brh2 could help bring the Rad51-ssDNA filament to the plasmid DNA through physical association with both the plasmid DNA and Rad51. Thus, the recruitment would require functional DNA binding activity and ability to associate with Rad51.
Here we asked which DNA-binding domain in Brh2 was responsible for the activity in stimulating Rad51 D-loop formation. Brh2 derivatives were added to Rad51 reactions containing ATP and divalent cation at a level such that autonomous D-loop formation by Brh2 was nearly eliminated. Rad51 promoted D-loop formation to a modest level, and this was stimulated about four times when Brh2 was included (compare Fig. 6B, lanes b and i) consistent with previous findings (24). That a functional BRC element was required for stimulating D-loop formation by Rad51 was evident when the Brh2F294A T296A mutant was used. In this case no stimulation was noted (Fig. 6B, lane d). Stimulation was higher when Brh2NT was used in place of Brh2 (Fig. 6B, lane f). This shows that the DNA binding activity associated with the N-terminal domain is most likely capable of directing interaction with the recipient plasmid DNA and that the associated BRC element works to contribute the Rad51 interaction activity. Brh2CT was not effective in promoting D-loop formation with Rad51 (Fig. 6B, lane h). In conclusion, the stimulation of the Rad51-promoted strand invasion reaction requires a functional BRC element and the N-terminal DNA-binding domain but is independent of the canonical C-terminal DBD.
DISCUSSION
There are two principal conclusions to be drawn from this study. First, the N-terminal region of Brh2 contributes substantially to the biological function of Brh2. It houses a DNA-binding domain able to recognize specific structural conformations of DNA and to promote DNA-DNA intermolecular transactions that were previously attributed to the full-length protein. The clear implication of these findings is that the N-terminal region of Brh2 constitutes the primary DNA-interacting point of the protein and in concert with the BRC element can deliver Rad51 to sites in the genome in need of repair and enable the process. The second conclusion, which is the opposite side of the coin of the first, is that the highly conserved C-terminal region containing the presumed canonical Dss1/DNA-binding domain does not appear to comprise the primary DNA-interacting point of the protein although it does have innate but weaker DNA binding capability. This raises the vexing question of what function the canonical DBD provides.
We showed previously that Dss1 is crucial for proficiency in DNA repair and recombination in U. maydis and suggested that it serves as a positive activator to regulate Brh2 function (14). Our interest in this current work stems from earlier efforts to identify variant forms of Brh2 that were active independent of the Dss1 requirement. In those studies we found that certain Brh2 C-terminal truncations deleted of the entire canonical DNA-binding domain were active in complementing to a substantial degree the UV sensitivity of the brh2 mutant, in enabling formation of subnuclear Rad51 foci in brh2 mutant cells following DNA damage, and in restoring recombination proficiency (17). The obvious biological activity associated with the N-terminal region of Brh2 opened the question of what could be the basis for such function. As is clear from the current work the N-terminal region of Brh2 harbors elements that make possible the DNA-interactive operations attributable to the full-length protein. With the additional feature of the Rad51-interacting BRC element, it would appear that the N-terminal region of Brh2 constitutes a module endowed with all of the abilities to engage Rad51 and to deliver it to DNA. This module seems fairly plastic in that the endogenous DNA-binding domain of the N-terminal region can be substituted by a heterologous single-strand binding domain. These observations are reminiscent of the findings from the Jasin laboratory, in which it was shown that a single BRC element fused to RPA70 could suppress DNA repair defects of BRCA2-deficient cells (6) and also call to mind the 394 residue Caenorhabditis elegans BRCA2 homolog, which contains a single BRC element and a single OB fold (29).
The positive and direct role of the N-terminal region of Brh2 in Rad51 presentation seems counterbalanced by forces imposed from the C-terminal region. This is most apparent in studies on recombination in which the frequency of gap repair as well as the rate of spontaneous allelic recombination is higher in cells expressing the Brh2NT as compared with cells expressing the full-length protein (17) suggesting the presence of a negative regulatory feature contributed by the C-terminal region. In previous work it was found that a second Rad51-interacting element, CRE, unrelated to BRC was present at the extreme C terminus of Brh2 (13). These elements appeared to communicate in some way to provide governance over recombinational activity, and Dss1 appeared to mediate the communication. Given the essential role of Dss1 in the context of the full-length Brh2 protein, it seems evident that interplay between Dss1 and its cognate interaction site within the Brh2 together with the innate DNA binding activity present within the C-terminal region likely provides means for fine-tuning recombination as directed through the N-terminal domain. In the absence of the C-terminal domain, one might expect that postnucleation aspects of Rad51-filament assembly such as stabilization as suggested for human BRCA2 (4, 19) and the worm homolog (30) would be missing. Equally plausible could be absence of a disassembly step. In this regard we have no decisive information at present about the integrity of Rad51 filaments in cells expressing the Brh2NT. It seems likely, however, that the filament quality is compromised somehow because cells expressing only the Brh2NT are less fit under certain conditions, for example, in suppressing UV sensitivity in the absence of Rec2 as reported here.
Finally, there is the broader question of how far the conclusions from the observations can be extended. There are numerous structural and functional parallels between Brh2 and BRCA2 homologs in animals. Most obvious is that inactivation or depletion of the proteins results in deficiencies in DNA repair, recombination, meiosis, and genome stability (1, 3, 31). In structural arrangement, as a rule, the overall organization of the proteins consists of a proximal region with variable numbers of BRC repeats followed by the distal, conserved DBD domain, featuring a tandem arrangement of a helix-rich domain and OB folds. Nevertheless, there are certain exceptions such as the BRCA2-related protein in Drosophila, which features several BRC elements but lacks any recognizable sequence corresponding to the canonical DBD (12, 32, 33). That the protein functions in vivo without any apparent DNA-binding domain would seem reminiscent of situation with the Brh2 N-terminal domain. In the vertebrate systems, BRCA2 is an essential gene, yet there are examples of viable cell lines obtained as well as live animals born with BRCA2 alleles deleted of the entire DBD (34–37). Residual function attributable to these hypomorphic alleles becomes easier to understand if indeed an unrecognized DNA-binding domain is still present in an upstream region of the protein.
It is remarkable to note parallels between functional design of Brh2 and Rad52 protein of budding yeast. Yeast appears to lack a protein structurally related to BRCA2, but instead relies on Rad52 to mediate loading of Rad51 on RPA-coated DNA (38). The N-terminal region of the protein harbors an oligomerization domain with DNA binding activity while the C-terminal region associates with Rad51. It had been generally assumed that the N-terminal DNA binding function was important for delivery of Rad51 to RPA-coated DNA (39), but remarkably, there is another DNA-binding domain in the C terminus that is able to promote Rad51 filament assembly and strand annealing more efficiently than the N-terminal domain (40).
The provocative finding revealed from this study is not so much that the N-terminal domain of Brh2 has an inherent ability to bind DNA and deliver Rad51. It is, on the contrary, that the highly conserved C-terminal DBD seems so expendable. What could be the meaning of the duality in DNA-binding domains. Illumination of this issue is sure to provide greater appreciation of the molecular mechanism by which Brh2 exerts control and governance of recombination.
Supplementary Material
Acknowledgments
We thank David S. Waugh, NCI, National Institutes of Health for providing the TEV protease-overproducing plasmid construct, and Jie Fan, Haijuan Yang, and Nikola Pavletich, Memorial Sloan-Kettering Cancer Center, for BRCA2 DBD. We are grateful to Lorraine Symington, Columbia University, for stimulating discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM42482 and GM79859. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: ss, single-stranded; CT, C-terminal; DBD, DNA-binding domain; ds, double-stranded; HD, helix-rich domain; MBP, maltose-binding protein; NT, N terminal; OB, oligonucleotide/oligosaccharide binding; oligo(dT), oligothymidylate; TEV, tobacco etch virus; aa, amino acids; HMM, hidden Markov model.
References
- 1.San Filippo, J., Sung, P., and Klein, H. (2008) Annu. Rev. Biochem. 77 229–257 [DOI] [PubMed] [Google Scholar]
- 2.Pellegrini, L., and Venkitaraman, A. (2004) Trends Biochem. Sci 29 310–316 [DOI] [PubMed] [Google Scholar]
- 3.Thorslund, T., and West, S. C. (2007) Oncogene 26 7720–7730 [DOI] [PubMed] [Google Scholar]
- 4.Esashi, F., Galkin, V. E., Yu, X., Egelman, E. H., and West, S. C. (2007) Nat. Struct. Mol. Biol. 14 468–474 [DOI] [PubMed] [Google Scholar]
- 5.Galkin, V. E., Esashi, F., Yu, X., Yang, S., West, S. C., and Egelman, E. H. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8537–8542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saeki, H., Siaud, N., Christ, N., Wiegant, W. W., van Buul, P. P., Han, M., Zdzienicka, M. Z., Stark, J. M., and Jasin, M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8768–8773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.San Filippo, J., Chi, P., Sehorn, M. G., Etchin, J., Krejci, L., and Sung, P. (2006) J. Biol. Chem. 281 11649–11657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shivji, M. K., Davies, O. R., Savill, J. M., Bates, D. L., Pellegrini, L., and Venkitaraman, A. R. (2006) Nucleic Acids Res. 34 4000–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang, H., Li, Q., Fan, J., Holloman, W. K., and Pavletich, N. P. (2005) Nature 433 653–657 [DOI] [PubMed] [Google Scholar]
- 10.Kojic, M., Zhou, Q., Lisby, M., and Holloman, W. K. (2006) Mol. Cell. Biol. 26 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pellegrini, L., Yu, D. S., Lo, T., Anand, S., Lee, M., Blundell, T. L., and Venkitaraman, A. R. (2002) Nature 420 287–293 [DOI] [PubMed] [Google Scholar]
- 12.Lo, T., Pellegrini, L., Venkitaraman, A. R., and Blundell, T. L. (2003) DNA Repair (Amst.) 2 1015–1028 [DOI] [PubMed] [Google Scholar]
- 13.Zhou, Q., Kojic, M., Cao, Z., Lisby, M., Mazloum, N. A., and Holloman, W. K. (2007) Mol. Cell. Biol. 27 2512–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kojic, M., Yang, H., Kostrub, C. F., Pavletich, N. P., and Holloman, W. K. (2003) Mol. Cell 12 1043–1049 [DOI] [PubMed] [Google Scholar]
- 15.Yang, H., Jeffrey, P. D., Miller, J., Kinnucan, E., Sun, Y., Thoma, N. H., Zheng, N., Chen, P. L., Lee, W. H., and Pavletich, N. P. (2002) Science 297 1837–1848 [DOI] [PubMed] [Google Scholar]
- 16.Kojic, M., Kostrub, C. F., Buchman, A. R., and Holloman, W. K. (2002) Mol. Cell 10 683–691 [DOI] [PubMed] [Google Scholar]
- 17.Kojic, M., Zhou, Q., Lisby, M., and Holloman, W. K. (2005) Mol. Cell. Biol. 25 2547–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davies, A. A., Masson, J. Y., McIlwraith, M. J., Stasiak, A. Z., Stasiak, A., Venkitaraman, A. R., and West, S. C. (2001) Mol. Cell 7 273–282 [DOI] [PubMed] [Google Scholar]
- 19.Davies, O. R., and Pellegrini, L. (2007) Nat. Struct. Mol. Biol. 14 475–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bochkarev, A., and Bochkareva, E. (2004) Curr. Opin. Struct. Biol. 14 36–42 [DOI] [PubMed] [Google Scholar]
- 21.Kojic, M., Mao, N., Zhou, Q., Lisby, M., and Holloman, W. K. (2008) Mol. Microbiol. 67 1156–1168 [DOI] [PubMed] [Google Scholar]
- 22.Mazloum, N., Zhou, Q., and Holloman, W. K. (2007) Biochemistry 46 7163–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kapust, R. B., Tozser, J., Fox, J. D., Anderson, D. E., Cherry, S., Copeland, T. D., and Waugh, D. S. (2001) Protein Eng. 14 993–1000 [DOI] [PubMed] [Google Scholar]
- 24.Mazloum, N., Zhou, Q., and Holloman, W. K. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopes, M., Foiani, M., and Sogo, J. M. (2006) Mol. Cell 21 15–27 [DOI] [PubMed] [Google Scholar]
- 26.Thorslund, T., Esashi, F., and West, S. C. (2007) EMBO J. 26 2915–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soding, J., Biegert, A., and Lupas, A. N. (2005) Nucleic Acids Res. 33 W244–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wold, M. S. (1997) Annu. Rev. Biochem. 66 61–92 [DOI] [PubMed] [Google Scholar]
- 29.Martin, J. S., Winkelmann, N., Petalcorin, M. I., McIlwraith, M. J., and Boulton, S. J. (2005) Mol. Cell. Biol. 25 3127–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petalcorin, M. I., Galkin, V. E., Yu, X., Egelman, E. H., and Boulton, S. J. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 8299–8304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shivji, M. K., and Venkitaraman, A. R. (2004) DNA Repair (Amst.) 3 835–843 [DOI] [PubMed] [Google Scholar]
- 32.Brough, R., Wei, D., Leulier, S., Lord, C. J., Rong, Y. S., and Ashworth, A. (2008) DNA Repair 7 10–19 [DOI] [PubMed] [Google Scholar]
- 33.Klovstad, M., Abdu, U., and Schupbach, T. (2008) PLoS Genet. 4 e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Connor, F., Bertwistle, D., Mee, P. J., Ross, G. M., Swift, S., Grigorieva, E., Tybulewicz, V. L., and Ashworth, A. (1997) Nat. Genet. 17 [DOI] [PubMed]
- 35.Friedman, L. S., Thistlethwaite, F. C., Patel, K. J., Yu, V. P., Lee, H., Venkitaraman, A. R., Abel, K. J., Carlton, M. B., Hunter, S. M., Colledge, W. H., Evans, M. J., and Ponder, B. A. (1998) Cancer Res. 58 1338–1343 [PubMed] [Google Scholar]
- 36.Patel, K. J., Yu, V. P., Lee, H., Corcoran, A., Thistlethwaite, F. C., Evans, M. J., Colledge, W. H., Friedman, L. S., Ponder, B. A., and Venkitaraman, A. R. (1998) Mol. Cell 1 347–357 [DOI] [PubMed] [Google Scholar]
- 37.Tarsounas, M., Davies, D., and West, S. C. (2003) Oncogene 22 1115–1123 [DOI] [PubMed] [Google Scholar]
- 38.Sung, P., and Klein, H. (2006) Nat. Rev. Mol. Cell Biol. 7 739–750 [DOI] [PubMed] [Google Scholar]
- 39.Sung, P., Krejci, L., Van Komen, S., and Sehorn, M. G. (2003) J. Biol. Chem. 278 42729–42732 [DOI] [PubMed] [Google Scholar]
- 40.Seong, C., Sehorn, M. G., Plate, I., Shi, I., Song, B., Chi, P., Mortensen, U., Sung, P., and Krejci, L. (2008) J. Biol. Chem. 283 12166–12174 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.