Abstract
Unlike the vast majority of flavoenzymes, bacterial luciferase requires an exogenous source of reduced flavin mononucleotide for bioluminescence activity. Within bioluminescent bacterial cells, species-specific oxidoreductases are believed to provide reduced flavin for luciferase activity. The source of reduced flavin in Escherichia coli-expressing bioluminescence is not known. There are two candidate proteins potentially involved in this process in E. coli, a homolog of the Vibrio harveyi Frp oxidoreductase, NfsA, and a luxG type oxidoreductase, Fre. Using single gene knock-out strains, we show that deletion of fre decreased light output by greater than two orders of magnitude, yet had no effect on luciferase expression in E. coli. Purified Fre is capable of supporting bioluminescence in vitro with activity comparable to that with the endogenous V. harveyi reductase (Frp), using either FMN or riboflavin as substrate. In a pull-down experiment, we found that neither Fre nor Frp co-purify with luciferase. In contrast to prior work, we find no evidence for stable complex formation between luciferase and oxidoreductase. We conclude that in E. coli, an enzyme primarily responsible for riboflavin reduction (Fre) can also be utilized to support high levels of bioluminescence.
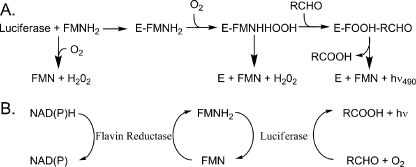
Bacterial luciferase is a heterodimeric (αβ) flavin monooxygenase that catalyzes the reaction of FMNH2, O2, and an aliphatic aldehyde to yield FMN, the corresponding carboxylic acid and blue-green light (1). In vitro, the reaction is usually initiated by injection of the reduced flavin into a vial containing enzyme, aldehyde, and oxygen. The enzyme binds to reduced flavin forming the E:FMNH2 complex, which reacts rapidly with molecular oxygen. The enzyme-flavin-oxygen complex then reacts with aldehyde, ultimately yielding the oxidized flavin, the carboxylic acid, and bioluminescence (see Fig. 1). Energy for the light production comes primarily from oxidation of the aldehyde to the carboxylic acid (2). In vivo, the aldehyde is supplied by an acid reductase comprised of the products of luxCDE, components of the lux regulon, which can reduce myristic acid to myristic aldehyde (3). However, the source of the reduced flavin in vivo is less clear.
FIGURE 1.
The bioluminescence reaction in vitro and in vivo. A, simplified in vitro bioluminescent reaction mechanism. The isoalloxazine ring of the flavin is oxygenated at position C-4a prior to formation of the aldehyde hemiacetal (47). The resolution of this tetrahedral intermediate leads to the population of an excited state emitter and ultimately light emission (2, 44). B, in vivo, and in the coupled assay in vitro, FMN is reduced by an oxidoreductase enzyme via the oxidation of NADH or NAD(P)H (10). Subsequently, FMNH2 is transferred to bacterial luciferase either by diffusion or by tunneling for the light emitting reaction.
When the bacterial luciferase genes were initially isolated from Vibrio harveyi (luxAB) in 1981, the expectation was that the recombinant cells would not express bioluminescence when exposed to aldehyde vapor because of the lack of a specific flavin oxidoreductase (4). However, we have never found conditions in which the intensity of bioluminescence from E. coli has been limited by the availability of reduced flavin mononucleotide. Essentially all living cells have been found competent to supply reduced flavin for the bioluminescence reaction (1). Within a given microbial cell, enzymes of varied function require electrons in the form of reduced flavin mononucleotide (5–8). Maintaining a steady supply of reduced flavin may be difficult during aerobic growth, as diffusion of free reduced flavin may be compromised because of reaction with molecular oxygen (9). Based on the extensive work of Tu and co-workers (10, 11), it has been proposed that a complex is formed involving luciferase and oxidoreductase in which FMNH2 is transferred directly from the reductase to the luciferase (see Fig. 1). This model obviates the problem of reaction of reduced flavin with molecular oxygen in solution. Implicit in this model is the requirement of molecular specificity in the transient complex. Bacterial luciferase is highly active in a variety of recombinant bacteria that lack the specific oxidoreductase from the native bioluminescent bacteria (12, 13). This observation is surprising in light of the proposal of a luciferase-oxidoreductase complex (10, 11), suggesting a lack of molecular specificity in the protein-protein interaction involved in the flavin transfer in vivo.
The three most extensively characterized flavin reductases are Frp from V. harveyi, Frase-I from V. fischeri, and Fre from E. coli (see Table 1, Ref. 14). These enzymes lack extensive sequence similarity, despite functional similarity (14). Frp and Frase-I appear to be related while Fre and Frase-I are distantly related (15). The nearest known homolog to Frp is NfsA from E. coli, which has 51% sequence identity with Frp (14).
TABLE 1.
Bioluminescence-supporting flavin oxidoreductases
The V. harveyi reductase, Frp, preferentially utilizes NADPH and a tightly bound FMN cofactor (16), and can be purified using immobilized luciferase chromatography, suggesting complex formation (17, 18). The V. fischeri reductase, Frase-I, also preferentially utilizes NADPH as a source of reducing equivalents and also has a bound FMN cofactor (19, 20). The E. coli reductase, Fre, lacks nicotinamide nucleotide specificity and has a slight preference for riboflavin rather than FMN (21). Genes designated luxG encode Fre homologs in four species of luminous bacteria (22, 23). This group of enzymes is thought to act as the primary source of reduced flavin for bioluminescent bacterial species lacking Frp or Frase-I type enzymes (23). In V. fischeri, the luxG encoded enzyme represents less than 10% of the total FMN reductase activity and thus, luxG is thought to be a minor player in vivo (19). Fre enzymes from various bioluminescent bacteria, such as V. harveyi, Photobacterium phosphoreum, and Xenorhabdus luminescens, appear to be capable of supplying FMNH2 for the bioluminescence reaction in vitro for luciferases from the same bacterial species (24–27). For species with more than one type of oxidoreductase, it is unknown if the enzymes are functionally redundant in vivo.
The oxygen-insensitive nitroreductase proteins from E. coli, NfsA and NfsB, appear to be unrelated (14). NfsA and NfsB can utilize both NADH and NADPH. NfsA, which has an FMN cofactor, can reduce exogenous FMN with only ∼1.9% the catalytic efficiency of Frase-I (14). NfsB resembles Frase-I, with 34% sequence identity, but has minimal FMN reductase activity (28, 29). Random PCR mutagenesis of NfsB demonstrated that a single amino acid substitution, F124S, resulted in an enzyme with three times the FMN reductase activity of the Frase-I enzyme (29). Likewise, a single amino acid change in NfsA, E99G, resulted in an enzyme with twice the FMN reductase activity of its homolog, Frp (30).
The aim of this study was identification of the oxidoreductase that supports the bioluminescent reaction in vivo in E. coli. Using deletion strains for Fre and NfsA, we found that expression of luciferase protein was unaffected by strain type, but the expression of bioluminescence in vivo was remarkably lower in the Fre deletion strain. Purified Fre had kinetic properties in the luciferase coupled assay similar to the non-homologous Frp oxidoreductase from V. harveyi.
EXPERIMENTAL PROCEDURES
Chemicals—All chemicals were obtained from Sigma Aldrich unless otherwise noted and were of reagent grade or better. Cells were cultured in standard Luria-Bertani broth on agar supplemented with 100 μg/ml ampicillin and 30 μg/ml kanamyacin.
Strains and Vectors—Single in-frame deletion strains were obtained from the Keio collection (31). These mutations were produced in E. coli K-12 BW25113 (lacIq, rrnBT14, Δlac-ZWJ16, hsdR514, ΔaraBADAH33, ΔrhaBA, LD78. Luciferase was expressed from pJHD500 (32).
In Vivo and Crude Lysate Activity Assays—Cells were grown until midlog phase (OD of 600 nm: 0.5) and induced by addition of 3-oxo n-hexanoyl homoserine lactone (A.I.-1) to a final concentration of 0.1 μm for 8 h at 25 °C. Aliquots consisting of 0.5 ml or less, depending on the cell density were withdrawn. Bioluminescence activity in vivo was measured using the sonicated aldehyde injection assay as previously described (33, 34). In the crude lysate assay, cells were concentrated by brief centrifugation at 3,000 rpm and resuspended in 100 μl of B-PER bacterial protein extraction reagent (Pierce). These samples were then frozen at –20 °C and slowly thawed prior to the flavin injection assay technique (34).
Western Blotting—A small volume of each culture was withdrawn and adjusted for cell density as above. Cells were collected by centrifugation and lysed with 2× SDS loading buffer (200 mm Tris-HCl, 200 mm dithiothreitol, 4% SDS, 0.2% bromphenol blue, and 25% glycerol). Samples were then boiled for 5 min and briefly centrifuged. These samples were subjected to electrophoresis on a 12.5% SDS-PAGE gel prior to transfer to a nitrocellulose membrane (35). The membrane was incubated overnight at 4 °C in blocking buffer (1% powdered milk with 0.05% Tween 20). The membrane was washed repeatedly in phosphate-buffered saline containing 0.05% Tween 20. Luciferase was detected with a polyclonal antiserum (1:5000 dilution in blocking buffer) followed by exposure to a Cy dye conjugated anti-rabbit secondary antibody (Odyssey IRDye). Unbound secondary antibody was removed by repeated washing with phosphate-buffered saline. Fluorescence excitation was carried out using an Odyssey Li-COR IR imager at 680 nm. Emission was detected at 700 nm and automatically corrected for background.
Protein Expression and Purification—Fre was amplified using pfu turbo DNA polymerase (Stratagene) from E. coli strain K-12 BW25113 with the nucleotide primer pair 5′-GGATGGCATATGACAACCTTAAGCTGTAAAGTGACC and 5′-GGATGGCTCGAGGATAAATGCAAACGCATCGCCAAAC (IDT). The resulting fragment was purified using the Qiaex II gel extraction kit (Qiagen) prior to digestion with NdeI and XhoI (New England Biolabs). This insert was ligated into a pET21b vector (Novagen). Single colony transformants in XL10 cells (Stratagene) were sequenced at the University of Arizona ARL core facility. A single isolate was found to lack any mutations and was designated pZCFRE1. The Fre protein was expressed from pZCFRE1 in a BL21 (λDE3) cell line after growth to an OD600 of 0.5 (Stratagene). Recombinant expression was initiated with the addition of isopropyl-1-thio-β-d-galactopyranoside to 1 mm. Expression continued for ∼6.5 h at 25 °C with constant agitation. The clarified lysate was applied to a custom nickel affinity column (Amersham Biosciences) and purified to >90% purity assessed by SDS-PAGE analysis. Purified protein was dialyzed extensively into buffer containing 100 mm Na+/K+ phosphate and 100 mm NaCl, pH 7.0. Using a high fidelity polymerase (pfu turbo, Stratagene), V. harveyi luciferase was amplified from the pJHD500 plasmid using the nucleotide primers 5′-gagcccctcgagcgagtgatatttg and 5′-ccatatgaaattcggaaacttccttc (IDT). The resulting insert was prepared in the same manner as before and ligated into a pET21b vector (Novagen). The vector resulted in the addition of six histidine residues to the C terminus of the β-subunit. Sequencing of the entire insert was used to verify fidelity (ARL sequencing facility, University of Arizona). This construct was designated pZCH2. Recombinant luciferase was expressed and purified as described above for Fre. For the co-precipitation experiments, luciferase was subcloned from pZCH2 into a pASKIBA-3c (IBA-GO) vector with the restriction sites XbaI and XhoI. The resulting luciferase contained a strep-II tag on the C terminus of the β-subunit. This construct was designated pZCB4. Protein was expressed as before except using anhydrotetracycline to a final concentration of 1 μg/liter (Sigma). The lysate was clarified by centrifuged at a high speed (15,000 rpm) but not purified prior to application to the strep tactin resin (IBA-GO).
Enzyme Assays—Coupled enzyme assays were carried out in buffer containing 100 mm Na+/K+ phosphate and 100 mm NaCl, pH 7.0. Assays were performed in two ways. For investigation of the effect of luciferase concentration in a coupled assay, reactions contained different amounts of luciferase, 1 μm Fre oxidoreductase, 10 mm decanal, 10 μm NADPH, and 2 μm FMN. For determination of the flavin Michaelis constants (Km), reactions contained different amounts of flavin, 1 μm Fre oxidoreductase, 10 μm decanal, 10 μm NADPH, and 5 μm luciferase. In both cases, substrates were placed in separate locations on the bottom of the glass assay vials as droplets. Reactions were initiated with the rapid injection of 1.0 ml of buffer. This step was necessary to ensure consistent mixing and to promote optimal aeration. Luciferase activity was also measured using the FMNH2 injection technique in which enzyme is incubated in 1.0 ml of 100 mm Na+/K+ phosphate containing 0.5 mg/ml bovine serum albumin and 0.001% aldehyde. Reactions were initiated by rapid injection of 1.0 ml of photoreduced FMNH2 (50 μm) (34).
Co-precipitation Experiment—Strep tactin resin (IBA-GO) was equilibrated in 1 mm EDTA, 100 mm Na+/K+ phosphate pH 7.0 (henceforth buffer A) by a 1:1000 dilution from buffer containing 50% ethanol. Following a brief incubation at 0 °C, beads were resuspended in buffer A. Clarified lysate (1.0 ml) containing strep-II-tagged luciferase and 1 mg of purified reductase (∼0.5 ml) was added to the beads, the final volume adjusted to 10 ml with buffer A, and allowed to incubate for a period of 1 h at 0 °C. The supernatant was decanted, and the beads were resuspended in 10 ml of buffer A. Aliquots of 100 μl were removed and assayed using the FMNH2 injection and the coupled assay. This process was repeated three times with 30-min incubations at 0 °C after each resuspension. After the final wash the beads were resuspended in 100 μl of buffer A containing 5 mm desthiobiotin (IBA-GO).
RESULTS AND DISCUSSION
The experiments reported here were undertaken to determine the source of the reducing equivalents required to drive the bacterial luciferase reaction in E. coli carrying the luciferase genes. We were also interested in the mechanism of flavin transfer from the oxidoreductase to luciferase. It has been appreciated for many years that bioluminescent bacteria possess pyridine nucleotide-dependent flavin oxidoreductases that can supply reduced flavin to bacterial luciferase in vitro, and it has been assumed that the same enzymes would also supply FMNH2 in vivo (17). Because bacterial luciferases function in recombinant E. coli in the absence of their endogenous oxidoreductases, it appears reasonable to assume that there must be analogous activities in E. coli that can also supply reducing equivalents to recombinant luciferase.
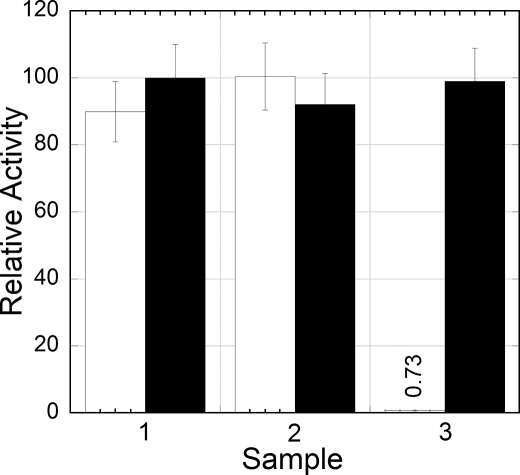
In V. harveyi, Frp has been reported to be the primary source of FMNH2 for luciferase, while in V. fischeri, Frase-I has been proposed to play the primary oxidoreductase role (15). Comparisons of the amino acid sequences show that these proteins are substantially different from each other and from E. coli flavin reductases. Expression of bioluminescence in E. coli lacking NfsA was the same as in the isogenic wildtype, but bioluminescence in the strain lacking Fre was reduced by over 99% (Fig. 2). These observations supply compelling evidence that bioluminescence in recombinant E. coli is dependent primarily on the activity of Fre. Furthermore, the results of these experiments demonstrate that NfsA is not competent to supply reduced flavin to recombinant luciferase to any significant extent. This observation is not surprising, as it is known that NfsA is a nitroreductase, not a flavin reductase, even though it can be converted to a flavin reductase by a single amino acid substitution, F124S (30). Bioluminescence from the Δfre strain was ∼1% that from the isogenic wildtype fre+ strain, suggesting that there may be other reductase activities in E. coli also competent to supply reduced flavin to the luciferase.
FIGURE 2.
Luciferase activity in both liquid culture (open bars) and crude lysate (filled bars). The three samples correspond to cultures of E. coli expressing luciferase from pJHD500 in: 1) K-12 BW25113, 2) K-12 BW25113 ΔNfsA, and 3) K-12 BW25113 Δfre. In the first set of measurements in vivo, aliquots (0.5 ml) of cells were assayed in triplicate using a bench top luminometer by the aldehyde injection technique (33). Measurements of luciferase activity in crude cell lysates were obtained using the flavin injection technique (34). The normalized value for K-12 BW25113 Δfre in the in vivo assay is given in the figure.
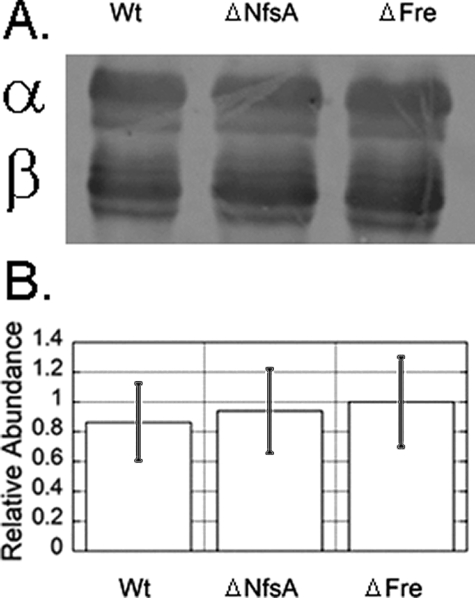
To exclude the possibility that the reduction in bioluminescence activity from the Δfre strain was due to a decrease in luciferase synthesis or accumulation in the cell, a Western blot was performed on a crude cell lysate (Fig. 3). The Western blot indicates no difference in the levels of luciferase expression in either the Δfre or ΔNsfA strains.
FIGURE 3.
Relative protein expression level of pJHD500 in the K-12 BW25113 deletion backgrounds. A, proteins in crude cell extracts were separated by SDS-PAGE prior to exposure to a dilute polyclonal rabbit anti-luciferase serum. The primary antibody preparation reacts preferentially with the β-subunit and does not reflect true differences in the relative expression of α- and β-subunits. Lane 1, pJHD500 expressed in K-12 BW25113; lane 2, pJHD500 expressed in K-12 BW25113 ΔNfsA; lane 3: pJHD500 expressed in K-12 BW25113 Δfre. B, relative quantity of each protein was determined using LiCor densitometry software.
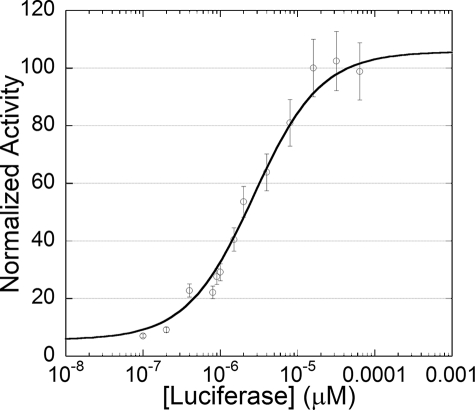
Experiments in vitro demonstrated that purified Fre is also competent to support bioluminescence in assays with purified reductase and luciferase with NADPH, either FMN or riboflavin, and n-decanal in the presence of atmospheric O2. The biochemical properties of purified Fre enzyme are different from the endogenous luminescent bacterial oxidoreductases. In the oxidoreductase coupled system, the influence of luciferase concentration on initial velocity was determined (see Fig. 4). Half-maximal activity occurred at a luciferase concentration of ∼1 μm, comparable to that found with the Frp enzyme (10). The bioluminescence activity in the coupled assay with Fre was, within experimental error, the same as that with Frp as the oxidoreductase (data not shown). The light emission from the reaction with Frp was ∼80% under similar but non-identical reaction conditions to that reported by Tu and co-workers (10).
FIGURE 4.
Influence of luciferase concentration on initial light intensity in the coupled reaction with Fre. Assays contained luciferase, 1 μm oxidoreductase, 10 μm decanal, 10 μm NADPH, and 2 μm FMN. Reactions were initiated with the rapid injection of 1.0 ml of buffer. Measurements were recorded once steady state light emission was reached, ∼5 s after injection. Data were fit to the Michaelis-Menten equation as described (50). This plot was generated with kaleidagraph (Synergy Software).
The influence of flavin substrate on oxidoreductase activity was also determined using the coupled assay (Table 2). Both FMN and riboflavin appear to have lower Michaelis constants relative to values determined spectrophotometrically (36). Consistent with the known substrate specificity of luciferase, riboflavin was capable of supporting bioluminescence but with a significantly lower quantum yield than FMN (37). The Fre oxidoreductase substrate specificity constant (kcat/Km) for riboflavin is 1257 m–1 · min–1 versus 73 m–1 · min–1 for FMN using NADPH as an electron donor (36). Even though the specificity constant of Fre strongly favors riboflavin as a substrate, reduced riboflavin is a very poor substrate for bacterial luciferase (Table 2). This difference may reflect evolutionary selection, as the other bioluminescence-supporting reductase enzymes are highly selective for FMN (15).
TABLE 2.
Kinetic parameters obtained from the Fre-luciferase coupled assay
Reactions were initiated with the rapid injection of buffer containing 100 mm Na+/K+ phosphate and 100 mm NaCl, pH 7.0.
| Flavin substrate | Light intensitya | Relative light intensity | Km coupledb | Km uncoupledc |
|---|---|---|---|---|
| (1011 × q·s-1) | μm | μm | ||
| FMN | 250 | 1 | 0.9 | 2.2 |
| Riboflavin | 7 | 0.028 | 1.3 | 2.5 |
Values determined 5 s after initiation of the reaction.
Michaelis constants were determined with different flavin concentrations, 1 μm Fre oxidoreductase, 10 mm decanal, and 10 μm NADPH and 5 μm luciferase. Values based upon plots of velocity measured by light output versus flavin concentration.
Taken from spectroscopic measurements previously reported (36).
Tu and co-workers (10) have proposed a flavin transfer mechanism involves the monomeric oxidoreductase interacting with luciferase. The well characterized oxidoreductase enzymes undergo a reversible monomer-dimer equilibrium (15), whereas, Fre exists exclusively as a monomer (15, 38). The V. harveyi oxidoreductase, Frp, has optimal activity at low micromolar concentrations in luciferase coupled assays, which have been suggested to correlate with a micromolar monomer-dimer equilibrium dissociation constant (11). These facts highlight significant differences in quaternary structure between these proteins.
The sequence identity between the Fre from E. coli and Fre from V. fischeri is 51% (28), indicative of an homologous relationship. The lack of sequence similarity between Fre and the endogenous oxidoreductase Frp indicates an ancient evolutionary relatedness or convergent evolution of the two proteins. Comparison of our results with previous studies with purified Frp from V. harveyi demonstrated that both enzymes catalyze FMN reduction with comparable efficiency in vitro (10). Based on our finding that Fre from E. coli is capable of supporting bioluminescence, we believe that Fre and Frp may be functionally redundant in vivo.
During maximal bioluminescence, V. harveyi maintains concentrations in vivo of Frp and luciferase of 3 μm and 172 μm, respectively, (11). This large concentration difference would necessitate a transient interaction between donor and acceptor to permit one Frp molecule to service multiple luciferase molecules. The high catalytic turnover number of Frp (2160 min–1) relative to that of luciferase (2–20 min–1) is consistent with the difference in concentrations regardless of the process by which reduced flavin is transferred from the reductase to luciferase (16, 39).
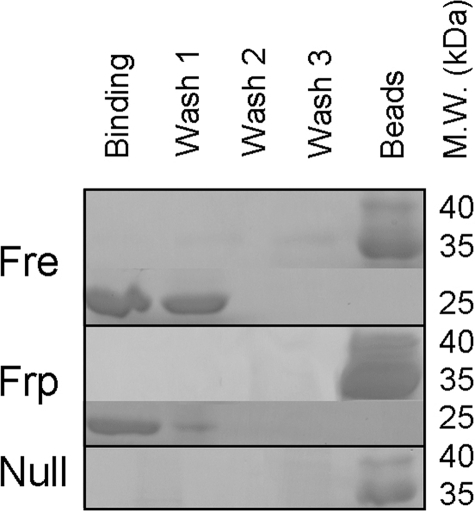
It is unknown if Fre interacts with luciferase. We generated a version of luciferase containing a C-terminal strep tag on the β-subunit for a co-purification experiment (Fig. 5). Because of the remarkable sensitivity of the instrumentation we use to observe light emission, residual reductase activity should be readily detected in a coupled assay. We used both Fre and Frp in these experiments, and in both cases, after a single wash step all reductase activity was lost based on the coupled assay (data not shown). This result agrees with the absence of a band at this step detected by SDS-PAGE. It has been reported that luciferase forms a stable complex with Frp (18). Our laboratory has been unable to reproduce this finding using crosslinking in vivo, crosslinking in vitro, analytical ultracentrifugation, and now a pull-down assay. These negative results suggest that either luciferase does not form a complex with either oxidoreductase, or the complex is extremely transient.
FIGURE 5.
SDS-PAGE analysis of oxidoreductase co-purification with luciferase. Approximately 1 mg of each oxidoreductase was incubated in a 10-ml solution containing immobilized luciferase. Oxidoreductase proteins were incubated with luciferase and repeatedly washed. Aliquots were visualized directly by SDS page and separately assayed using both the flavin injection and coupled bioluminescence assay (33, 34). All activity measured by the coupled assay was lost following the first wash step concurrent with the results obtained by SDS-PAGE. Approximate molecular weights are shown on the right portion of the panel.
It has previously been noted that under standard Frp coupled assay conditions, ∼70% of the reduced flavin can be provided by free diffusion (40). These authors demonstrate that in the coupled assay, bioluminescence is quenched by increasing O2, a result consistent with the free diffusion model. However, these authors chose to interpret their result to include a mix of free diffusion and complex formation. It is conceivable that the E. coli oxidoreductase Fre does not interact directly with luciferase and thus bypasses any mechanism involving protein-protein interaction or electron transfer. This hypothesis is consistent with the lack of structural homology between Fre and other oxidoreductase proteins (41). Fre is monomeric, with an all beta strand Greek key fold. The crystal structure of Frp reveals a similar protein fold to that of FRase-1, a strand-exchanged dimer with a mixed three layer α/β/α sandwich core (42). This is also consistent with the results of the co-precipitation experiment.
Significant attention has been paid to the bacterial luciferase mechanism in vitro (2, 43, 44). Two assay methods are commonly used to quantitate enzymatic activity. In the first, aldehyde is incubated with luciferase and reduced flavin is rapidly injected (33, 34). In the second assay, luciferase is incubated with FMN, and a small amount of sodium dithionite is added to reduce the flavin immediately prior to aldehyde injection (34). The second assay method gives a specific activity value approximately double that of the FMNH2 injection method. Aldehyde binding to free enzyme prevents binding of FMNH2, leading to inhibition (45, 46). The proposed kinetic mechanism dictates ordered sequential binding of reduced flavin prior to aldehyde (47). Binding of aldehyde to the enzyme-reduced flavin complex slows the rate of reaction with O2. The rate of formation of 4a-hydroperoxyflavin is two orders of magnitude more rapid prior to aldehyde binding (47). If the proposed mechanism for the bioluminescent reaction in vitro is analogous to that inside the cell, then the kinetically preferred pathway of flavin transfer or binding would favor aldehyde-free luciferase. If direct transfer of reduced flavin from the reductase to the luciferase is the primary mechanism for supplying FMNH2 to luciferase in vivo, then three rather divergent oxidoreductase proteins must be capable of recognizing flavin-free luciferase (41, 42, 48).
Summary—Several reports propose a stable complex between luciferase and a flavin oxidoreductase involved in flavin transfer. We support the opposing mechanism whereby luciferase obtains reduced flavin by free-diffusion (15). Our argument is 5-fold.
First, the oxidoreductase enzymes from non-bioluminescent bacteria are fully competent to provide the reduced flavin substrate in vivo. This reaction is approximately as efficient with purified enzyme using the endogenous reductase enzyme. If there were direct flavin transfer via a protein-protein complex, there must be specificity in the encounter complex between a reductase and ligand-free luciferase. Given the lack of sequence similarity between Fre and Frp, this appears highly unlikely.
Second, despite numerous attempts, we have never been able to reproduce the finding that any oxidoreductase enzyme forms either a stable or transient complex with luciferase. It has been suggested that the homodimerization of Frp inhibits both interaction and transfer between the reductase-luciferase pair (11). Furthermore, several techniques that were interpreted to demonstrate complex formation have been justified based on incorrect assumptions (11, 40, 49). Traditional techniques (gel filtration, analytical centrifugation etc.) necessitate protein concentrations above the Frp oxidoreductase dissociation constant for homodimerization. Therefore, they are supposedly unable to detect a complex between the monomeric FRP and the luciferase. This interpretation conflicts with basic thermodynamics. While at concentrations above the equilibrium dissociation constant, the fraction monomer decreases, the absolute concentration of monomer increases at increasing protein concentration. The reductase characterized in this work (Fre) is strictly a monomer and we did not detect any interaction with luciferase using a pull-down assay (41).
Third, when provided with aldehyde, luciferase is a highly active reporter in vivo in diverse organisms ranging from plants to fungi (1, 3). The existence of a bioluminescence-specific reductase in such a diverse range of non-bioluminescent organisms appears unlikely.
Fourth, the substrate specificity of Fre is remarkably broad. Many highly active reductase enzymes possess broad substrate specificity (15). It is likely that luciferase simply borrows this metabolite from the available pool of reduced flavin. The source of flavin appears to be inconsequential.
And finally, the cluster of luciferase-related genes known to be related co-transcriptionally lacks a highly active flavin reductase (3). This suggests that the supply of reduced flavin is not limiting for bioluminescence, unlike the supply of aldehyde substrate. Genes of common function are often clustered in bacteria as are genes that encode proteins which physically interact. The lack of conservation of gene context within bioluminescent bacteria suggests that the reductase enzymes also perform physiological tasks unrelated to bioluminescence.
To the best of our knowledge, this study represents the first demonstration of a specific flavin oxidoreductase from a non-bioluminescent organism capable of supporting bacterial bioluminescence in vivo and in vitro. This work supports the free-diffusion model for obtaining reduced flavin for the bioluminescence reaction.
Acknowledgments
We thank Dr. Miriam Ziegler for detailed analysis and stylistic suggestions in preparing this manuscript.
This work was supported by National Science Foundation Grant MCB 0078363. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
References
- 1.Baldwin, T. O., and Ziegler, M. M. (1992) in Chemistry and Biochemistry of Flavoenzymes, (Müller, F., ed) pp. 467–530, CRC Press, Boca Raton
- 2.Eberhard, A., and Hastings, J. W. (1972) Biochem. Biophys. Res. Commun. 47 348–353 [DOI] [PubMed] [Google Scholar]
- 3.Meighen, E. A. (1991) Microbiol. Rev. 55 123–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin, T. O., Berends, T., Bunch, T. A., Holzman, T. F., Rausch, S. K., Shamansky, L., Treat, M. L., and Ziegler, M. M. (1984) Biochemistry 23 3663–3667 [DOI] [PubMed] [Google Scholar]
- 5.Galan, B., Diaz, E., Prieto, M. A., and Garcia, J. L. (2000) J. Bacteriol. 182 627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanc, V., Lagneaux, D., Didier, P., Gil, P., Lacroix, P., and Crouzet, J. (1995) J. Bacteriol. 177 5206–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker, D., Schrader, T., and Andreesen, J. R. (1997) Eur J Biochem. 249 739–747 [DOI] [PubMed] [Google Scholar]
- 8.Parry, R. J., and Li, W. (1997) J. Biol. Chem. 272 23303–23311 [DOI] [PubMed] [Google Scholar]
- 9.Gibson, Q. H., and Hastings, J. W. (1962) Biochem. J. 83 368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei, B., and Tu, S. C. (1998) Biochemistry 37 14623–14629 [DOI] [PubMed] [Google Scholar]
- 11.Jeffers, C. E., Nichols, J. C., and Tu, S. C. (2003) Biochemistry 42 529–534 [DOI] [PubMed] [Google Scholar]
- 12.Liu, Y., Golden, S. S., Kondo, T., Ishiura, M., and Johnson, C. H. (1995) J. Bacteriol. 177 2080–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon, S., Chung, G., and Andrew, P. (1998) in Mycobacteria Protocols (Parish, T., and Stoker, N. G., eds) pp. 235–244, Humana Press
- 14.Zenno, S., Koike, H., Kumar, A. N., Jayaraman, R., Tanokura, M., and Saigo, K. (1996) J. Bacteriol. 178 4508–4514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu, S. C. (2001) Antioxid. Redox Signal. 3 881–897 [DOI] [PubMed] [Google Scholar]
- 16.Lei, B., Liu, M., Huang, S., and Tu, S. C. (1994) J. Bacteriol. 176 3552–3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jablonski, E., and DeLuca, M. (1976) Proc. Natl. Acad. Sci. U. S. A. 73 3848–3851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tu, S. C., and Hastings, J. W. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 249–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zenno, S., Saigo, K., Kanoh, H., and Inouye, S. (1994) J. Bacteriol. 176 3536–3543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inouye, S. (1994) FEBS Lett. 347 163–168 [DOI] [PubMed] [Google Scholar]
- 21.Spyrou, G., Haggard-Ljungquist, E., Krook, M., Jornvall, H., Nilsson, E., and Reichard, P. (1991) J. Bacteriol. 173 3673–3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zenno, S., and Saigo, K. (1994) J. Bacteriol. 176 3544–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, J. W., Chao, Y. F., and Weng, S. F. (1998) Biochem. Biophys. Res. Commun. 246 446–452 [DOI] [PubMed] [Google Scholar]
- 24.Duane, W., and Hastings, J. W. (1975) Mol. Cell Biochem. 6 53–64 [DOI] [PubMed] [Google Scholar]
- 25.Jablonski, E., and DeLuca, M. (1977) Biochemistry 16 2932–2936 [DOI] [PubMed] [Google Scholar]
- 26.Puget, K., and Michelson, A. M. (1972) Biochimie (Paris) 54 1197–1204 [DOI] [PubMed] [Google Scholar]
- 27.Schmidt, T. M., Kopecky, K., and Nealson, K. H. (1989) Appl. Environ. Microbiol. 55 2607–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990) J. Mol. Biol. 215 403–410 [DOI] [PubMed] [Google Scholar]
- 29.Zenno, S., Koike, H., Tanokura, M., and Saigo, K. (1996) J. Bacteriol. 178 4731–4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zenno, S., Kobori, T., Tanokura, M., and Saigo, K. (1998) J. Bacteriol. 180 422–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baba, T., Ara, T., Hasegawa, M., Takai, Y., Okumura, Y., Baba, M., Datsenko, K. A., Tomita, M., Wanner, B. L., and Mori, H. (2006) Mol. Syst. Biol. 2 2006 0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Devine, J. H., Shadel, G. S., and Baldwin, T. O. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 5688–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu, S., and Hastings, J. W. (1975) Biochemistry 14 4310–4316 [DOI] [PubMed] [Google Scholar]
- 34.Hastings, J. W., Baldwin, T. O., and Nicoli, M. Z. (1978) in Methods in Enzymology (DeLuca, M., ed) Academic Press, New York
- 35.Schagger, H., and von Jagow, G. (1987) Anal. Biochem. 166 368–379 [DOI] [PubMed] [Google Scholar]
- 36.Fieschi, F., Niviere, V., Frier, C., Decout, J. L., and Fontecave, M. (1995) J. Biol. Chem. 270 30392–30400 [DOI] [PubMed] [Google Scholar]
- 37.Meighen, E. A., and MacKenzie, R. E. (1973) Biochemistry 12 1482–1491 [DOI] [PubMed] [Google Scholar]
- 38.Gaudu, P., Touati, D., Niviere, V., and Fontecave, M. (1994) J. Biol. Chem. 269 8182–8188 [PubMed] [Google Scholar]
- 39.Wang, H., Lei, B., and Tu, S. C. (2000) Biochemistry 39 7813–7819 [DOI] [PubMed] [Google Scholar]
- 40.Li, X., and Tu, S. C. (2006) Arch. Biochem. Biophys. 454 26–31 [DOI] [PubMed] [Google Scholar]
- 41.Ingelman, M., Ramaswamy, S., Niviere, V., Fontecave, M., and Eklund, H. (1999) Biochemistry 38 7040–7049 [DOI] [PubMed] [Google Scholar]
- 42.Tanner, J. J., Tu, S. C., Barbour, L. J., Barnes, C. L., and Krause, K. L. (1999) Protein Sci. 8 1725–1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balny, C., and Hastings, J. W. (1975) Biochemistry 14 4719–4723 [DOI] [PubMed] [Google Scholar]
- 44.Raushel, F. M., and Baldwin, T. O. (1989) Biochem. Biophys. Res. Commun. 164 1137–1142 [DOI] [PubMed] [Google Scholar]
- 45.Lei, B., Cho, K. W., and Tu, S. C. (1994) J. Biol. Chem. 269 5612–5618 [PubMed] [Google Scholar]
- 46.Francisco, W. A., Abu-Soud, H. M., Baldwin, T. O., and Raushel, F. M. (1993) J. Biol. Chem. 268 24734–24741 [PubMed] [Google Scholar]
- 47.Francisco, W. A., Abu-Soud, H. M., DelMonte, A. J., Singleton, D. A., Baldwin, T. O., and Raushel, F. M. (1998) Biochemistry 37 2596–2606 [DOI] [PubMed] [Google Scholar]
- 48.Koike, H., Sasaki, H., Kobori, T., Zenno, S., Saigo, K., Murphy, M. E., Adman, E. T., and Tanokura, M. (1998) J. Mol. Biol. 280 259–273 [DOI] [PubMed] [Google Scholar]
- 49.Tu, S. C., Lei, B., Liu, M., Tang, C. K., and Jeffers, C. (2000) J Nutr 130 Suppl. 2S, 331S–332S [DOI] [PubMed] [Google Scholar]
- 50.Sparks, J. M., and Baldwin, T. O. (2001) Biochemistry 40 15436–15443 [DOI] [PubMed] [Google Scholar]
- 51.Murzin, A. G., Brenner, S. E., Hubbard, T., and Chothia, C. (1995) J. Mol. Biol. 247 536–540 [DOI] [PubMed] [Google Scholar]