Abstract
In immune cells, generation of sustained Ca2+ levels is mediated by the Ca2+ release-activated Ca2+ (CRAC) current. Molecular key players in this process comprise the stromal interaction molecule 1 (STIM1) that functions as a Ca2+ sensor in the endoplasmic reticulum and ORAI1 located in the plasma membrane. Depletion of endoplasmic reticulum Ca2+ stores leads to STIM1 multimerization into discrete puncta, which co-cluster with ORAI1 to couple to and activate ORAI1 channels. The cytosolic C terminus of STIM1 is sufficient to activate ORAI1 currents independent of store depletion. Here we identified an ORAI1-activating small fragment (OASF, amino acids 233–450/474) within STIM1 C terminus comprising the two coiled-coil domains and additional 50–74 amino acids that exhibited enhanced interaction with ORAI1, resulting in 3-fold increased Ca2+ currents. This OASF, similar to the complete STIM1 C terminus, displayed the ability to homomerize by a novel assembly domain that occurred subsequent to the coiled-coil domains. A smaller fragment (amino acids 233–420) generated by a further deletion of 30 amino acids substantially reduced the ability to homomerize concomitant to a loss of coupling to as well as activation of ORAI1. Extending OASF by 35 amino acids (233–485) did not alter homomerization but substantially decreased efficiency in coupling to and activation of ORAI1. Expressing OASF in rat basophilic leukemia (RBL) mast cells demonstrated its enhanced plasma membrane targeting associated with 2.5-fold larger CRAC currents in comparison with the complete STIM1 C terminus. In aggregate, we have identified two cytosolic key regions within STIM1 C terminus that control ORAI1/CRAC activation: a homomerization domain indispensable for coupling to ORAI1 and a modulatory domain that controls the extent of coupling to ORAI1.
Store-operated Ca2+ entry is key to cellular regulation of short term responses such as contraction and secretion as well as long term processes like proliferation and cell growth (1). The prototypic and best characterized store-operated channel is the Ca2+ release-activated Ca2+ (CRAC)5 channel (2–6). However, its molecular components have remained elusive until 3 years ago; the stromal interacting molecule 1 (STIM1) (7, 8) and later on ORAI1 (9–11) have been identified as the two limiting components for CRAC activation. STIM1 is an ER-located Ca2+ sensor (7, 8, 12), and store depletion triggers its aggregation into puncta close to the plasma membrane, resulting in stimulation of CRAC currents (13, 14). Its N terminus is located in the ER lumen and contains an EF-hand Ca2+ binding motif that senses the ER Ca2+ level and a sterile α motif that is suggested to mediate homomeric STIM1 aggregation (15, 16). In the cytosolic STIM1 C terminus, two coiled-coil regions overlapping with the ezrin-radixin-moesin (ERM)-like domain and a lysine-rich region have been proposed as essential for CRAC activation (15, 17, 18). ORAI1 has been assumed to act in concert with STIM1 (10, 19, 20), activating inward Ca2+ currents after store depletion. We and others have recently provided evidence that store depletion leads to a dynamic coupling of STIM1 to ORAI1 (21–23), probably involving the putative coiled-coil domain in the C terminus of ORAI1 (22). Furthermore, the C terminus of STIM1 has been established as the key fragment for CRAC as well as ORAI1 activation because its expression alone, without the necessity to deplete ER store, is sufficient for constitutive current activation (18, 22, 24).
In this study, we focused on identifying a small ORAI1-activating fragment (OASF) of STIM1 C terminus. Such a functional fragment (aa 233–450/474) that contained an indispensable homomerization domain fully coupled to and activated ORAI1 channels. Moreover, OASF extension up to aa 485 resulted in reduced constitutive currents and coupling efficiency, uncovering a small domain that modulates ORAI1 channel activity in an inhibitory manner.
MATERIALS AND METHODS
Molecular Cloning and Mutagenesis—Human ORAI1 (ORAI1; accession number NM_032790) was kindly provided by the A. Rao laboratory (Harvard Medical School). Human ORAI2 (ORAI2; accession number NM_032831.1) and ORAI3 (ORAI3; accession number NM_152288.1) were courtesy of the Lutz Birnbaumer laboratory (NIEHS, National Institutes of Health, Research Triangle Park, NC). N-terminally tagged ORAI1 constructs were cloned via SalI and SmaI restriction sites of pECFP-C1 and pEYFP-C1 expression vectors (Clontech). For N-terminally tagged ORAI2 constructs, the restriction sites KpnI and XbaI were used, and for ORAI3, BamHI and XbaI were used.
pECFP/pEYFP-C1/ORAI1 served as a template for the generation of the coiled-coil mutant L273S. Suitable primers exchanged the corresponding codon from GAG to TCG (L273S) using the QuikChange XL site-directed mutagenesis kit (Stratagene).
Human STIM1 (STIM1; accession number NM_003156) N-terminally enhanced CFP- and enhanced YFP-tagged was kindly provided by the T. Meyer laboratory, Stanford University. pEYFP-C1 Stim1 was used as template for the generation of pEYFP-C1 Stim1 1–420, 1–450, and 1–485 by introducing a stop codon at amino acid positions 421, 451, and 485, respectively.
STIM1 C terminus (aa 233–685) and the coiled coil deletion mutants (aa 353–685 and 400–685) were cloned into the T/A site of pcDNA3.1V5 His TOPO by PCR and subcloned into pECFP-C1 and pEYFP-C1 via their internal restriction sites KpnI and XbaI. pECFP-C1 and pEYFP-C1 STIM1 C terminus was used as template for the generation of the STIM1 fragments by introducing a stop codon at position 400 (aa 233–399), position 421 (aa 233–420), position 451 (aa 233–450), position 475 (aa 233–474), position 486 (aa 233–485), and position 536 (aa 233–535) using the QuikChange XL site-directed mutagenesis kit (Stratagene). STIM1 C terminus fragment aa 400–474 was generated by PCR, cloned into the T/A site of pcDNA3.1V5 His TOPO, and subcloned into pECFP-C1 and pEYFP-C1 via their internal restriction sites KpnI and XbaI. The integrity of all resulting clones was confirmed by sequence analysis.
Expression of STIM1 C-terminal Fragments—YFP-tagged STIM1 C terminus fragments were obtained from transiently transfected HEK293 cells. The cells were harvested 24 h after transfection, washed with 1× phosphate-buffered saline, and resuspended in Dulbecco's modified Eagle's medium high glucose without supplements. Afterward the cells were broken up by sonification, cell debris were pelleted by centrifugation at 14,000 rpm for 10 min, and the supernatants were used for PFO-PAGE (ABCR GmbH & Co. KG).
PFO-PAGE—The methods employed for PFO-PAGE (25) were similar to those employed for SDS-PAGE. Freshly poured 12% gels without SDS were used. Cell lysates were mixed with doubly concentrated sample buffer, incubated for 30 min at room temperature, and then applied to the gel. The double-concentrated sample buffer contained 100 mm Tris base, 0.8% NaPFO, 20% glycerol, 0.005% bromphenol blue, pH 8.0, adjusted with NaOH. The running buffer contained 25 mm Tris, 192 mm glycine, and 0.1% PFO, pH 8.1. Protein bands were analyzed by Western blot analysis. The transfer buffer contained 25 mm Tris, 192 mm glycine, and 20% methanol. Protein detection was carried out using a primary anti-green fluorescent protein mouse IgG antibody (Roche Applied Science) and a secondary anti-mouse IgG peroxidase conjugate (Sigma). Proteins were detected by developing with the ECL detection kit (GE Healthcare).
Electrophysiology and Cell Transfection—Electrophysiological recordings comparing characteristics of 2–3 constructs were carried out in paired comparison on the same day. Expression pattern and the levels of the various constructs were carefully monitored by confocal fluorescence microscopy and were not significantly changed by the introduced mutations. Experiments were performed at 20–24 °C, using the patch clamp technique in the whole-cell recording configuration. For STIM1/ORAI as well as STIM1 C terminus/ORAI current measurements, voltage ramps were usually applied every 5 s from a holding potential of 0 mV, covering a range of –90 to 90 mV over 1 s. The internal pipette solution contained (in mm) 3.5 MgCl2, 145 cesium methane sulfonate, 8 NaCl, 10 HEPES, 10 EGTA, pH 7.2. Extracellular solution consisted of (in mm) 145 NaCl, 5 CsCl, 1 MgCl2, 10 HEPES, 10 glucose, 10 CaCl2, pH 7.4. Currents were leak-corrected by subtracting the leak current obtained in the presence of 10 μm LaCl3. Bio-Rad TransFectin reagent was used for transient transfection, and following 12–48 h, cells were employed for experiments. For reducing cell density, cells were sometimes reseeded >7 h before the experiments started.
Confocal Förster Resonance Energy Transfer (FRET) Fluorescence Microscopy—Confocal FRET microscopy was performed similarly as in (26). In brief, a QLC100 real-time confocal system (VisiTech International) was used for recording fluorescence images connected to two Photometrics CoolSNAPHQ monochrome cameras (Roper Scientific) and a dual port adapter (dichroic, 505 long pass; cyan emission filter, 485/30; yellow emission filter, 535/50; Chroma Technology Corp.). This system was attached to an Axiovert 200M microscope (Zeiss) in conjunction with an argon ion multiwavelength (457, 488, 514 nm) laser (Spectra Physics). The wavelengths were selected by an Acousto optical tunable filter (VisiTech International). MetaMorph 5.0 software (Universal Imaging Corp.) was used to acquire images and to control the confocal system. Illumination times of about 900–1500 ms were typically used for CFP, FRET, and YFP images that were consecutively recorded with a minimum delay. Prior to the calculation, the images had to be corrected due to cross-talk as well as cross-excitation. For this, the appropriate cross-talk calibration factors were determined for each of the constructs on the day the FRET experiments were performed. The corrected FRET image (NFRET) was calculated on a pixel-to-pixel basis after background subtraction and threshold determination using a custom-made software (27) integrated in MatLab 7.0.4 according to the method published by Ref. 28. The local ratio between CFP and YFP might vary due to different localizations of diverse protein constructs, which could lead to the calculation of false FRET values (29). Accordingly, the analysis was limited to pixels with a CFP:YFP molar ratio between 1:10 and 10:1 to yield reliable results (29).
Statistics—Mean ± S.E. values were shown throughout this study. Significance analysis was performed with the two-tailed Mann-Whitney test.
RESULTS AND DISCUSSION
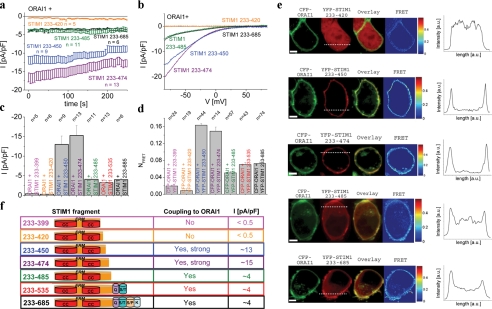
A 217-amino Acid Fragment of STIM1 C Terminus Allows for Maximum Activation of ORAI1—Recently we and others demonstrated that the cytosolic portion of STIM1 (STIM1 C terminus, aa 233–685) is sufficient for constitutive activation of ORAI1 (22, 30–32) and CRAC (18) channels. Here, we aimed at the identification of an OASF of the STIM1 C terminus by generating several constructs with increasing deletions of the C-terminal end. These fragments were analyzed (Fig. 1) for their constitutive coupling to and activation of ORAI1 channels monitoring their in vivo functionality by a combined approach of confocal FRET microscopy and electrophysiology. Both N-terminally labeled YFP-STIM1 fragments and CFP-ORAI1 fragments were co-expressed with YFP-labeled STIM1 fragments in HEK 293 cells. Deletion of up to 200 amino acids from the C terminus (233–535, 233–485) retained (Fig. 1, a–c) constitutive activation of ORAI1 to a similar extent as obtained by the complete STIM1 C terminus (233–685). Further 11–35 aa deletion (233–474, 233–450) generated two remarkably more potent fragments that generated 3-fold higher levels of ORAI1 activity, whereas continued 30–51 aa deletion (233–420, 233–399) drastically abolished stimulating activity of these fragments (Fig. 1, a–c). Confocal fluorescence microscopy revealed three distinct patterns of distribution of these fragments when co-expressed with ORAI1, as additionally displayed in intensity profiles (Fig. 1e). The two shortest STIM1 C-terminal fragments (233–399, 233–420) exhibited uniform distribution even within the cell nucleus, whereas the functionally most potent fragments (233–450, 233–474) displayed impressive targeting to the plasma membrane. The three longer fragments with comparable efficiency (233–485, 233–535, 233–658) exhibited a clear enrichment close to the plasma membrane yet together with some cytosolic expression, possibly reflecting a reduced affinity for the coupling to ORAI1. Moreover, confocal FRET analysis revealed the strongest coupling to ORAI1 with the 233–450/474 fragments, intermediate with 233–485, 233–535, and 233–685 fragments and minor coupling with 233–399 and 233–420 fragments (Fig. 1, d and e), perfectly in line with the extent of constitutive currents recorded in electrophysiological experiments (Fig. 1c). Deletion of coiled-coil domains at the N-terminal side of STIM1 C terminus (Δ first coiled-coil, 353–685; Δ first + second coiled-coil, 400–685) resulted in fragments that both failed to activate ORAI1 currents (data not shown).
FIGURE 1.
a, time course of constitutive whole-cell inward currents at –74 mV of HEK293 cells expressing the following C-terminal STIM1 fragments: 233–420, 233–450, 233–474, 233–485, and 233–685 (STIM1 C terminus wild-type) with ORAI1. b, respective I/V curves of a from representative cells taken at t = 0 s. c, block diagram summarizing constitutive STIM1 C terminus-mediated current densities at t = 0 s from a. d, block diagram displaying FRET of STIM1 fragments with ORAI1. e, localization, overlay, and calculated FRET life cell image series of YFP-STIM1 fragments and CFP-ORAI1. Additional intensity plots represent localization of STIM1 fragments across the cell as indicated by the dashed line. f, a model depicting STIM1 fragments in correlation with their function regarding stimulation of as well as coupling to ORAI1. a.u., arbitrary units.
Consistent results were obtained by co-expression of corresponding deletion mutants of full-length STIM1 (1–420, 1–450, and 1–485,) with ORAI1 reaching current densities (pA/pF) at 200 s following store depletion of –0.23 ± 0.04 (n = 16), –12.31 ± 2.29 (n = 13), and –8.67 ± 2.36 (n = 13) in comparison with full-length STIM1 (–7.99 ± 1.42 (n = 7)), respectively. Thus, we uncovered the 233–450/474 strands of STIM1 C terminus as OASF together with the extended 475–485 portion probably playing a modulatory (inhibitory) role for the constitutive activation of ORAI1. Furthermore, a mandatory key domain for ORAI1 activation was represented by the 30-amino acid stretch between 420 and 450, the presence of which was essential for both coupling to and activation of ORAI1 channels. Based on the distinctly different cellular distributions of the shorter (up to 420) STIM1 C-terminal fragments that showed both cytosolic and clear nuclear localization, we suspected formation of at least dimers or larger assemblies from the longer STIM1 fragments possibly preventing passive nuclear entry, which has been reported to occur until a molecular mass between <40 and 60 kDa (33).
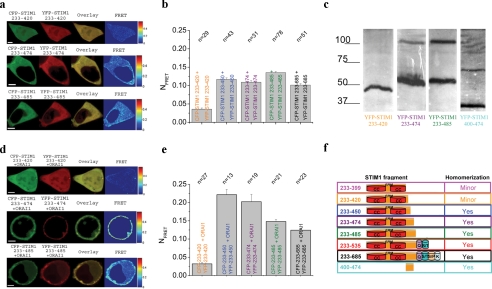
Minor Homomerization of the Shorter STIM1 (up to 420) Fragments Correlates with Their Inability to Activate ORAI1 Channels—Recently, single molecule fluorescence microscopy has suggested that activation in a tetrameric functional ORAI1 channel (30, 31) occurs by two STIM1 molecules (31). Based on the known dimerization/homomerization potential of STIM1, we examined next whether the failure of the two shorter STIM1 C-terminal fragments (233–399, 233–420) to couple to and activate ORAI1 might possibly reflect an alteration in their ability to homomerize in comparison with the longer STIM1 C-terminal fragments. When co-expressing CFP/YFP-tagged STIM1 fragments in HEK 293 cells, a similar picture emerged in that the shorter STIM1 fragments (up to 420) displayed (Fig. 2a) an almost uniform distribution both in the cytosol and in the nucleus, whereas the longer stretches (up to 450–685) mainly localized to the cytosol. The STIM1 C-terminal fragment 233–450 exhibited in about 40% of the cells a clustered, cytosolic expression pattern (data not shown), whereas the similarly effective 233–474 fragment showed more homogenous expression throughout the cytosol. Confocal FRET microscopy revealed a significantly reduced FRET for the two shorter STIM1 C terminus 233–399 and 233–420 fragments, suggesting diminished homomerization in contrast to the longer stretches ranging from 233–450 to 233–685 constructs (complete STIM1 C terminus) (Fig. 2c) that all showed comparably larger FRET. Employing an independent in vitro biochemical assay, we examined homomerization of expressed STIM1 C-terminal key fragments in a PFO-gel. Homomerization as judged from dimer formation was clearly evident for 233–474 and 233–485 fragments, whereas the shorter 233–420 construct displayed only a clear monomer band (Fig. 2c). However, a 400–474 fragment lacking both coiled-coil domains clearly retained the ability to homomerize, suggesting the presence of a novel assembly domain within STIM1 C terminus subsequent to the coiled-coil domains. Thus, it is tempting to speculate that nearly abolished homomerization of the shorter STIM1 C-terminal fragments accounted for the lack of ORAI1 activation evident both by a minor coupling to as well as by negligible constitutive activation of currents through ORAI1 channels. The longer STIM1 fragments that displayed functional coupling and stimulated ORAI1 currents showed a comparable level of robust homomerization FRET (Fig. 2, a–c). Thus, the difference in the extent of coupling to ORAI1 observed between 233–450/474 and 233–485 (Fig. 1d) is not reflected in the FRET determined from homomerized STIM1 fragments, suggesting that an increased homomerization per se might not account for this gain in function, particularly in the context that preferential dimers of STIM1 C terminus are proposed to couple to ORAI1 (31). Another explanation for the increased coupling to and activation of ORAI1 by OASF might be found in an enhanced affinity. In an attempt to address that point, we examined whether co-expression of the key STIM1 C-terminal fragments together with ORAI1 would alter their homomerization (Fig. 2, d and e). It indeed turned out that the most potent OASF 233–450/474 exhibited a clear localization close to the plasma membrane together with a significantly higher homomerization FRET than the longer 233–485/685 fragments compatible with an enhanced affinity for the former in their coupling to ORAI1. The shorter fragment 233–420 displayed neither localization to the plasma membrane nor a substantial homomerization FRET similar as observed when expressed in the absence of ORAI1 (compare Fig. 2, d and e, with Fig. 2, a and b). Therefore, in line with our biochemical data showing dimer formation for the 400–474 fragment, the 30/54- amino acid stretch between aa 420 and 450/474 might represent a novel cytosolic homomerization domain, the presence of which is required for both STIM1 C terminus homomultimerization as well as coupling to ORAI1. Hence, our results support the concept of an indispensable, cytosolic homomeric assembly domain assumedly located between 420 and 450 of STIM1 C terminus and a modulatory domain between 475 and 485 that additionally controls the extent of coupling to ORAI1. Consistent with our previous report on the requirement of an intact C terminus of ORAI1 for the coupling to STIM1 (22), OASF 233–450 similarly failed to couple to as well as activate the ORAI1 L273S mutant with a disrupted coiled-coil C-terminal motif (data not shown).
FIGURE 2.
a, localization, overlay, and calculated FRET life cell image series of YFP- and CFP-STIM1 fragments 233–420, 233–474, and 233–485. b, a block diagram summarizing homomerization FRET of STIM1 fragments: 233–420, 233–450, 233–474, 233–485, and 233–685 (complete STIM1 C terminus). c, PFO plot depicting monomers and possible dimers for the indicated STIM1 fragments. d, localization, overlay, and calculated FRET life cell image series of YFP- and CFP-STIM1 fragments 233–420, 233–474, and 233–485 in the presence of co-expressed ORAI1. e, block diagram summarizing homomerization FRET of STIM1 fragments as in b but now in the presence of co-expressed ORAI1. f, model depicting STIM1 fragments in correlation with their ability to homomultimerize.
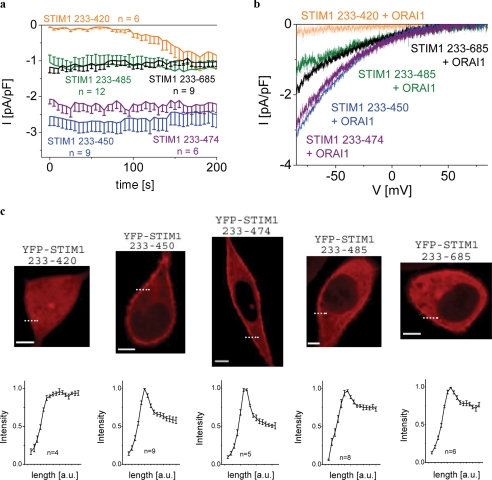
Native CRAC of RBL Mast Cells Responds to STIM1 C-terminal Fragments in a Similar Manner as Heterologously Expressed ORAI1 Channels—We used RBL mast cells to examine the STIM1 C-terminal fragments including the OASF on a native cell that exhibits robust endogenous CRAC activity (Fig. 3). Expression of STIM1 233–485 as well as full-length STIM1 C terminus (233–685) led to comparable, constitutive current densities of about 1 pA/pF, which are in a similar range as those observed from endogenous CRAC activation upon store depletion by 10 mm EGTA. Expression of STIM1 C terminus OASF 233–450/474 yielded 2.5-fold enhanced Ca2+ currents comparable with the potency on expressed ORAI1 in HEK cells (Figs. 1, a and c, and 3, a and b). Down-regulation of endogenous STIM1 in RBL cells by an small interfering RNA directed against a STIM1 N-terminal sequence revealed similar CRAC current densities induced by STIM1 C terminus 233–450 OASF (data not shown), indicating that endogenous STIM1 did not contribute to or affect its action. However, the smaller STIM C-terminal fragment 233–420 failed to constitutively activate endogenous CRAC currents, which, however, activated with the expected time delay upon store depletion in an undisturbed manner.
FIGURE 3.
a, time course of constitutive whole-cell inward currents at –74 mV of RBL cells expressing the following C-terminal STIM1 fragments: 233–420, 233–450, 233–474, 233–485, and 233–685 (STIM1 C terminus wild-type). b, respective I/V curves from a from representative cells taken at t = 0 s density activation. c, confocal fluorescence microscopy images of RBL cells expressing YFP-labeled STIM1 fragments: 233–420, 233–450, 233–474, 233–485, and 233–685 (complete STIM1 C terminus) and intensity plots representing localization of STIM1 fragments in regions close to the cell membrane as indicated by the dashed line. a.u., arbitrary units.
All the longer fragments displayed plasma membrane targeting, which was correlated with constitutive activation of native CRAC currents in RBL mast cells (Fig. 3c). Their relative efficiency for endogenous CRAC current activation was coherently reflected by the extent of their plasma membrane localization reaching a maximum with OASF 233–450/474 of STIM1 C terminus as depicted by corresponding fluorescence intensity plots (Fig. 3c, lower panel).
In summary, we resolved two cytosolic domains within STIM1 C terminus that are essential for the communication with ORAI1. The first is a cytosolic assembly domain that is indispensable for STIM1 homomerization and coupling to ORAI1, whereas the second domain exerts an inhibitory impact on STIM1/ORAI1 communication and may thus play a role in noise reduction or signal fine tuning. As a cluster of negative amino acids is located within this second inhibitory domain, it is tempting to speculate on their involvement in Ca2+-dependent feedback processes (34–36), although its sequence does not fully represent a canonical EF-hand (37). Our results are consistent with a recent report (32) demonstrating that the C-terminal polybasic region of STIM1 C terminus is essential for gating of TRPC channels but not for ORAI1. Moreover, OASF 233–450 constitutively activated in addition to ORAI1 also ORAI2 and ORAI3 with a potency comparable with full-length STIM1 (supplemental Fig. 1). In extension of recent results demonstrating STIM1 oligomerization as a critical transduction event of the CRAC activation cascade that involves an ER-luminally located assembly domain of STIM1 (38), our data provide evidence for an additional cytosolic homomerization domain, which is suggested as an essential determinant of STIM1 C terminus oligomerization and coupling to ORAI1.
Supplementary Material
Acknowledgments
We thank S. Buchegger and B. Kenda for excellent technical assistance.
This work was supported by the Austrian Science Foundation (FWF) Project P18280 (to K. G.) and Projects P18169 and P21118 as well as subproject 11 within W1201 (to C. R.).
The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure.
Footnotes
The abbreviations used are: CRAC, Ca2+ release-activated Ca2+; STIM, stromal interaction molecule; ER, endoplasmic reticulum; OASF, ORAI1-activating small fragment; aa, amino acids; YFP, yellow fluorescent protein; CFP, cyan fluorescent protein; PFO, perfluoro-octanoic acid; FRET, Förster resonance energy transfer; pF, picofarads; RBL, rat basophilic leukemia.
References
- 1.Berridge, M. J., Bootman, M. D., and Roderick, H. L. (2003) Nat. Rev. Mol. Cell Biol. 4 517–529 [DOI] [PubMed] [Google Scholar]
- 2.Hoth, M., and Penner, R. (1992) Nature 355 353–356 [DOI] [PubMed] [Google Scholar]
- 3.Zweifach, A., and Lewis, R. S. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 6295–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis, R. S., and Cahalan, M. D. (1990) Annu. Rev. Physiol. 52 415–430 [DOI] [PubMed] [Google Scholar]
- 5.Parekh, A. B., and Putney, J. W., Jr. (2005) Physiol. Rev. 85 757–810 [DOI] [PubMed] [Google Scholar]
- 6.Prakriya, M., and Lewis, R. S. (2003) Cell Calcium 33 311–321 [DOI] [PubMed] [Google Scholar]
- 7.Roos, J., DiGregorio, P. J., Yeromin, A. V., Ohlsen, K., Lioudyno, M., Zhang, S., Safrina, O., Kozak, J. A., Wagner, S. L., Cahalan, M. D., Velicelebi, G., and Stauderman, K. A. (2005) J. Cell Biol. 169 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liou, J., Kim, M. L., Heo, W. D., Jones, J. T., Myers, J. W., Ferrell, J. E., Jr., and Meyer, T. (2005) Curr. Biol. 15 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feske, S., Gwack, Y., Prakriya, M., Srikanth, S., Puppel, S. H., Tanasa, B., Hogan, P. G., Lewis, R. S., Daly, M., and Rao, A. (2006) Nature 441 179–185 [DOI] [PubMed] [Google Scholar]
- 10.Vig, M., Peinelt, C., Beck, A., Koomoa, D. L., Rabah, D., Koblan-Huberson, M., Kraft, S., Turner, H., Fleig, A., Penner, R., and Kinet, J. P. (2006) Science 312 1220–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, S. L., Yeromin, A. V., Zhang, X. H., Yu, Y., Safrina, O., Penna, A., Roos, J., Stauderman, K. A., and Cahalan, M. D. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 9357–9362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng, L., Stathopulos, P. B., Li, G. Y., and Ikura, M. (2008) Biochem. Biophys. Res. Commun. 369 240–246 [DOI] [PubMed] [Google Scholar]
- 13.Luik, R. M., Wu, M. M., Buchanan, J., and Lewis, R. S. (2006) J. Cell Biol. 174 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu, M. M., Buchanan, J., Luik, R. M., and Lewis, R. S. (2006) J. Cell Biol. 174 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baba, Y., Hayashi, K., Fujii, Y., Mizushima, A., Watarai, H., Wakamori, M., Numaga, T., Mori, Y., Iino, M., Hikida, M., and Kurosaki, T. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 16704–16709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stathopulos, P. B., Li, G. Y., Plevin, M. J., Ames, J. B., and Ikura, M. (2006) J. Biol. Chem. 281 35855–35862 [DOI] [PubMed] [Google Scholar]
- 17.Dziadek, M. A., and Johnstone, L. S. (2007) Cell Calcium 42 123–132 [DOI] [PubMed] [Google Scholar]
- 18.Huang, G. N., Zeng, W., Kim, J. Y., Yuan, J. P., Han, L., Muallem, S., and Worley, P. F. (2006) Nat. Cell Biol. 8 1003–1010 [DOI] [PubMed] [Google Scholar]
- 19.Mercer, J. C. (2006) J. Biol. Chem. 281 24979–24990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frischauf, I., Schindl, R., Derler, I., Bergsmann, J., Fahrner, M., and Romanin, C. (2008) Channels (Austin) 2 261–268 [DOI] [PubMed] [Google Scholar]
- 21.Barr, V. A., Bernot, K. M., Srikanth, S., Gwack, Y., Balagopalan, L., Regan, C. K., Helman, D. J., Sommers, C. L., Oh-Hora, M., Rao, A., and Samelson, L. E. (2008) Mol. Biol. Cell 19 2802–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muik, M., Frischauf, I., Derler, I., Fahrner, M., Bergsmann, J., Eder, P., Schindl, R., Hesch, C., Polzinger, B., Fritsch, R., Kahr, H., Madl, J., Gruber, H., Groschner, K., and Romanin, C. (2008) J. Biol. Chem. 283 8014–8022 [DOI] [PubMed] [Google Scholar]
- 23.Navarro-Borelly, L., Somasundaram, A., Yamashita, M., Ren, D., Miller, R. J., and Prakriya, M. (2008) J. Physiol. (Lond.) 586 5383–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, S. L., Kozak, J. A., Jiang, W., Yeromin, A. V., Chen, J., Yu, Y., Penna, A., Shen, W., Chi, V., and Cahalan, M. D. (2008) J. Biol. Chem. 283 17662–17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramjeesingh, M., Huan, L. J., Garami, E., and Bear, C. E. (1999) Biochem. J. 342 119–123 [PMC free article] [PubMed] [Google Scholar]
- 26.Singh, A., Hamedinger, D., Hoda, J. C., Gebhart, M., Koschak, A., Romanin, C., and Striessnig, J. (2006) Nat. Neurosci. 9 1108–1116 [DOI] [PubMed] [Google Scholar]
- 27.Derler, I., Hofbauer, M., Kahr, H., Fritsch, R., Muik, M., Kepplinger, K., Hack, M. E., Moritz, S., Schindl, R., Groschner, K., and Romanin, C. (2006) J. Physiol. (Lond.) 577 31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xia, Z., and Liu, Y. (2001) Biophys. J. 81 2395–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berney, C., and Danuser, G. (2003) Biophys. J. 84 3992–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Penna, A., Demuro, A., Yeromin, A. V., Zhang, S. L., Safrina, O., Parker, I., and Cahalan, M. D. (2008) Nature 456 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji, W., Xu, P., Li, Z., Lu, J., Liu, L., Zhan, Y., Chen, Y., Hille, B., Xu, T., and Chen, L. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 13668–13673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeng, W., Yuan, J. P., Kim, M. S., Choi, Y. J., Huang, G. N., Worley, P. F., and Muallem, S. (2008) Mol. Cell 32 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weis, K. (2003) Cell 112 441–451 [DOI] [PubMed] [Google Scholar]
- 34.Malli, R., Naghdi, S., Romanin, C., and Graier, W. F. (2008) J. Cell Sci. 121 3133–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamashita, M., Navarro-Borelly, L., McNally, B. A., and Prakriya, M. (2007) J. Gen. Physiol. 130 525–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lis, A., Peinelt, C., Beck, A., Parvez, S., Monteilh-Zoller, M., Fleig, A., and Penner, R. (2007) Curr. Biol. 17 794–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gifford, J. L., Walsh, M. P., and Vogel, H. J. (2007) Biochem. J. 405 199–221 [DOI] [PubMed] [Google Scholar]
- 38.Luik, R. M., Wang, B., Prakriya, M., Wu, M. M., and Lewis, R. S. (2008) Nature 454 538–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.