Abstract
Amyloid fibril formation of mutant transthyretin (TTR) that causes familial amyloid polyneuropathy occurs in the extracellular space. Thus, secretion of TTR variants contributes to the pathogenesis of amyloidosis. However, the molecular mechanisms underlying the endoplasmic reticulum (ER) exit or retention and subsequent degradation of TTR variants remain unclear. Here, we demonstrated that the nonsecreted TTR variants, such as D18G TTR and amyloidogenic TTRs with introduced monomeric mutation (M-TTRs), stably interact with the ER chaperone BiP in mammalian cells. These proteins were co-secreted with the secreted form of BiP in which the KDEL signal was removed, indicating that BiP partially contributes to the ER retention of nonsecreted TTR variants. More interestingly, the degradation efficiency of nonsecreted TTRs was increased when BiP was down-regulated by small interfering RNA. Thus, BiP protects the TTR variants from immediate degradation. Additionally, we showed that the stability of nonsecreted TTR variants is not disturbed in the coat complex II-deficient conditions, which are enough to inhibit the ER export of secreted TTR variants, including wild-type TTR. Therefore, the post-ER retrieval mechanism might not contribute to the ER-associated degradation of nonsecreted TTR variants. These findings suggest that the affinity to the ER-resident protein BiP regulates the fate of TTR variants in the ER.
Familial transthyretin (TTR)4 disease, which is autosomal dominant, is associated with a point mutation in the TTR gene. TTR is a soluble nonglycosylated secretory protein that functions as a homotetramer in the extracellular space (1). Mutant TTR forms amorphous aggregates and amyloid fibrils, which cause familial transthyretin disease (2). To date, over 100 TTR variants have been reported, and these variants can be classified according to their amyloidogenicity as either amyloidogenic TTR, such as V30M, D18G, and A25T TTRs, or nonamyloidogenic TTR, such as T119M TTR. It is known that onset of the disease, tissue selectivity, and severity are different with TTR variants (2–4), and these variations result from differences in secretion efficiency and amyloidogenicity of TTR variants. D18G and A25T TTRs, which exhibit late onset central nervous system-selective amyloidosis and are present at very low concentrations in the blood (3, 5), have low secretion efficiencies that might be attributed to a combination of low thermodynamic and kinetic stabilities in vitro (4). These TTRs are recognized by the endoplasmic reticulum (ER) quality control system and subjected to proteasomal degradation, which is known as ER-associated degradation (ERAD). Therefore, ERAD leads to low secretion of highly destabilized TTR variants, protecting against severe early onset of systemic amyloidosis.
Secretory proteins are translocated co-translationally or post-translationally to the ER. These newly synthesized proteins interact with ER molecular chaperones to become properly folded and assembled into a mature protein complex for transport along the secretory pathway (6). When maturation of proteins is aborted or inefficient, they are retained in the ER with ER chaperones. Thus, ER chaperones also participate in a quality control mechanism to prevent transport-incompetent proteins from being exported out of the ER. Moreover, ER chaperones act on those retained proteins either for further maturation or for ERAD, which protects the ER from the destructive consequence of protein aggregation (7, 8). Some ERAD substrates require transport between the ER and Golgi (9). These proteins are exported from the ER and they can be retrieved to the ER by retrograde transport at post-ER compartments (10, 11), indicating that the post-ER retrieval system also participates in quality control mechanisms for proper disposal of some proteins. It is likely that the ER quality control of TTR variant proteins is also regulated by such mechanism(s).
We have previously investigated the secretion pattern of TTR variants and their monomeric counterparts (M-TTRs), which are TTRs with introduced monomeric mutations F87M/L110M. In this report, we demonstrated that D18G TTR and amyloidogenic M-TTRs are retained in the ER and subsequently degraded by ERAD (12). Recent report shows that D18G TTR is significantly captured by BiP in the Escherichia coli system (13), suggesting that BiP probably facilitates D18G TTR degradation. However, the interaction of BiP with TTR variants in mammalian cells and the role of BiP in ER quality control for TTR variants remain unclear.
Here, we investigated which molecular chaperones bind to TTR variants in mammalian cells and showed that only BiP interacts with D18G TTR and amyloidogenic (D18G and V30M) M-TTRs, which are retained in the ER (referred to as nonsecreted TTRs). This interaction partially contributes to the ER retention of these TTR variants. Moreover, our results indicated that BiP negatively regulates the ERAD of these variants. Furthermore, we suggested that a post-ER retrieval mechanism might not be required for the efficient degradation of the nonsecreted TTR variants. Thus, our findings suggest that the fate of TTR variants is regulated by BiP in mammalian cells.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection—HeLa cells were cultured in minimal essential medium (Sigma) supplemented with 10% fetal bovine serum. Cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 and 95% air. Transient transfections of plasmid DNA were performed with TransIT-LT-1 (Mirus Corp., Madison, WI), as described previously (12).
Plasmid Constructs and Materials—Human wild-type TTR and TTR variant constructs have been described previously (12). Human BiP/GRP78 cDNA was inserted into the BamHI and XbaI sites of pEF6/Myc-His vector (Invitrogen). BiP-myc and BiPs-myc were generated using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA). Human Sar1-GTP (H79G) in pIRES2-DsRed2 vector was described previously (14). Antibodies used were as follows. Rabbit polyclonal anti-human TTR (anti-prealbumin) antibody was from Dako (Glostrup, Denmark); goat polyclonal anti-GRP78/BiP (C-20; sc-1051), goat polyclonal anti-calregulin (calreticulin; T-19), mouse monoclonal anti-c-Myc (9E10), and goat polyclonal anti-actin (I-19; sc-1616) antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA); rabbit polyclonal anti-Hsp70 (SPA-812), rabbit polyclonal anti-calreticulin (anti-CRT; SPA-600), mouse monoclonal anti-KDEL (SPA-827), and rabbit polyclonal anti-protein-disulfide isomerase (anti-PDI; SPA-890) antibodies were from Stressgen (San Diego, CA); mouse monoclonal anti-human cystic fibrosis transmembrane conductance regulator C terminus antibody was from R&D Systems, Inc. (clone 24-1; Minneapolis, MN); mouse monoclonal anti-ubiquitinylated protein antibody (clone FK2) was from BIOMOL International (Plymouth Meeting, PA); horseradish peroxidase-conjugated anti-rabbit, anti-goat, and anti-mouse antibodies were from Jackson Immuno-Research Laboratories (West Grove, PA). Cycloheximide (CHX) was purchased from Sigma. H89 was from BIOMOL. MG-132 was from Calbiochem.
Small Interfering RNA (siRNA) Preparation—The target sequence of BiP siRNA 1, BiP siRNA 2, and GL2-LUC siRNA (control siRNA) were reported previously (15, 16). Transient transfection with siRNA was performed using TransIT-TKO (Mirus Corp.) following the protocol recommended by the manufacturer.
Immunoprecipitation—For immunoprecipitation of the TTR-BiP complex and ubiquitinylated TTR, cell lysates were isolated from cells, as described previously (12), except that we used here 1% Triton X-100 buffer (1% Triton X-100, 50 mm Tris-HCl, pH 7.5, and 150 mm NaCl) to detect the TTR-BiP complex. Medium and lysate samples were incubated for 6 h at 4 °C with each antibody described in the figure legends, followed by incubation with protein G-Sepharose 4 Fast Flow (GE Healthcare) for 2 h at 4 °C. The immunoprecipitates were washed four times with protein recovery buffer and prepared for SDS-PAGE.
SDS-PAGE and Western Blotting—Media and cell lysate samples were prepared as described previously (12). Samples were analyzed by SDS-PAGE on 6, 10, 12, or 15% polyacrylamide gels. Proteins were transferred onto polyvinylidene difluoride membranes (Millipore Corp., Bedford, MA) and then probed with the indicated antibodies. Immunological bands were identified with horseradish peroxidase-conjugated secondary antibody followed by reaction with SuperSignal chemiluminescence reagent (Pierce).
Metabolic Labeling and Chase Experiment—Cells were transiently transfected with DNAs as described in the figure legends. Forty-eight hours after, cells were preincubated with labeling medium (methionine and cysteine-free Dulbecco's modified Eagle's medium; Invitrogen) for 15 min at 37 °C and labeled with [35S]methionine and [35S]cysteine (>1000 Ci/mmol; PerkinElmer Life Sciences) at 50 μCi/ml for 15 min. For chase, the cells were washed twice with minimal essential medium, and the labeling medium was replaced with complete minimal essential medium containing 50 μm CHX. Both the intracellular and extracellular forms of radiolabeled TTR were isolated by immunoprecipitation. Media were immunoprecipitated as described above. The cells were lysed in radioimmunoprecipitation assay buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mg/ml sodium deoxycholate, and 1% Nonidet P-40) containing 1% protease inhibitor mixture (Sigma) and subjected to immunoprecipitation with anti-human TTR antibody. Immune complexes were precipitated, washed four times with radioimmunoprecipitation assay buffer, boiled for 10 min in sample buffer (final 2% SDS), and separated by 15% SDS-PAGE under reducing conditions. The gels were then analyzed by a BAS imaging plate scanner (BAS-2000; Fujifilm) and quantified using Image Gauge software (version 3.4; Fujifilm).
RESULTS
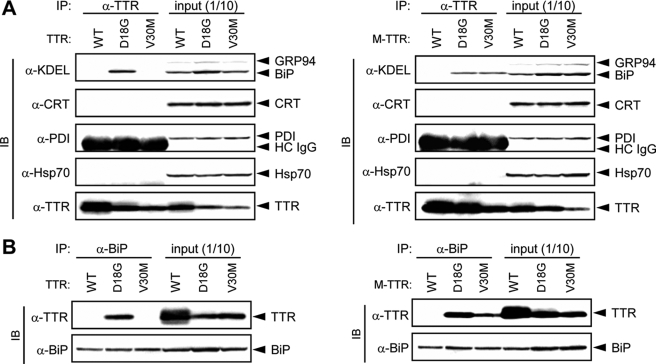
ER-resident Chaperone BiP Preferentially Associates with D18G TTR and Amyloidogenic M-TTRs—ER-resident chaperones are known to participate in a quality control mechanism to prevent transport-incompetent proteins from being exported out of the ER (17, 18). Moreover, a recent report (13) showed that D18G TTR binds to BiP in E. coli. To investigate the role of molecular chaperones, including BiP, on the ER quality control of TTR variants, we first examined whether nonsecreted TTR variants are bound to BiP or other ER-resident chaperones in mammalian cells. Wild-type TTR and variant (D18G and V30M) TTRs or their corresponding M-TTRs were transiently transfected into HeLa cells, which do not express TTR, and cell lysates were immunoprecipitated with anti-TTR antibody prior to analysis by SDS-PAGE. Western blotting of co-immunoprecipitates with the indicated antibodies showed that BiP but not other ER-resident (CRT and PDI) or cytosolic (heat shock protein 70 (Hsp70)) chaperones were co-immunoprecipitated with D18G TTR and amyloidogenic M-TTRs, which are not secreted (Fig. 1A). In contrast, none of the chaperones that we analyzed were co-immunoprecipitated with the secreted TTRs: wild-type TTR, V30M TTR, and wild-type M-TTR (Fig. 1A). These results are consistent with a previous report using recombinant proteins (13). Furthermore, immunoprecipitation analysis using anti-BiP antibody confirmed that D18G TTR and amyloidogenic M-TTRs but not wild-type TTR, V30M TTR, and wild-type M-TTR interacted with BiP (Fig. 1B). The failure to co-immunoprecipitate BiP with wild-type TTR, V30M TTR, and wild-type M-TTR was not due to lower expression levels of these proteins, since similar levels of TTR and M-TTR variants were found in the total cell lysates (Fig. 1, input). Therefore, these results imply that stable interaction of ER-resident protein BiP with D18G TTR and amyloidogenic M-TTR variants possibly contributes to the ER retention of these proteins.
FIGURE 1.
D18G TTR and amyloidogenic M-TTRs associate with BiP. TTRs and M-TTRs were transiently transfected in HeLa cells. After 48 h, cells were lysed and immunoprecipitated (IP) with anti-TTR (A) or anti-BiP (B) antibody. Co-immunoprecipitates were blotted (IB) with the indicated antibodies. HC IgG, heavy chain IgG. WT, wild type.
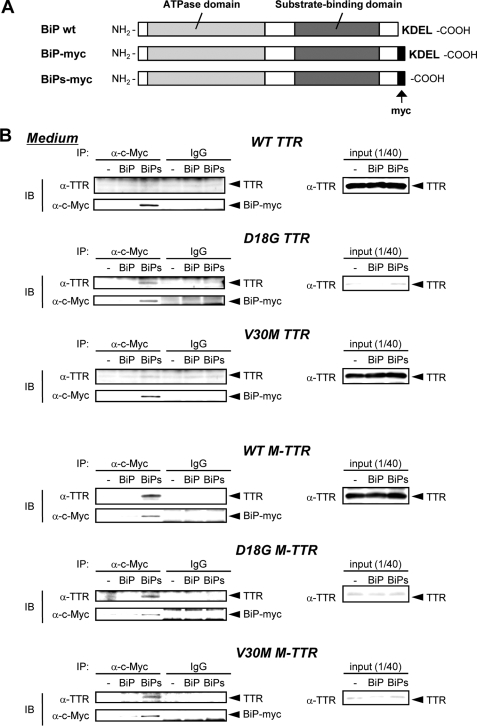
BiP Is Responsible for the ER Retention of D18G TTR and Amyloidogenic M-TTRs—If BiP is the dominant molecule that contributes to the ER retention of nonsecreted TTR variants, overexpression of the secreted form of BiP in which the ER-retention signal, KDEL, was removed and replaced with Myc tag (hereafter referred to as BiPs-myc; Fig. 2A) would be expected to cause D18G TTR and amyloidogenic M-TTRs to leave the ER and move with BiPs-myc to the culture medium (19). Cell media from HeLa cells that were transfected with BiPs-myc and TTRs or M-TTRs were collected and immunoprecipitated with anti-c-Myc antibody. Western blotting confirmed that, similar to previous reports (19), BiPs-myc was secreted into the media (Fig. 2B). On the other hand, BiP-myc, which has an intact KDEL sequence (Fig. 2A), was not present in the media (Fig. 2B). Because the intracellular levels of BiP-myc and BiPs-myc were comparable and both were efficiently recognized by anti-c-Myc antibody, these results could not be attributed to differences in intracellular level or antibody recognition (data not shown).
FIGURE 2.
D18G TTR and amyloidogenic M-TTRs are co-secreted with ΔKDEL BiP (BiPs). A, BiP constructs used in this study. B, BiP-myc, BiPs-myc, or control vector was transiently co-transfected with TTRs or M-TTRs in HeLa cells. After 24 h, medium was changed, and cells were maintained for another 24 h. Media were immunoprecipitated (IP) with anti-c-Myc antibody and analyzed by Western blotting (IB) with anti-TTR and anti-c-Myc antibodies. WT, wild type.
Wild-type and V30M TTRs were not co-secreted with BiPs-myc, but interestingly, D18G TTR and all M-TTRs were co-immunoprecipitated with BiPs-myc and detected in the media (Fig. 2B), indicating a stable interaction of D18G TTR and amyloidogenic M-TTRs with BiP. These results collectively suggest that the strong interaction of BiP with D18G TTR and amyloidogenic M-TTRs is one of the mechanisms by which these TTRs are retained in the ER.
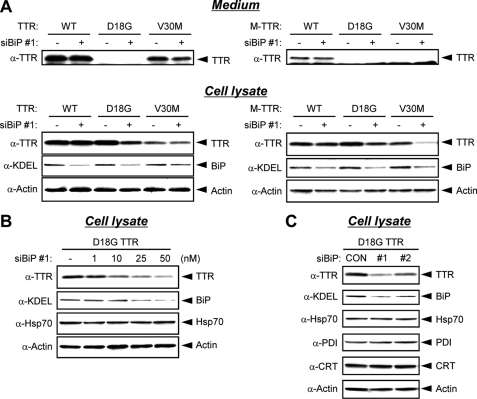
BiP Down-regulation Promotes the Degradation of D18G TTR—We further investigated the role of BiP in the fate of D18G TTR and amyloidogenic M-TTRs in the ER by knocking down the endogenous BiP expression using siRNAs, BiP siRNA 1 or 2 (15). We found no significant difference in the secretion levels of wild-type TTR, V30M TTR, and wild-type M-TTR in cells transfected with BiP siRNA 1 when compared with that in cells transfected with control siRNA (GL2-Luc siRNA) (Fig. 3A, Medium). Consistently, the intracellular levels of wild-type TTR, V30M TTR, and wild-type M-TTR were barely affected by BiP siRNA 1 (Fig. 3A, Cell lysate). In contrast, the intracellular levels of D18G TTR and amyloidogenic M-TTRs, which interact with BiP, were decreased in BiP siRNA 1-transfected cells compared with that in control siRNA-transfected cells (Fig. 3A, Cell lysate). Moreover, intracellular levels of D18G TTR were decreased in a BiP siRNA 1 dose-dependent manner (Fig. 3B). Similarly, co-transfection of D18G TTR with another siRNA against BiP (BiP siRNA 2) reduced the intracellular levels of D18G TTR (Fig. 3C). No change in protein levels of other chaperones, such as PDI, CRT, and Hsp70, a protein homologous to BiP, was observed upon transfection of BiP siRNA 1 or 2 (Fig. 3C and Fig. S1). Control siRNA did not affect the expression levels of either BiP or Hsp70 (data not shown), demonstrating the specificity of BiP siRNA in our system. The down-regulation of the intracellular levels of D18G TTR and amyloidogenic M-TTRs by BiP siRNA implies that BiP modulates the ERAD of these proteins.
FIGURE 3.
Expression levels of D18G TTR and amyloidogenic M-TTRs are decreased by BiP siRNA. A, 25 nm BiP siRNA (siBiP) 1 or GL2-LUC siRNA (Control siRNA) was transiently co-transfected with TTRs (left) or M-TTRs (right) in HeLa cells. After 44 h, medium was changed, and cells were maintained for another 4 h. Media (top) and cell lysates (bottom) were analyzed by Western blotting with anti-TTR, anti-KDEL, and anti-actin antibodies. B, BiP siRNA (siBiP) 1 was transiently co-transfected with D18G TTR in HeLa cells at the indicated concentration. After 48 h, cells were lysed and analyzed by Western blotting with anti-KDEL, anti-Hsp70, and anti-actin antibodies. C, 25 nm BiP siRNA 1, BiP siRNA 2, or control siRNA (CON) was transiently co-transfected with D18G TTR in HeLa cells. After 48 h, cell lysates were analyzed by Western blotting with anti-TTR, anti-KDEL, anti-Hsp70, anti-PDI, anti-CRT, and anti-actin antibodies. WT, wild type.
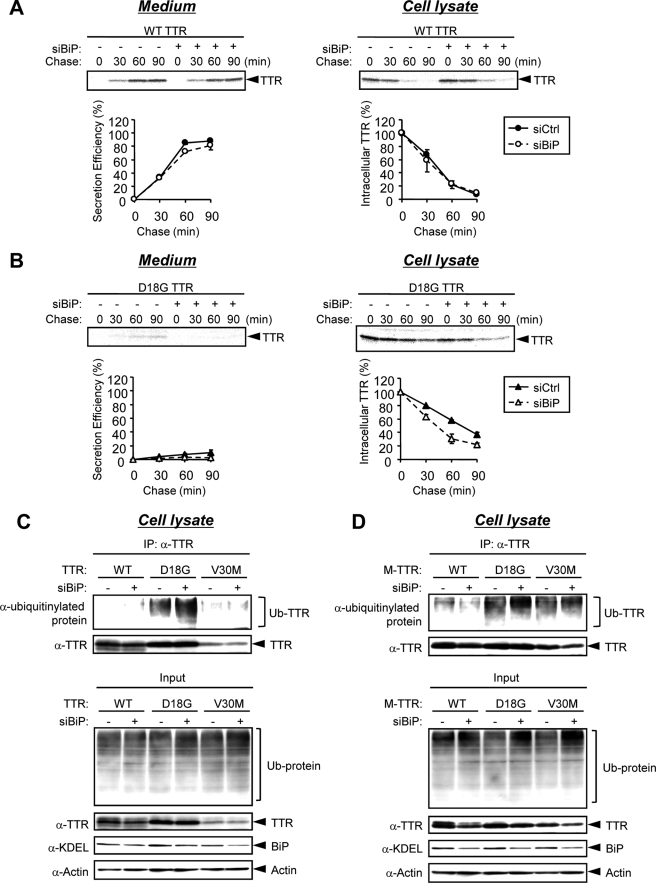
To clarify this possibility, we next investigated the effect of BiP down-regulation on the protein stability of D18G TTR by performing pulse-chase experiments with or without BiP siRNA. In these experiments, we only used BiP siRNA 1, because this was enough to knock down BiP. HeLa cells co-transfected with wild-type or D18G TTR and BiP siRNA or control siRNA were pulse-labeled with [35S]methionine and chased at the indicated time. Intracellular and extracellular TTR was immunoprecipitated and then loaded onto SDS-polyacrylamide gel, and the amount of protein was quantified using Image Gauge software. Wild-type TTR was slightly secreted at 30 min after pulse labeling and was almost completely secreted into the medium at 90 min in control siRNA-transfected cells (Fig. 4A, Medium, closed circles). BiP siRNA did not affect the secretion efficiency of wild-type TTR (Fig. 4A, Medium, open circles). The secretion of D18G TTR was barely detectable in HeLa cells transfected with control or BiP siRNA (Fig. 4B, Medium), indicating that BiP siRNA did not increase the secretion efficiency of D18G TTR. The intracellular level of labeled D18G TTR in control siRNA-transfected cell lysates was decreased time dependently with t½ ∼ 70 min (Fig. 4B, Cell lysate, closed triangles). Interestingly, in BiP siRNA-transfected cells, the intracellular level of labeled D18G TTR at the 60 min time point was already less than 50% (t½ ∼ 40 min) (Fig. 4B, Cell lysate, open triangles). These results suggest that the degradation efficiency of D18G TTR was enhanced by BiP down-regulation. BiP siRNA did not affect the expression level of wild-type cystic fibrosis transmembrane conductance regulator (Fig. S2), which is degraded by ERAD through the BiP-independent pathway (20–22). Moreover, BiP siRNA treatment did not alter the steady-state levels of other chaperones (PDI, CRT, and Hsp70) and ubiquitinylated proteins in the lysates of cells that were transfected with control vector pEF6 (Fig. S3). These results indicated that BiP siRNA treatment did not substantially affect the total (global) ERAD efficiency in the cells under our experimental conditions. Taken together, we conclude that BiP negatively regulates the ERAD of D18G TTR (and possibly that of the amyloidogenic M-TTRs) by interacting stably with this variant TTR.
FIGURE 4.
BiP siRNA promotes the ERAD of D18G TTR and amyloidogenic M-TTRs. A and B, 25 nm BiP siRNA (siBiP) or control siRNA (siCtrl) was transiently co-transfected with wild-type TTR (A) or D18G TTR (B) in HeLa cells. After 48 h, cells were labeled with [35S]methionine/cysteine for 15 min and chased at the indicated times. Intracellular and secreted TTRs were immunoprecipitated and loaded onto SDS-polyacrylamide gel, followed by autoradiography (top). The signals for TTR were quantified and plotted (bottom). Secretion efficiency (%) = 100 × ((secreted TTR at given time t)/(intracellular TTR at t = 0 min)). The mean and S.E. values were calculated from two independent experiments. C and D, TTRs (C) or M-TTRs (D) were transiently co-transfected with 25 nm BiP siRNA or control siRNA in HeLa cells. After 42 h, cells were treated with 30 μm proteasome inhibitor MG-132 for 6 h. Cell lysates were immunoprecipitated (IP) with anti-TTR antibody. The immunoprecipitates and cell lysates (Input) were resolved by SDS-PAGE and analyzed by Western blotting with anti-ubiquitinylated protein, anti-TTR, anti-KDEL, and anti-actin antibodies. Ub-TTR and Ub-protein, ubiquitinylated TTR and ubiquitinylated proteins, respectively. WT, wild type.
BiP Down-regulation Promotes D18G TTR Polyubiquitination—D18G TTR and amyloidogenic M-TTRs were previously shown to be degraded by the proteasomal pathway (4, 12). Since most ERAD substrates are ubiquitinylated prior to proteasome targeting, D18G TTR and amyloidogenic M-TTRs may be ubiquitinylated before proteasomal degradation (23). Because our results above (Figs. 3 and 4) suggested that BiP siRNA enhanced the ERAD of D18G TTR and amyloidogenic M-TTRs, we next investigated whether the ubiquitination levels of D18G TTR and amyloidogenic M-TTRs are increased by BiP siRNA. Ubiquitinylated TTRs and M-TTRs in the presence or absence of BiP siRNA were determined by immunoprecipitation using anti-TTR antibody and Western blotting with anti-ubiquitinylated protein antibody. Low levels of ubiquitinylated wild-type TTR or M-TTR and ubiquitinylated V30M TTR were detected within 6 h of MG-132 treatment, but these levels were not affected by BiP siRNA co-transfection (Fig. 4, C and D). In contrast, the ubiquitination levels of the nonsecreted TTRs, D18G TTR, D18G M-TTR, and V30M M-TTR, were increased in BiP siRNA-transfected cells compared with control siRNA-transfected cells (Fig. 4, C and D). These results support our hypothesis that BiP down-regulation promotes the ERAD of nonsecreted TTR variants.
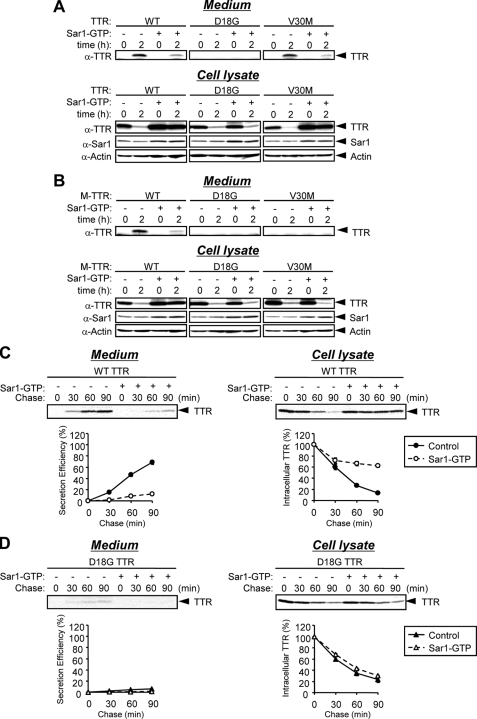
Post-ER Retrieval Pathway Is Not Required for ERAD of TTR Variants—Some ERAD substrates require transport between the ER and Golgi, which is mediated by the KDEL receptor (9). BiP is one of the proteins that have been shown to leak out to the post-ER compartments and to be retrieved to the ER through its C-terminal KDEL motif (24). To investigate whether the post-ER retrieval mechanism contributes to the ER quality control of D18G TTR, we co-transfected Sar1 (H79G) (referred to as Sar1-GTP) with TTR or M-TTR variants in HeLa cells. Sar1-GTP is a constitutively active mutant locked in the GTP-bound form that inhibits COPII coat disassembly, resulting in inhibition of ER to Golgi transport (25–27). Cell media and lysates were collected at 0 or 2 h after CHX treatment. At the 0 h time point, samples were taken immediately after adding CHX. In cells co-transfected with Sar1-GTP, the secretion of wild-type TTR, V30M TTR, and wild-type M-TTR was decreased compared with that in cells transfected with empty vector (control cells) (Fig. 5, A and B, Medium). Consistently, the intracellular levels of these TTRs at 2 h after adding CHX were higher in Sar1-GTP-transfected cells than in control cells, in which the TTR intracellular levels were substantially reduced due to their efficient secretion into the media (Fig. 5, A and B, Cell lysate) (12). Sar1-GTP co-transfection had no effect on the decreased levels of intracellular D18G TTR, D18G M-TTR, and V30M M-TTR at 2 h after adding CHX (Fig. 5, A and B, Cell lysate). To confirm the effect of Sar1-GTP on the secretion and degradation efficiencies of TTR, HeLa cells co-transfected with wild-type TTR or D18G TTR and Sar1-GTP or empty vector were pulse-labeled with [35S]methionine for 15 min, and then the media and cell lysates were collected at the indicated time. Intracellular and extracellular TTRs were immunoprecipitated and then loaded onto SDS-polyacrylamide gel, and the amount of protein was quantified using Image Gauge software. Pulse-chase analysis indicated that the secretion efficiency of wild-type TTR was substantially decreased by co-transfection of Sar1-GTP compared with control cells (Fig. 5C, Medium). On the other hand, the degradation efficiency of nonsecreted variants D18G TTR, D18G M-TTR, and V30M M-TTR was not affected by Sar1-GTP (Fig. 5D, Cell lysate), indicating that the ERAD of this variant (and possibly that of the amyloidogenic M-TTRs) could occur efficiently under COPII-deficient conditions.
FIGURE 5.
Degradation of TTR is not COPII-dependent. A and B, Sar1-GTP (H79G) was transiently co-transfected with TTRs (A) or M-TTRs (B) in HeLa cells. After 24 h, media and cell lysates were collected immediately (0 h) or 2 h after the addition of 50 μm CHX. Media and cell lysates were analyzed by Western blotting with anti-TTR, anti-Sar1, and anti-actin antibodies. C and D, Sar1-GTP or empty vector was transiently co-transfected with wild-type TTR (C) or D18G TTR (D) in HeLa cells. After 24 h, cells were labeled with [35S]methionine/cysteine for 15 min and chased at the indicated times. Intracellular and secreted TTRs were immunoprecipitated and loaded onto SDS-polyacrylamide gel, followed by autoradiography (top). The signals for TTR were quantified and plotted (bottom). Secretion efficiency (%) = 100 × ((secreted TTR at given time t)/(intracellular TTR at t = 0 min)). The mean and S.E. values were calculated from two independent experiments. WT, wild type.
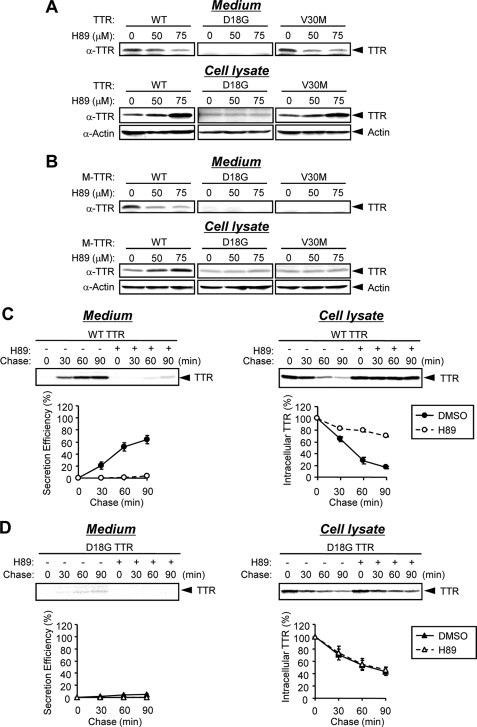
To confirm these observations, we used the protein kinase inhibitor H89 that was previously shown to inhibit Sar1 recruitment and COPII coat assembly (28). The addition of H89 and CHX for 3 h to the cells transfected with TTR and M-TTR variants strongly inhibited the secretion of wild-type TTR, V30M TTR, and wild-type M-TTR in a dose-dependent manner (Fig. 6, A and B, Medium). In agreement with these results, the secretion efficiency of wild-type TTR was highly inhibited in H89-treated cells relative to control cells (DMSO-treated) in pulse-chase analysis (Fig. 6C, Medium). However, the levels of nonsecreted TTR variants D18G TTR, D18G M-TTR, and V30M M-TTR in the media and cell lysates were not affected by H89 treatment (Fig. 6, A and B). These results were confirmed by pulse-chase analysis of D18G TTR (Fig. 6D). Taken together, these findings indicate that the conditions that interfere with COPII function prevent the secretion of wild-type TTR, V30M TTR, and wild-type M-TTR but have little effect on the nonsecretion or the degradation of D18G TTR, D18G M-TTR, and V30M M-TTR. Thus, the secretion pathway of wild-type TTR and V30M TTR (the secreted TTRs) is largely dependent on COPII machinery, whereas the ERAD pathway of nonsecreted TTR variants does not require the post-ER mechanism.
FIGURE 6.
Secretion of TTR but not degradation is inhibited by the kinase inhibitor H89. A and B, HeLa cells were transiently transfected with TTRs (A) and M-TTRs (B). After 24 h, cells were treated with 50μm CHX together with H89 (50 or 75 μm) or DMSO (as control) for 3 h. Media and cell lysates were analyzed by Western blotting with anti-TTR and anti-actin antibodies. C and D, HeLa cells were transiently transfected with wild-type TTR (C) or D18G TTR (D). After 24 h, cells were treated with 75μm H89 for 3 h and then were labeled with [35S]methionine/cysteine for 15 min and chased at the indicated times. Intracellular and secreted TTRs were immunoprecipitated and loaded onto SDS-polyacrylamide gel, followed by autoradiography (top). The signals for TTR were quantified and plotted (bottom). Secretion efficiency (%) = 100 × ((secreted TTR at given time t)/(intracellular TTR at t = 0 min)). The mean and S.E. values were calculated from two independent experiments.
DISCUSSION
In this study, we investigated the mechanism of the ER retention and ERAD of TTR variants in mammalian cells by examining the molecular chaperones that associate with these proteins. As determined by immunoprecipitation analyses, BiP specifically interacted with nonsecreted TTR variants D18G TTR and amyloidogenic M-TTRs. The present study first shows D18G TTR binding to endogenous BiP in mammalian cells (Fig. 1). The stable interaction of BiP with D18G TTR and amyloidogenic M-TTRs was attested by the fact that these variant TTRs were secreted only in the presence of BiPs-myc, the secreted form of BiP in which the ER retention KDEL signal was removed (Fig. 2B). This strong interaction with BiP may partly explain the reason why these variants were retained in the ER. However, because their secretion efficiencies were not improved when the BiP expression level was down-regulated (Fig. 3A, Medium), it might be possible that the nonsecretion of these variants was due to their inability to be recognized by COPII components. By overexpressing Sar1-GTP, which is a dominant-negative mutant that promotes cargo collection into stable COPII-coated intermediates, the nonsecreted TTRs would have been protected from ERAD if they were recruited to the assembling COPII polymer at the ER exit sites, like wild-type cystic fibrosis transmembrane conductance regulator (29). However, degradation efficiency of D18G TTR was comparable between the control- and Sar1-GTP-transfected cells (Fig. 5D), indicating that D18G TTR failed to engage the COPII machinery. Thus, the ER retention of nonsecreted TTR variants may be doubly caused by binding with BiP and the nonrecognition by COPII machinery. These two regulatory mechanisms are not necessarily mutually exclusive (6).
More importantly, we indicated here that the ubiquitination level of non-secreted TTR variants and the ERAD of D18G TTR were increased by BiP down-regulation (Fig. 4, B–D). These results suggest that interaction of nonsecreted TTR variants with BiP may suppress their ERAD pathway including retrotranslocation to the cytoplasm and subsequent ubiquitination of these proteins. This is in contrast to previous studies showing that BiP is required for ERAD (7, 30–32). Although the underlying factors crucial for the decision of BiP to lead to or to retain its substrates from ERAD remain undetermined, the regulation by BiP is probably dependent on BiP substrate features. The ERAD process is constituted by several steps: 1) recognition of substrate to prevent from aggregation and immediate degradation (recognition step), 2) delivery of substrate to the retrotranslocation machinery (delivery step), 3) retrotranslocation and ubiquitination, and 4) proteasomal targeting and degradation (23). In mammalian cells, it is reported that the calnexin cycle and the BiP/PDI chaperone system may act sequentially not only to assist protein folding but also to promote ERAD of BACE457, which is a membrane glycoprotein, and its soluble variant BACEΔ457 (7). Additionally, treatment of SubAB, which specifically cleaves BiP, depleting cellular stores of BiP, extended the half-life of type I membrane glycoprotein, T-cell antigen receptor α (30). On the other hand, the half-life of unfolded immunoglobulin light chains, which are soluble nonglycosylated proteins, is probably determined by the physical stability of BiP association with an unfolded region of immunoglobulin light chain (33). In addition, we first showed here that BiP down-regulation promotes the ERAD of D18G TTR and amyloidogenic M-TTRs, which are soluble nonglycosylated proteins. Based on these findings, we propose some possibilities about ERAD regulation by BiP. One is that the ERAD promotion (or inhibition) by BiP may depend on whether or not its substrate is glycosylated. Another possibility is that the effect of BiP on ERAD of substrate may depend on the step of the ERAD process in which BiP is involved. These possibilities may or may not be interdependent. It is likely that when BiP works at the delivery step of the ERAD process, BiP positively regulates the degradation of substrate, such as in the case of BACE and T-cell antigen receptor α. On the other hand, when BiP works at the recognition step of the ERAD process, BiP may negatively regulate the degradation of substrate, such as D18G TTR. These substrates may need to be subsequently transferred from BiP to another molecule like XTP3-B or OS-9, which will deliver the substrate to the retrotranslocation machinery. This seems probable, since XTP3-B is known to interact with D18G TTR (34).
In contrast to a previous report, which suggests that the post-ER retrieval pathway through KDEL proteins, such as BiP, has an important role in the ERAD of T-cell antigen receptor α (9), our results showed that inhibiting ER export (under COPII-deficient conditions) had no significant effect on the ERAD of D18G TTR (Figs. 5 and 6). Therefore, undergoing post-ER retrieval may also be one of the major differences between the substrates whose ERAD is promoted by BiP and the substrates whose ERAD is inhibited by BiP.
In conclusion, we suggest here that the ER retention of D18G TTR and amyloidogenic M-TTRs is partially regulated by their affinity for the ER-resident protein, BiP. Moreover, we first showed that the binding of BiP to TTR probably functions to delay the ERAD of nonsecreted TTR variants. These findings suggest that complicated molecular networks in the ER, including BiP, may control the ER retention or ERAD of TTR variants.
Supplementary Material
Acknowledgments
We thank Drs. Toru Mizushima (Kumamoto University) and Dieter C. Gruenert (California Pacific Medical Center Research Institute) for supplying BiP/GRP78 and cystic fibrosis transmembrane conductance regulator cDNA constructs, respectively, and Dr. Tomohiko Aoe (Chiba University) for helpful discussion of the manuscript. We also thank Y. Suganuma and M. Kugimiya for technical assistance.
This work was supported by grants from the Amyloidosis Research Committee, the Pathogenesis and Therapy of Hereditary Neuropathy Research Committee, Surveys and Research on Specific Disease, the Ministry of Health and Welfare of Japan, the Charitable Trust Clinical Pathology Research Foundation of Japan, and grants-in-aid for scientific research from the Ministry of Education, Science, Sports and Culture (MEXT) of Japan and from the Global Centers of Excellence Program (Cell Fate Regulation Research and Education Unit), MEXT, Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: TTR, transthyretin; ER, endoplasmic reticulum; ERAD, ER-associated degradation; M-TTR, amyloidogenic TTR with introduced monomeric mutation; CHX, cycloheximide; siRNA, small interfering RNA; PDI, protein-disulfide isomerase; CRT, calreticulin.
References
- 1.Hamilton, J. A., and Benson, M. D. (2001) Cell. Mol. Life Sci. 58 1491–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connors, L. H., Lim, A., Prokaeva, T., Roskens, V. A., and Costello, C. E. (2003) Amyloid 10 160–184 [DOI] [PubMed] [Google Scholar]
- 3.Hammarstrom, P., Sekijima, Y., White, J. T., Wiseman, R. L., Lim, A., Costello, C. E., Altland, K., Garzuly, F., Budka, H., and Kelly, J. W. (2003) Biochemistry 42 6656–6663 [DOI] [PubMed] [Google Scholar]
- 4.Sekijima, Y., Wiseman, R. L., Matteson, J., Hammarstrom, P., Miller, S. R., Sawkar, A. R., Balch, W. E., and Kelly, J. W. (2005) Cell 121 73–85 [DOI] [PubMed] [Google Scholar]
- 5.Sekijima, Y., Hammarstrom, P., Matsumura, M., Shimizu, Y., Iwata, M., Tokuda, T., Ikeda, S., and Kelly, J. W. (2003) Lab. Invest. 83 409–417 [DOI] [PubMed] [Google Scholar]
- 6.Anelli, T., and Sitia, R. (2008) EMBO J. 27 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molinari, M., Galli, C., Piccaluga, V., Pieren, M., and Paganetti, P. (2002) J. Cell Biol. 158 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodsky, J. L. (2007) Biochem. J. 404 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto, K., Fujii, R., Toyofuku, Y., Saito, T., Koseki, H., Hsu, V. W., and Aoe, T. (2001) EMBO J. 20 3082–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allan, B. B., and Balch, W. E. (1999) Science 285 63–66 [DOI] [PubMed] [Google Scholar]
- 11.Letourneur, F., Gaynor, E. C., Hennecke, S., Demolliere, C., Duden, R., Emr, S. D., Riezman, H., and Cosson, P. (1994) Cell 79 1199–1207 [DOI] [PubMed] [Google Scholar]
- 12.Sato, T., Susuki, S., Suico, M. A., Miyata, M., Ando, Y., Mizuguchi, M., Takeuchi, M., Dobashi, M., Shuto, T., and Kai, H. (2007) EMBO J. 26 2501–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sorgjerd, K., Ghafouri, B., Jonsson, B. H., Kelly, J. W., Blond, S. Y., and Hammarstrom, P. (2006) J. Mol. Biol. 356 469–482 [DOI] [PubMed] [Google Scholar]
- 14.Okiyoneda, T., Harada, K., Yamahira, K., Wada, I., Hashimoto, Y., Ueno, K., Suico, M. A., Shuto, T., and Kai, H. (2004) J. Pharmacol. Sci. 95 471–475 [DOI] [PubMed] [Google Scholar]
- 15.Hegde, N. R., Chevalier, M. S., Wisner, T. W., Denton, M. C., Shire, K., Frappier, L., and Johnson, D. C. (2006) J. Biol. Chem. 281 20910–20919 [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto, Y., Okiyoneda, T., Harada, K., Ueno, K., Sugahara, T., Yamashita, A., Shuto, T., Suico, M. A., and Kai, H. (2008) Biochim. Biophys. Acta 1783 153–162 [DOI] [PubMed] [Google Scholar]
- 17.Ellgaard, L., Molinari, M., and Helenius, A. (1999) Science 286 1882–1888 [DOI] [PubMed] [Google Scholar]
- 18.Trombetta, E. S., and Parodi, A. J. (2003) Annu. Rev. Cell Dev. Biol. 19 649–676 [DOI] [PubMed] [Google Scholar]
- 19.Swanton, E., High, S., and Woodman, P. (2003) EMBO J. 22 2948–2958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan, M. L., Youker, R. T., Watkins, S. C., and Brodsky, J. L. (2003) J. Histochem. Cytochem. 51 545–548 [DOI] [PubMed] [Google Scholar]
- 21.Varga, K., Goldstein, R. F., Jurkuvenaite, A., Chen, L., Matalon, S., Sorscher, E. J., Bebok, Z., and Collawn, J. F. (2008) Biochem. J. 410 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varga, K., Jurkuvenaite, A., Wakefield, J., Hong, J. S., Guimbellot, J. S., Venglarik, C. J., Niraj, A., Mazur, M., Sorscher, E. J., Collawn, J. F., and Bebok, Z. (2004) J. Biol. Chem. 279 22578–22584 [DOI] [PubMed] [Google Scholar]
- 23.Vembar, S. S., and Brodsky, J. L. (2008) Nat. Rev. Mol. Cell Biol. 9 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munro, S., and Pelham, H. R. (1987) Cell 48 899–907 [DOI] [PubMed] [Google Scholar]
- 25.Kuge, O., Dascher, C., Orci, L., Rowe, T., Amherdt, M., Plutner, H., Ravazzola, M., Tanigawa, G., Rothman, J. E., and Balch, W. E. (1994) J. Cell Biol. 125 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aridor, M., Weissman, J., Bannykh, S., Nuoffer, C., and Balch, W. E. (1998) J. Cell Biol. 141 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, M., Weissman, J. T., Beraud-Dufour, S., Luan, P., Wang, C., Chen, W., Aridor, M., Wilson, I. A., and Balch, W. E. (2001) J. Cell Biol. 155 937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aridor, M., and Balch, W. E. (2000) J. Biol. Chem. 275 35673–35676 [DOI] [PubMed] [Google Scholar]
- 29.Wang, X., Matteson, J., An, Y., Moyer, B., Yoo, J. S., Bannykh, S., Wilson, I. A., Riordan, J. R., and Balch, W. E. (2004) J. Cell Biol. 167 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lass, A., Kujawa, M., McConnell, E., Paton, A. W., Paton, J. C., and Wojcik, C. (2008) Int. J. Biochem. Cell Biol. 40 2865–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishikawa, S. I., Fewell, S. W., Kato, Y., Brodsky, J. L., and Endo, T. (2001) J. Cell Biol. 153 1061–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabani, M., Kelley, S. S., Morrow, M. W., Montgomery, D. L., Sivendran, R., Rose, M. D., Gierasch, L. M., and Brodsky, J. L. (2003) Mol. Biol. Cell 14 3437–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skowronek, M. H., Hendershot, L. M., and Haas, I. G. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 1574–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christianson, J. C., Shaler, T. A., Tyler, R. E., and Kopito, R. R. (2008) Nat. Cell Biol. 10 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.