Abstract
Yeast cell wall remodeling is controlled by the equilibrium between glycoside hydrolases, glycosyltransferases, and transglycosylases. Family 72 glycoside hydrolases (GH72) are ubiquitous in fungal organisms and are known to possess significant transglycosylase activity, producing elongated β(1–3) glucan chains. However, the molecular mechanisms that control the balance between hydrolysis and transglycosylation in these enzymes are not understood. Here we present the first crystal structure of a glucan transglycosylase, Saccharomyces cerevisiae Gas2 (ScGas2), revealing a multidomain fold, with a (βα)8 catalytic core and a separate glucan binding domain with an elongated, conserved glucan binding groove. Structures of ScGas2 complexes with different β-glucan substrate/product oligosaccharides provide “snapshots” of substrate binding and hydrolysis/transglycosylation giving the first insights into the mechanisms these enzymes employ to drive β(1–3) glucan elongation. Together with mutagenesis and analysis of reaction products, the structures suggest a “base occlusion” mechanism through which these enzymes protect the covalent protein-enzyme intermediate from a water nucleophile, thus controlling the balance between hydrolysis and transglycosylation and driving the elongation of β(1–3) glucan chains in the yeast cell wall.
The cell wall of fungal organisms is a dynamic structure, providing protection against hostile environments, yet also harboring many hydrolytic and toxic molecules required for the fungus to invade its ecological niche (1). Polysaccharides account for over 90% of the cell wall. The central skeletal component of the cell wall common to the vast majority of fungal species is a branched core of β(1,3) glucan, linked to chitin via a β(1,4) linkage (1). Interchain β(1,6) glucosidic linkages account for 3 and 4% of the total glucan linkages in Saccharomyces cerevisiae and Aspergillus fumigatus, respectively (2–4). This core is embedded in a complex of amorphous proteins and/or polysaccharide whose composition is highly species-dependent. The core β(1,3) glucan is subjected to continuous synthetic elaboration, degradation, and remodeling by a large arsenal of enzymes, whose activities must be appropriately balanced to provide the cell wall with adequate elasticity to allow growth, budding, or branching and yet sufficient strength to guard against cell lysis (1).
Glucan synthase is a protein complex located at the plasma membrane, synthesizing β(1,3) glucan from UDP-glucose (65–90% of the total glucan). In cell wall remodeling, glycoside hydrolases and glycosyltransferases/transglycosylases play a crucial role (1, 5). Pure glycoside hydrolases degrade glycans mainly to regulate the plasticity of the cell wall under different circumstances, such as cell division, cell separation, and sporulation (5), whereas glycoside hydrolases with significant transglycosylase activity are capable of forming new glycosidic bonds between oligosaccharides, generating longer or branched polymers. Previous studies have shown that several proteins anchored to the plasma membrane by a glycosylphosphatidylinositol (GPI)3 anchor have transglycosylase activities (6–10). Among them are the Gas (in S. cerevisiae)/Gel (in A. fumigatus) proteins that belong to the GH72 family in the CAZy data base (11). For laminarioligosaccharides with >10 sugars, these enzymes are able to cleave a β(1–3) bond and transfer the newly formed reducing end (the “donor”) to the nonreducing end of another oligosaccharide (the “acceptor”) (6, 12, 13). This transferase reaction generates a new β(1,3) linkage, resulting in the elongation of β(1,3) glucan chains, offering a mechanism for the synthesis of longer glucan chains as alternative to, or in synergy with, glucan synthase. The Gas/Gel proteins consist of a signal sequence, a catalytic core, and either a cysteine-rich domain (classified as a carbohydrate-binding module, CBM43) (11) or a Ser-Thr-rich motif, followed by a GPI anchor (Fig. 1A). Based on the presence or absence of the C-terminal cysteine-rich domain, the family is subdivided into GH72+ (with a CBM43 domain) and GH72– (without a CBM43 domain) (14). The genome of S. cerevisiae contains five proteins (Gas1–Gas5), two of which (Gas1 and Gas2) belong to the GH72+ subfamily. With the exception of Gas3, transglycosylase activity has been reported for all these enzyme (14, 15). A. fumigatus contains seven genes (gel1–gel7), with only Gel1p, Gel2p (both GH72–), and Gel4p (GH72+) being expressed during mycelial growth in rich media (12). In Candida albicans five GH72 enzymes, PHR1–3 (known as pH-regulated enzymes) and PGA4–5, have been detected. PHR1, PHR2, and PGA5 belong to GH72+, whereas PHR3 and PGA4 belong to the GH72– subfamily. It was shown that all these proteins, irrespectively of the presence/absence of a CBM43 domain, display the same glycosyltransferase activity (14).
FIGURE 1.
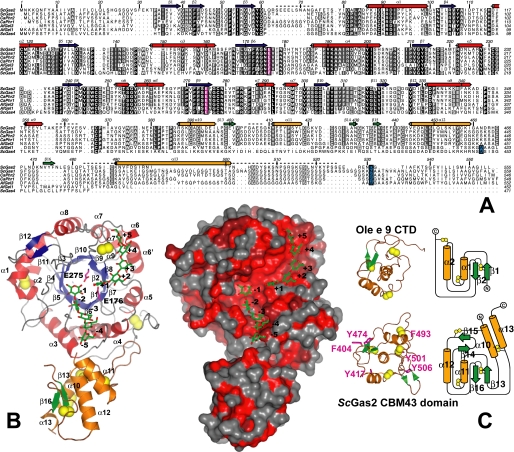
Overall structure of ScGas2 and comparison with structurally related proteins. A, multiple sequence alignment of the GH72 family members ScGas2, ScGas1, CaPHR2, CaPHR1, AfGel3, AfGel1, and ScGas4. Secondary structure elements from the ScGas2 structure are shown, with α-helices in red and orange for the catalytic and cysteine-rich domains, respectively, and β-strands correspondingly in blue and green. Regions that are disordered in some or all of the ScGas2 structures are marked with a dashed line. Conserved catalytic glutamate residues are highlighted in pink boxing, and the (predicted) GPI-anchor attachment site is indicated in blue boxing. B, overall crystal structure of ScGas2 in complex with laminaripentaose. The Glu176 and Glu275 are shown with pink carbon atoms and labeled. The seven disulfide bridges are highlighted in yellow. Helix α13 was not built in the laminaripentaose complex structure and thus is absent from this figure. Ligand molecules are shown as sticks with green carbon atoms and the sugar-binding sites are labeled –5 to +5, following standard nomenclature. Other colors as in A. Also shown is a surface representation of the ScGas2, colored by sequence conservation (red (100% identity) to gray (<50% identity)). C, comparison of the CBM43 domains of ScGas2 (bottom; E176Q mutant) and Ole e 9 (top; PDB ID 2JON (39)). Disulfide bridge sulfur atoms are shown as yellow spheres. Secondary structure elements are colored as in B and labeled. Unique features of either structure are shown in lighter colors in the picture (left). The topology diagram was drawn with Topdraw (50). Surface-exposed aromatic amino acids of the CBM43 domain of ScGas2 (Phe404, Tyr417, Tyr474, Phe493, Tyr501, and Tyr506) are shown as sticks with pink carbon atoms.
The essential function of these enzymes in fungal morphogenesis has been shown by gene disruption studies in a number of organisms. For instance, a gene knock-out of Scgas1 led to aberrant cell morphology, reduced growth rate, cell aggregation, and different cell wall composition (16, 17). A double knock-out of Scgas2 and Scgas4 showed a severe reduction in the efficiency of sporulation, an increased permeability of the spores to exogenous substances, and production of unviable spores (15). Single and double knockouts of Afge12 and double knockouts of Afge11/Afge12 resulted in slower growth, abnormal conidiogenesis, and an altered cell wall composition (12). Caphr1 and Caphr2 single knock-outs show defects in growth and morphogenesis, reduction in β(1,3) glucan-associated β(1,6) glucans, and a 5-fold increase in the chitin content of the walls (18–20).
Despite considerable interest in the molecular mechanisms of these transglycosylases, it is currently not understood what structure these enzymes adopt, how they interact with the substrate, what mechanism they adopt for the initial nucleophilic attack, and crucially, how they are able to drive the reaction toward transglycosylation and not hydrolysis in the presence of a high concentration of a more available nucleophile, water.
Here we present the first crystal structure of a GH72+ glucanosyltransferase enzyme, ScGas2, in complex with laminaripentaose and a complex with the hydrolysis products of laminariheptaose. The crystal structure reveals a (βα)8 catalytic core, tightly interacting with the C-terminal CBM43 glucan binding domain. The active site is located in an unusual tyrosine-rich groove, possessing two glutamic acids as catalytic residues. Site-directed mutagenesis data together with crystal structures of the ScGas2-oligosaccharide complexes shows that product binding in the acceptor site is crucial for tuning the balance between hydrolysis and transglycosylation.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification—The DNA sequence encoding amino acid residues 26–525 of the S. cerevisiae ScGas2 (Swiss-Prot, locus ScGas2_YEAST; Swiss-Prot accession number Q06135; GeneID, 851056), defined as scgas2, was obtained by PCR from S. cerevisiae strain W303 genomic DNA, using the forward primer, 5′-ATCTCGAGAAAAGAGAGGCTGAAGCTTCAGGTGTCAGCTTTGAAAAAACCCCTG, containing a recognition sequence for XhoI (underlined) and a KEX2 cleavage signal (italic), and the reverse primer, 5′-GCTCTAGACTAACTCGATGGGTACTTTACGTTCAAACTTTCC-3′, containing a recognition sequence for XbaI (underlined), and cloned into the pSC-B vector (Stratagene). Following digestion with XhoI and XbaI, the cloned sequence was subcloned into the Pichia pastoris protein expression and secretion vector pPICZαA (Invitrogen), resulting in the expression plasmid pPICZαAscgas2 (Ser26–Ser525). Subsequently, the codons for the asparagine residues at positions 498 and 510 in ScGas2 were simultaneously changed by site-directed mutagenesis to aspartic acid codons (removing N-linked glycosylation sites) using the forward primer, 5′-GAATGTTTTTGATTCTATAAGGGATATCACATACAATCATGGCGATTATTC-3′, and the reverse primer, 5′-CTTTCCTTGCTACGCGATGGATCTGATTTTGAATAATCGCCATG-3′.
The resulting plasmid, pPICZαAgas2 (Ser26–Ser525) N498D/N510D, referred to here as the wild type, was used as template for introducing the following single amino acid changes by site-directed mutagenesis as follows: Q62A, Y107F, Y107Q, D132N, N175A, E176Q, Y244F, Y244Q, E275Q, Y307Q, F404A, and Y474A, such that each of the resulting 12 plasmids carried the indicated mutation in addition to the previously introduced asparagine to aspartic acid mutations at positions 498 and 510. Site-directed mutagenesis was carried out following the QuikChange protocol (Stratagene), using the KOD HotStart DNA polymerase (Novagene). All plasmids were verified by sequencing (DNA Sequencing Service, College of Life Sciences, University of Dundee, Scotland, UK).
All plasmids were isolated from Escherichia coli strain DH5α, linearized with PmeI, and used to transform P. pastoris strain into X-33 following the LiCl method (Invitrogen) or using the Pichia EasyComp™ transformation kit (Invitrogen). Transformants were selected on YPD plates (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) dextrose) containing 100 μg/ml Zeocin (Invitrogen). Batch cultures were performed in a 100-ml volume of BMGY medium (1% (w/v) yeast extract, 2% (w/v) peptone, 100 mm potassium phosphate (pH 6.0), 1.34% (w/v) yeast nitrogen base, and 1% (v/v) glycerol). 50 ml were used to grow 500 ml of BMGY medium overnight at 30 °C, and expression was induced by methanol (1%, v/v) for 72 h at room temperature in a shaking incubator (270 rpm). Yeast cells were harvested by centrifugation at 3480 × g for 30 min. The supernatants containing soluble ScGas2 were filtered to 0.2 μm, concentrated to 50 ml using a Vivaflow 200 cassette (10,000 Mr weight cutoff, PES membrane; Vivascience), and dialyzed against water.
The samples were then loaded onto a 2 × 5-ml HiTrap Q FF column (Amersham Biosciences) that had been equilibrated with 10 column volumes of 25 mm Tris, pH 7.0, on an AKTA purifier system. Following loading, the column was washed with 10 column volumes of 25 mm Tris, pH 7.0. The protein was eluted with a salt gradient (0–500 mm NaCl) over 20 column volumes, collecting 2-ml fractions. The fractions containing the proteins were then pooled and concentrated to 5 ml using Vivaspin 10,000 Mr weight cutoff. Subsequently, gel filtration was carried out using a Superdex 75 XK26/60 column in 25 mm Tris, 150 mm NaCl, pH 7.0. The concentrated ScGas2 proteins were used for both kinetic analysis and crystallization trials.
Enzymology—To test for β(1,3) glucanosyltransferase/hydrolase activity, the purified proteins (10 μg) were incubated with the linear reduced laminarioligosaccharides rG5, rG7, and rG19 at a concentration of 4 mm in 50 mm sodium acetate buffer, pH 5.5, at 37 °C. Aliquots of 2.5 μl were withdrawn at different times (0, 1, and 3 h and overnight), supplemented with 47.5 μl of 50 mm NaOH, and then analyzed by high performance anion-exchange chromatography through a CarboPAC-PA1 column (Dionex 4.6 mm inner diameter × 250 mm), as described by Hartland et al. (21).
Crystallization and Data Collection—ScGas2 was spin-concentrated to 21 mg/ml. Crystals were grown by sitting drop experiments at 20 °C through mixing 1 μl of protein with an equal volume of a reservoir solution (20% 1,4-butanediol, 5% acetone, 0.1 m sodium acetate, pH 4.5). Under these conditions, crystals appeared within 3–7 days. They were cryoprotected with 0.1 m sodium acetate, 30% 1,4 butanediol, 5% acetone, pH 4.5, and flash-cooled prior to data collection at 100 K.
(NH4)3IrCl6 derivative, as well as oligosaccharide complexes, were generated by soaking apo-crystals in cryoprotectant supplemented with heavy atom salt (10–100 mm), laminaripentaose (200 mm), or laminariheptaose (100 mm), respectively, for 10–20 min prior to data collection. Data for heavy atom derivative was collected at beamline ID23-2 (ESRF, Grenoble, France), all other data were collected in-house on a Rigaku RU-200 rotating anode with an R-AXIS IV detector. All data were processed and scaled using the HKL suite (22) and CCP4 software (23), relevant statistics are given in Table 1.
TABLE 1.
Data collection and refinement statistics
Values in parentheses refer to the highest resolution shell. Ramachandran plot statistics were determined with PROCHECK (29). NA means not applicable.
| E176Q apo-structure | (NH4)3IrCl6 derivative | Laminaripentaose complex | Laminariheptaose complex | |
|---|---|---|---|---|
| Wavelength | 0.912 Å | 0.873 Å | 1.54 Å | 1.54 Å |
| Resolution | 20.00 to 1.62 Å (1.68 to 1.62 Å) | 25.00 to 2.10 Å (2.15 to 2.10 Å) | 20.00 to 1.85 Å (1.92 to 1.85 Å) | 20.00 to 1.90 Å (1.92 to 1.85 Å) |
| Cell dimensions | a = 051.88, b = 064.48, c = 152.04 Å | a = 050.37, b = 070.57, c = 150.91 Å | a = 050.04, b = 070.84, c = 149.15 Å | a = 050.02, b = 070.27, c = 148.91 Å |
| Unique reflections | 65,435 | 31,981 | 46,135 | 42,459 |
| Completeness | 98.2 (96.6) | 99 (100) | 99.7 (100) | 94.7 (97.8) |
| Rsym | 0.054 (0.250) | 0.077 (0.162) | 0.045 (0.293) | 0.043 (0.228) |
| I/σ(I) | 18.4 (4.1) | 21.0 (10.9) | 23.3 (5.5) | 20.2 (5.7) |
| Redundancy | 3.6 (2.9) | 8.2 (8.1) | 5.3 (5.2) | 4.4 (4.2) |
| No. of protein residues | 470 | 436 | 432 | |
| N0. of sugar residues | 000 | 010 | 007 | |
| Solvent molecules | 677 | 313 | 293 | |
| Rwork/Rfree | 0.175/0.217 | 0.186/0.209 | 0.182/0.190 | |
| r.m.s.d. from ideal geometry, bonds | 0.012 Å | 0.012 Å | 0.013 Å | |
| r.m.s.d. from ideal geometry, angles | 1.28° | 1.33° | 1.33° | |
| Wilson B | 28.3 Å2 | 25.6 Å2 | 37.4 Å2 | 38.5 Å2 |
| 〈B〉 overall | 27.9 Å2 | 42.4 Å2 | 41.2 Å2 | |
| 〈B〉 ligand | NA | 44.7 Å2 | 54.1 Å2 | |
| 〈B〉 solvent | 32.1 Å2 | 45.1 Å2 | 44.1 Å2 | |
| Ramachandran plot | ||||
| Most favored | 90.3% | 91.1% | 90.2% | |
| Additionally allowed | 9.5% | 8.9% | 9.8% | |
| Generously allowed | 0.2% | 0.0% | 0.0% | |
| PDB ID | 2W61 | 2W62 | 2W63 |
Structure Determination and Refinement—Using SHELXC/D/E we were able to find 14 ordered iridium sites. By SIRAS, using data from a crystal soaked with (NH4)3IrCl6 as well as a data set on a crystal of the E176Q mutant (Table 1), an initial model for the apo-structure was generated with ARP/warp (25) (initially building 412 residues of the single protein monomer in the asymmetric unit) and improved through cycles of manual model building in Coot (26) and refinement with REFMAC5 (27). Molecular replacement with this structure as a search model was used to generate phases and starting models for the remaining data sets, which were refined similarly. Topologies for the oligosaccharide ligands were generated with PRODRG (28). The final models were validated with PROCHECK (29) and WHATCHECK (30), and model statistics are given in Table 1. Coordinates and structure factors have been deposited in the Protein Data Bank. The structure has several disordered regions as follows: one in the poorly conserved loop between β3 and α1, one covering a short stretch of the interdomain region between α9 and α10, and one (absent in three of the present four structures) preceding α11. In addition, the C terminus of ScGas2 (from at least Ser507, but in some cases as early as Leu483) is completely disordered. Although density for the α13 helix could be observed in all three structures, its quality was too poor for it to be modeled in the oligosaccharide complexes (Fig. 1B).
RESULTS AND DISCUSSION
Structure of ScGas2 Reveals a Two-domain Fold—A truncated form of ScGas2 (amino acids 26–525, excluding the signal/GPI-anchor sequences, Fig. 1A) was expressed as a secreted protein in P. pastoris and purified by ion-exchange and gel filtration chromatography. The structure of ScGas2 was solved by SIRAS with an iridium derivative, complexes with laminarioligosaccharides and a structure of a E176Q mutant were solved by molecular replacement, and all structures were refined to 1.6–2.1 Å with Rfree <0.22 (Table 1).
The structure of ScGas2 is composed of two interacting domains as follows, a (βα)8 catalytic domain, abundantly found in other carbohydrate active enzymes; and a C-terminal cysteine-rich domain of the CBM43 family (11) (Fig. 1). The (β/α)8 domain deviates from the canonical topology by kinks in the sixth and seventh α-helix (α6/α6′ and α7/α7′, Fig. 1A). Similar to many other (β/α)8 barrel proteins, the first strand of the barrel (β3) is preceded by a two-stranded antiparallel β-sheet, which seals the “bottom” of the barrel. A second short two-stranded sheet (β11/β12) is inserted into the last βα loop (31), placing it on the opposite side of the barrel and within 20 Å of the active site (Fig. 1).
Not surprisingly, a DALI (32) search with the ScGas2 catalytic domain yields over 300 proteins with significant structural similarity; among the most similar structures are numerous glucosidases. The two crystal structures with the highest Z scores are domain 3 from human β-glucuronidase (PDB ID 1BHG (33)) and Cellvibrio mixtus mannosidase 5A (PDB ID 1UUQ (34)), which superimpose with r.m.s.d. of 2.7 Å and 3.0 Å for ≈260 C-α atoms, respectively.
The ScGas2 catalytic domain contains three disulfide bridges, one between Cys89 and Cys118 connecting α1 and α2, and another between Cys231 from the fifth αβ loop and Cys367 from the interdomain loop. It is noteworthy that both these disulfides occur in the vicinity of disordered loops, and it is possible that they help to limit flexibility. The third disulfide of the catalytic domain is formed by Cys247 and Cys278 from the sixth and seventh βα loop (preceding α6 and α8, respectively). Both these loops are involved in forming the acceptor saccharide-binding site, and it is possible that the disulfide bridge helps to correctly position them.
When a sequence alignment of GH72 enzymes (Fig. 1A) is interpreted in the context of the ScGas2 structure (Fig. 1B), it is clear that most of the sequence conservation locates to the catalytic core, whereas the C-terminal cysteinerich CBM43 domain of ScGas2 is less conserved. In particular the active site of ScGas2 is highly conserved (Fig. 1B), from the –2 to the +2 site, suggesting that ScGas2 may be a good representative of the GH72 family for further mechanistic studies.
CBM43 Domain Contains a Conserved Cysteine Structure and Exposed Aromatic Residues—Based on its sequence, the C-terminal domain of ScGas2 (Fig. 1) has been assigned to the CBM43 family of carbohydrate-binding modules (11). It assumes a predominantly α-helical structure; a core formed by four α-helixes is augmented by two small antiparallel two-stranded β-sheets (Fig. 1C). Although the amino acid sequence of the catalytic domain is reasonably well conserved among GH72 family members (32–61% identity), there is considerably more variation in the C-terminal domain, where, even among GH72+ subfamily members, sequence identity can drop below 20% (Fig. 1A). The few amino acids that are completely conserved include a number of hydrophobic residues and eight cysteines, which, in the ScGas2 structure, form four disulfide bridges (Cys390–Cys442; Cys399–Cys466; Cys419–Cys424; and Cys457–Cys489, Fig. 1, A–C), in agreement with a recent mass spectrometry-based assignment (35). The C-terminal ≈40 residues of the expressed protein, which in the full-length protein would lead up to the GPI anchor, are (mostly) disordered in our structures; it is likely that the poorly conserved stretch between α15 and the GPI anchor site functions as a flexible tether.
Structural homology searches of this domain with DALI (32) yielded no significant hits. As CBM43 domains are most commonly associated with β(1,3) glucan-processing domains from CAZy families GH17/72, it is possible that they would possess β(1,3) glucan binding activity. It has been shown that carbohydrate binding to CBM domains is generally effected by surface-exposed tyrosine, tryptophan, and phenylalanine residues (36). The ScGas2 cysteine-rich domain contains six such surface aromatic amino acids: Phe404, Tyr417, Tyr474, Phe493, Tyr501, and Tyr506 (Fig. 1C), which could play a role binding to β(1,3) glucan, although none of them are conserved between different GH72+ family members (Fig. 1A). To test this hypothesis, Phe404 and Tyr474 were mutated to alanines. Only the F404A mutant showed a small difference in transglycosylation/hydrolysis activity of a G19 laminarioligosaccharide compared with the wild type enzyme (Table 2). Earlier studies also showed that the presence or absence of the CBM43 domain in the GH72 enzymes does not appear to significantly affect transglycosylation activity (13, 14).
TABLE 2.
Percentage of degraded reduced G19 after 1 h of incubation of the ScGas2 mutants compared with the wild type enzyme. ND means not determined
| ScGas2 (S26-S525) | Percentage of degraded G19 | Transglycosylation product |
|---|---|---|
| % | ||
| ScGas2 (wildtype) | 93 ± 4 | 100 |
| Q62A | 88 ± 4 | ND |
| Y107F | 49 ± 2 | 20 ± 8 |
| Y107Q | ND | ND |
| D132N | 85 ± 4 | ND |
| N175A | 1 ± 2 | ND |
| E176Q | 1 ± 2 | ND |
| Y244Q | 39 ± 2 | 9 ± 7 |
| Y244F | ND | ND |
| E275Q | 1 ± 2 | ND |
| Y307Q | 33 ± 4 | 11 ± 2 |
| F404A | 49 ± 2 | ND |
| Y474A | 92 ± 4 | 100 ± 12 |
So far, only two CBM43 proteins, both olive pollen allergens, have been biochemically characterized in some detail. The olive pollen-derived Ole e 10 (an isolated CBM43 domain) and the GH17 family member Ole e 9 (with a C-terminal CBM43 domain) have been shown to possess the ability to bind β-1,3 glucan structures (37, 38). Although alignment between the olive pollen CBM43 domains and that of ScGas2 shows poor sequence conservation (identity of ≈17%), six of the GH72+ cysteines appear to be conserved. Very recently, an NMR structure of the C-terminal domain (CTD) of Ole e 9 has become available (39). A superposition with the ScGas2 cysteine-rich domain is relatively poor, giving an r.m.s.d. of 2.7 Å (with only 64 out of the 101 possible equivalenced C-α atoms; Fig. 1C). Although the two CBM43 structures share the “core” formed by α11, α12, β13, and β16 (ScGas2 numbering), the Ole e 9 CTD lacks the second β-sheet (β14–β15) of ScGas2 as well as the α10 and α13 helices. Of the six equivalent cysteine residues, only four participate in structurally conserved disulfide bridges, whereas the other two form a third disulfide in the Ole e 9 CTD but participate in two separate disulfide bridges in ScGas2 (Fig. 1C). Altogether, the two available structures of CBM43 domains suggest that GH17-associated “plant” and GH72-linked “fungal” CBM43 domains, although sharing some structural motifs, are overall not similar enough that their functional equivalence can be assumed. Some of the ScGas2 structural elements absent from the Ole e 9 CTD participate in interactions with the catalytic domain (see below), and their absence from the plant protein may indicate differences in the interaction between the CBM and the two classes of catalytic domains. It is noteworthy that the part of the CBM43 domains incorporating the shared features is closest to the ScGas2 active site, whereas the dissimilar side faces away from it (Fig. 1B). It is thus possible that the CBMs of ScGas2 and Ole e 9 bind glucans on that side of the domain, using similar binding sites. Ole e 9 exposes a cluster of four surface aromatic residues on this side, most of which are conserved in Ole e 10 (39). Only one of these residues is conserved between Ole e 9 and ScGas2 (Tyr417 in ScGas2 numbering).
One of the characteristics of CBM motifs/modules is that they are frequently separate domains and indeed can occur as individual proteins (40). In contrast, the cysteine-rich domain of ScGas2 shares extensive contacts with the catalytic core, incorporating hydrophobic interactions as well as seven strong direct hydrogen bonds and burying ≈2650 Å2, compatible with a stable domain interface (41). The catalytic subunit contributes residues from around the N termini of helices α3 and α4 as well as the loop preceding α10 to the interface, whereas the CBM uses residues from the N-terminal ends of α10 and α12 and, most importantly, the small β-sheet formed by β14 and β15 that is absent from the Ole e 9 structure.
Binding Mode of a Transglycosylation Acceptor Substrate—Recent landmark studies toward the structure and mechanism of a plant xylosyltransferase PttXET16A has revealed how a large, fully ordered, oligosaccharide is bound to the acceptor site, tightly interacting with the catalytic base (Fig. 2), whereas elegant kinetic studies have demonstrated a remarkably long lived covalent enzyme-donor intermediate (42, 43).
FIGURE 2.
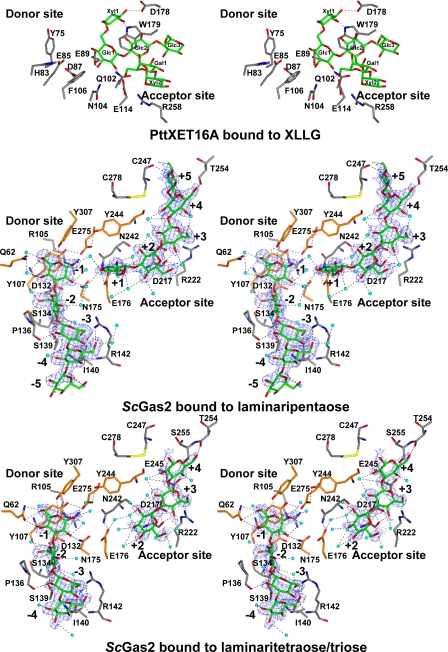
Structures of ScGas2-laminarioligosaccharide complexes. Stereo view of the active site of ScGas2 in complex with laminaripentaose and the hydrolysis products of laminariheptaose (i.e. laminaritetraose + laminaritriose), and comparison with PttXET16A bound to XLLG. The active site oriented to facilitate identification of the donor (left) and acceptor (right) subsites. The amino acids placed in the donor site and acceptor sites are shown as sticks with gray carbons. The residues targeted by site-directed mutagenesis, Gln62, Tyr107, Asp132, Asn175, Glu176, Tyr244, Glu275, Tyr307, Phe404, and Tyr474, are shown with orange carbon atoms. XLLG, laminaripentaose, laminaritetraose, and laminaritriose are represented as stick models with green carbon atoms. Protein-ligand and water-ligand hydrogen bonds are shown as dotted black lines. Water molecules involved in hydrogen bonds with the ligands are shown as cyan spheres. For clarity purposes, protein-water hydrogen bonds are not shown. Unbiased (i.e. before inclusion of any ligand model) |Fo| – |Fc|, φcalc electron density maps are shown at 2.5 σ.
To allow trapping of an ScGas2-acceptor transglycosylation complex, we sought to identify the minimal (and therefore most soluble) β-glucan-derived laminarioligosaccharide that would not undergo hydrolysis, yet still serves as an acceptor substrate given a suitable donor in a transglycosylation reaction. Hydrolysis experiments show that laminaripentaose is not hydrolyzed by ScGas2 (Fig. 3A), although a previous study of the A. fumigatus Gel1 ScGas2 orthologue showed that laminaripentaose was the smallest laminarioligosaccharide acceptor used by the enzyme (21). Surprisingly, soaking experiments with ScGas2 revealed not only an ordered laminaripentaose bound to the acceptor site, but also a (nonconvalently bound) laminaripentaose bound in the donor site (Fig. 2). The active site is defined by two catalytic residues, Glu176 and Glu275, previously identified by mutagenesis (13), and three tyrosines, Tyr107, Tyr244, and Tyr307. These residues are all conserved in the GH72 family, as well as several other residues lining the binding groove (Fig. 1A and Fig. 2). Together, the two laminaripentaose molecules appear to occupy the –5 to –1 and +1 to +5 binding sites, without any evidence of bond formation between the –1/+1 sugars, but with the –1 O-1 and +1 O-3 hydroxyls only 4.3 Å apart. Thus, this arrangement may represent the position of transglycosylation reactants, with the –5 to –1 sugars representing the donor site, and the +1 to +5 sugars representing the acceptor site.
FIGURE 3.
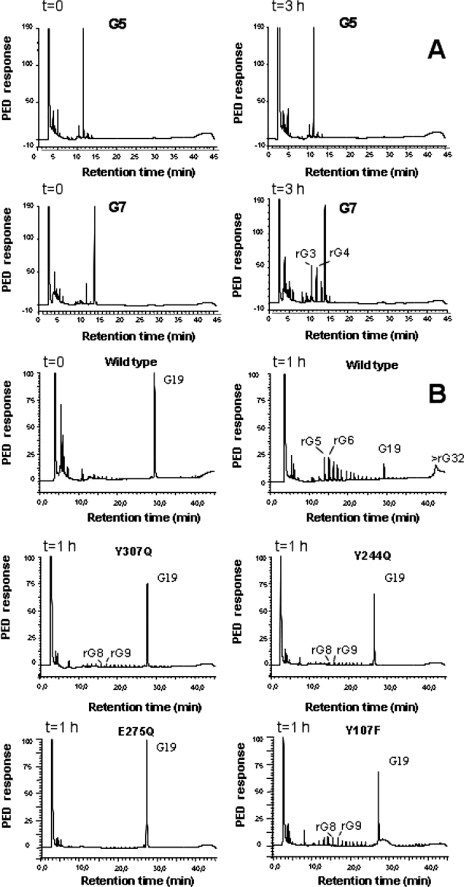
High pressure liquid chromatography analysis of β(1,3) glucanosyltransferase/hydrolysis products. A, comparison of wild type ScGas2 kinetics against laminaripentaose and laminariheptaose, identifying laminaritetraose and laminaritriose as the main two degradation products of hydrolyzed laminariheptaose. B, product analysis from the incubation of the recombinant wild type ScGas2 and the following single mutant enzymes, Y107F, Y244Q, E275Q, and Y307Q, with 4 mm reduced G19 samples taken at the indicated time points.
The functions of the two catalytic residues, Glu176 and Glu275, can be inferred from the ScGas2-laminaripentaose complex (Fig. 2). Glu176, the catalytic acid/base, hydrogen bonds O-3 of the +1 sugar in the acceptor site, occupying a position equivalent to the catalytic base (Glu89) in the PttXET16A structure (Fig. 2) (44). Glu275, on the opposite side of the binding groove, approaches the anomeric the carbon of the –1 sugar to within 4 Å under a geometry compatible with nucleophilic attack. Mutation of either of these glutamates to glutamine abrogates catalytic activity (Table 2 and Fig. 3). GH families can be classified as inverting or retaining enzymes based on the distance between the two catalytic residues, with inverting enzymes giving a distance of 10 ± 2 Å on average, although retaining enzymes have the two residues located ∼5.5 Å apart (45). In the ScGas2 crystal structure, Glu176 and Glu275 are 5.1 Å apart, suggesting the active site structure is compatible with a retaining mechanism, in agreement with previously published product analysis of ScGas2 (21). Given the sequence conservation of these residues, this will extend to all GH72 members. In addition to the two glutamates, there are three conserved tyrosines lining the active site. Two of these (Tyr107/Tyr244) interact with Glu275, positioning it for nucleophilic attack (Fig. 2). Mutation of these residues to phenylalanines shows small effects on hydrolysis (Table 2 and Fig. 3). A plethora of further interactions between protein and substrate is found in the elongated binding groove, involving both hydrogen bonds and stacking interactions with aromatic residues (Fig. 2). For instance, two conserved residues, Tyr107 and Pro136, are involved in hydrophobic stacking interactions with sugars in the donor site, whereas the conserved Arg142 gives the donor site a groove-like character (Fig. 2). Residues Asn175 and Tyr307 hydrogen bond the –1 sugar, and mutation of these severely to moderately affects hydrolysis and transglycosylation, respectively (Table 2 and Fig. 3). Negative control mutations of residues that do not directly interact with the sugars (Gln62 and Asp132) do not show significant effects on activity (Table 2 and Fig. 3).
Product Binding as a “Base Occlusion” Mechanism to Protect against Hydrolysis—The ScGas2-laminaripentaose complex suggests that it is possible for both products of the initial step in hydrolysis/transglycosylation to remain associated with the enzyme, with an “occlusion” of the catalytic base by the product in the acceptor site, very similar to what has been observed for the PttXET16A-acceptor complex (Fig. 2). This suggests that the enzyme may use binding of products from the initial step to protect the newly formed enzyme-sugar intermediate from nucleophilic attack by a water molecule. The products can then only be displaced by longer (and tighter binding) oligosaccharides, perhaps involving an acceptor-induced conformational change as suggested by Hartland et al. (21).
We sought to gain further support for this base occlusion hypothesis through soaking studies with a laminarioligosaccharide that is a substrate for hydrolysis, yet cannot act as both a donor and acceptor. We identified laminariheptaose as a suitable substrate for hydrolysis (yielding trimers and tetramers), without any measurable transglycosylation activity (Fig. 3A). Strikingly, soaking experiments with laminariheptaose resulted in electron density revealing that the oligosaccharide was hydrolyzed by the enzyme, leaving laminaritetraose in the donor site and laminaritriose in the acceptor site, in agreement with the kinetic data (Fig. 2 and Fig. 3A). The active site conformation is essentially identical to the laminaripentaose complex, but although the laminaritetraose occupies the –4 to –1 subsites, surprisingly on the acceptor side the laminaritriose occupies sites +2 to +4 (0.75 Å maximum atomic shift with respect to the sugars in the laminaripentaose complex). Crucially, this leaves the +1 subsite, and thus the catalytic base, solvent-exposed. This allows for two important observations. First, the fact that laminaritriose, one of the hydrolysis products, occupies the +2 to +4 subsites, and not the +1 to +3 subsites, suggests that the leaving group of the initial nucleophilic attack can “slide” along the reducing end subsites, rather than randomly diffuse out of the active site. This may offer a transglycolysis mechanism with progressive exposure of subsites by a sliding out product with simultaneous occupation of these subsites by a new acceptor, while limiting access of water molecules near the covalent intermediate. Second, the observed arrangement in the acceptor side is in good agreement with the base occlusion model. Because the leaving group of the initial reaction with laminariheptaose moves to occupy the +2 to +4 subsites, this leaves the +1 subsite and the catalytic base fully exposed, allowing water to flood in, interact with the base, and act as a nucleophile to break the protein-enzyme intermediate, explaining why exclusively hydrolysis, and not transglycosylation, is observed with laminariheptaose (Fig. 3A). Indeed, an ordered water molecule (B = 35 Å2) is seen to occupy a position in the –1 subsite in the laminariheptaose product complex, forming a hydrogen bond with the catalytic base (Fig. 2).
The base occlusion hypothesis also suggests that if interactions between the +1 sugar and the protein are disrupted, this might shift the balance between hydrolysis and transglycosylation away from the latter. We attempted this by studying the effects on hydrolysis and transglycosylation of a mutant (Y244F) of a conserved tyrosine lining the +1 subsite (Figs. 2 and 3 and Table 2). Strikingly, although hydrolysis is only moderately affected, a 10-fold reduction in transglycosylation is observed.
Concluding Remarks—Cell wall remodeling is an essential process in fungal organisms. Several enzymes with transglycosylation activities have been proposed to be involved in this process, but only the GH72 enzymes have been shown in vitro to transglycosylate the main cell wall carbohydrate polymer, β(1,3) glucan (13, 14, 21). Genetic data in different organisms show the involvement of these enzymes in virulence, morphology, and growth, in some cases supporting an essential function in sporulation (12, 15, 17–20, 46).
Enzymes displaying significant transglycosylation are essentially glycoside hydrolases that have developed mechanisms to protect the (covalent) reaction intermediate from nucleophilic attack by a water molecule, although these mechanisms are not understood. The first crystal structure of a GH72 transglycosylase enzyme described here shows two interacting domains and a wide, conserved, and solvent-exposed active site. This includes the first crystal structure of an ordered CBM43 domain, which, although defining the CBM43 fold, does not immediately offer clues as to how it might contribute to glucan binding/hydrolysis/transglycosylation, as either truncation of the domain, or mutation of exposed aromatic residues does not have large effects on activity on (short) substrates.
Although recent work on xylosyltranferases has defined acceptor-protein interactions and has demonstrated that the donor-enzyme intermediate, generated after the initial nucleophilic attack, is long lived, this did not yet explain how transglycosylases might protect the intermediate from nucleophilic attack by water. The data described here provide the first insights into the mechanisms that may control the balance between hydrolysis and transglycosylation in a transglycosylase. The substrate-product trapped complexes, together with the mutagenesis data, suggest that product binding in the acceptor subsite might offer a method for occluding the catalytic base, and therefore prevent activation of an incoming water molecule. Compatible with this hypothesis, a substrate that yields a product that does not occupy the +1 site (leaving the catalytic base accessible to solvent), exclusively shows hydrolysis.
The fungal cell wall has been thought to be a treasure trove for novel drug targets to combat the increasing occurrence of invasive fungal infections. Indeed, the most recently developed anti-fungals of the echinocandin class target glucan synthase (1, 47), and there are efforts to develop inhibitors of fungal chitinases with anti-fungal activity (48, 49). Significant genetic validation now exists for the GH72 enzymes; they appear to be essential for proper development and morphogenesis of the fungal cell. Given the significant degree of sequence conservation in the GH72 family, it may be possible to develop chemical tools/probes that would inhibit all of the members of this multigene family in a single organism. The work here provides a useful scaffold for the future development and/or evaluation of such molecules.
Acknowledgments
We thank the European Synchrotron Radiation Facility, Grenoble, France, and Diamond Light Source, Oxford, United Kingdom, for beam time.
This work was supported by a Wellcome Trust senior research fellowship (to D. M. F. v. A.) and European Union FP6 STREP Fungwall Programme. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The atomic coordinates and structure factors (codes 2W61, 2W62, and 2W63) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Footnotes
The abbreviations used are: GPI, glycosylphosphatidylinositol; CTD, C-terminal domain; PDB, Protein Data Bank; r.m.s.d., root mean square deviation.
References
- 1.Latge, J. P. (2007) Mol. Microbiol. 66 279–290 [DOI] [PubMed] [Google Scholar]
- 2.Fleet, G. H. (1991) in The Yeasts (Rose, A. H., ed) pp. 199–277, Academic Press, London
- 3.Kollar, R., Petrakova, E., Ashwell, G., Robbins, P. W., and Cabib, E. (1995) J. Biol. Chem. 270 1170–1178 [DOI] [PubMed] [Google Scholar]
- 4.Fontaine, T., Simenel, C., Dubreucq, G., Adam, O., Delepierre, M., Lemoine, J., Vorgias, C. E., Diaquin, M., and Latge, J. P. (2000) J. Biol. Chem. 275 27594–27607; Correction (2000) J. Biol. Chem. 275, 41528 [DOI] [PubMed] [Google Scholar]
- 5.Adams, D. J. (2004) Microbiology 150 2029–2035 [DOI] [PubMed] [Google Scholar]
- 6.Mouyna, I., Fontaine, T., Vai, M., Monod, M., Fonzi, W. A., Diaquin, M., Popolo, L., Hartland, R. P., and Latge, J. P. (2000) J. Biol. Chem. 275 14882–14889 [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Pena, J. M., Cid, V. J., Arroyo, J., and Nombela, C. (2000) Mol. Cell. Biol. 20 3245–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitagaki, H., Wu, H., Shimoi, H., and Ito, K. (2002) Mol. Microbiol. 46 1011–1022 [DOI] [PubMed] [Google Scholar]
- 9.Tougan, T., Chiba, Y., Kakihara, Y., Hirata, A., and Nojima, H. (2002) Genes Cells 7 217–231 [DOI] [PubMed] [Google Scholar]
- 10.Cabib, E., Blanco, N., Grau, C., Rodriguez-Pena, J. M., and Arroyo, J. (2007) Mol. Microbiol. 63 921–935 [DOI] [PubMed] [Google Scholar]
- 11.Coutinho, P. M., and Henrissat, B. (1999) in Recent Advances in Carbohydrate Bioengineering (Gilbert, H. J., Davies, G., Henrissat, B., and Svensson, B., eds) pp. 3–12, Royal Society of Chemistry, Cambridge, UK
- 12.Mouyna, I., Morelle, W., Vai, M., Monod, M., Lechenne, B., Fontaine, T., Beauvais, A., Sarfati, J., Prevost, M. C., Henry, C., and Latge, J. P. (2005) Mol. Microbiol. 56 1675–1688 [DOI] [PubMed] [Google Scholar]
- 13.Mouyna, I., Monod, M., Fontaine, T., Henrissat, B., Lechenne, B., and Latge, J. P. (2000) Biochem. J. 347 741–747 [PMC free article] [PubMed] [Google Scholar]
- 14.Ragni, E., Fontaine, T., Gissi, C., Latge, J. P., and Popolo, L. (2007) Yeast 24 297–308 [DOI] [PubMed] [Google Scholar]
- 15.Ragni, E., Coluccio, A., Rolli, E., Rodriguez-Pena, J. M., Colasante, G., Arroyo, J., Neiman, A. M., and Popolo, L. (2007) Eukaryot. Cell 6 302–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popolo, L., Vai, M., Gatti, E., Porello, S., Bonfante, P., Balestrini, R., and Alberghina, L. (1993) J. Bacteriol. 175 1879–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ram, A. F., Kapteyn, J. C., Montijn, R. C., Caro, L. H., Douwes, J. E., Baginsky, W., Mazur, P., van den Ende, H., and Klis, F. M. (1998) J. Bacteriol. 180 1418–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saporito-Irwin, S. M., Birse, C. E., Sypherd, P. S., and Fonzi, W. A. (1995) Mol. Cell. Biol. 15 601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghannoum, M. A., Spellberg, B., Saporito-Irwin, S. M., and Fonzi, W. A. (1995) Infect. Immun. 63 4528–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muhlschlegel, F. A., and Fonzi, W. A. (1997) Mol. Cell. Biol. 17 5960–5967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartland, R. P., Fontaine, T., Debeaupuis, J. P., Simenel, C., Delepierre, M., and Latge, J. P. (1996) J. Biol. Chem. 271 26843–26849 [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307–326 [DOI] [PubMed] [Google Scholar]
- 23.Collaborative Computational Project, No. 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760–76315299374 [Google Scholar]
- 24.Pape, D., Seil, R., Kohn, D., and Schneider, G. (2004) Orthop. Clin. North Am. 35 293–303, viii [DOI] [PubMed] [Google Scholar]
- 25.Perrakis, A., Morris, R., and Lamzin, V. S. (1999) Nat. Struct. Biol. 6 458–463 [DOI] [PubMed] [Google Scholar]
- 26.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240–255 [DOI] [PubMed] [Google Scholar]
- 28.Schuttelkopf, A. W., and van Aalten, D. M. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 1355–1363 [DOI] [PubMed] [Google Scholar]
- 29.Laskowski, R. A., Moss, D. S., and Thornton, J. M. (1993) J. Mol. Biol. 231 1049–1067 [DOI] [PubMed] [Google Scholar]
- 30.Hooft, R. W., Vriend, G., Sander, C., and Abola, E. E. (1996) Nature 381 272. [DOI] [PubMed] [Google Scholar]
- 31.Wierenga, R. K. (2001) FEBS Lett. 492 193–198 [DOI] [PubMed] [Google Scholar]
- 32.Holm, L., and Sander, C. (1995) Trends Biochem. Sci. 20 478–480 [DOI] [PubMed] [Google Scholar]
- 33.Jain, S., Drendel, W. B., Chen, Z. W., Mathews, F. S., Sly, W. S., and Grubb, J. H. (1996) Nat. Struct. Biol. 3 375–381 [DOI] [PubMed] [Google Scholar]
- 34.Dias, F. M., Vincent, F., Pell, G., Prates, J. A., Centeno, M. S., Tailford, L. E., Ferreira, L. M., Fontes, C. M., Davies, G. J., and Gilbert, H. J. (2004) J. Biol. Chem. 279 25517–25526 [DOI] [PubMed] [Google Scholar]
- 35.Popolo, L., Ragni, E., Carotti, C., Palomares, O., Aardema, R., Back, J. W., Dekker, H. L., de Koning, L. J., de Jong, L., and de Koster, C. G. (2008) J. Biol. Chem. 283 18553–18565 [DOI] [PubMed] [Google Scholar]
- 36.Boraston, A. B., Bolam, D. N., Gilbert, H. J., and Davies, G. J. (2004) Biochem. J. 382 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barral, P., Suarez, C., Batanero, E., Alfonso, C., Alche Jde, D., Rodriguez-Garcia, M. I., Villalba, M., Rivas, G., and Rodriguez, R. (2005) Biochem. J. 390 77–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodriguez, R., Villalba, M., Batanero, E., Palomares, O., Quiralte, J., Salamanca, G., Sirvent, S., Castro, L., and Prado, N. (2007) J. Investig. Allergol. Clin. Immunol. 17 Suppl. 1, 4–10 [PubMed] [Google Scholar]
- 39.Trevino, M. A., Palomares, O., Castrillo, I., Villalba, M., Rodriguez, R., Rico, M., Santoro, J., and Bruix, M. (2008) Protein Sci. 17 371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shoseyov, O., Shani, Z., and Levy, I. (2006) Microbiol. Mol. Biol. Rev. 70 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bahadur, R. P., Chakrabarti, P., Rodier, F., and Janin, J. (2003) Proteins 53 708–719 [DOI] [PubMed] [Google Scholar]
- 42.Sulova, Z., Takacova, M., Steele, N. M., Fry, S. C., and Farkas, V. (1998) Biochem. J. 330 1475–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sulová, Z., Baran, R., and Farkas, V. (2001) Plant Physiol. Biochem. 39 927–932 [Google Scholar]
- 44.Johansson, P., Brumer, H., III, Baumann, M. J., Kallas, A. M., Henriksson, H., Denman, S. E., Teeri, T. T., and Jones, T. A. (2004) Plant Cell 16 874–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rye, C. S., and Withers, S. G. (2000) Curr. Opin. Chem. Biol. 4 573–580 [DOI] [PubMed] [Google Scholar]
- 46.de Medina-Redondo, M., Arnaiz-Pita, Y., Fontaine, T., Del Rey, F., Latge, J. P., and Vazquez de Aldana, C. R. (2008) Mol. Microbiol. 68 1283–1299 [DOI] [PubMed] [Google Scholar]
- 47.Kauffman, C. A., and Carver, P. L. (2008) Semin. Respir. Crit. Care Med. 29 211–219 [DOI] [PubMed] [Google Scholar]
- 48.Hurtado-Guerrero, R., and van Aalten, D. M. (2007) Chem. Biol. 14 589–599 [DOI] [PubMed] [Google Scholar]
- 49.Andersen, O. A., Nathubhai, A., Dixon, M. J., Eggleston, I. M., and van Aalten, D. M. (2008) Chem. Biol. 15 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bond, C. S. (2003) Bioinformatics (Oxf.) 19 311–312 [DOI] [PubMed] [Google Scholar]