Abstract
The disease protein in frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U) and amyotrophic lateral sclerosis (ALS) was identified recently as the TDP-43 (TAR DNA-binding protein 43), thereby providing a molecular link between these two disorders. In FTLD-U and ALS, TDP-43 is redistributed from its normal nuclear localization to form cytoplasmic insoluble aggregates. Moreover, pathological TDP-43 is abnormally ubiquitinated, hyperphosphorylated, and N-terminally cleaved to generate C-terminal fragments (CTFs). However, the specific cleavage site(s) and the biochemical properties as well as the functional consequences of pathological TDP-43 CTFs remained unknown. Here we have identified the specific cleavage site, Arg208, of a pathological TDP-43 CTF purified from FTLD-U brains and show that the expression of this and other TDP-43 CTFs in cultured cells recapitulates key features of TDP-43 proteinopathy. These include the formation of cytoplasmic aggregates that are ubiquitinated and abnormally phosphorylated at sites found in FTLD-U and ALS brain and spinal cord samples. Furthermore, we observed splicing abnormalities in a cell culture system expressing TDP-43 CTFs, and this is significant because the regulation of exon splicing is a known function of TDP-43. Thus, our results show that TDP-43 CTF expression recapitulates key biochemical features of pathological TDP-43 and support the hypothesis that the generation of TDP-43 CTFs is an important step in the pathogenesis of FTLD-U and ALS.
TDP-43 (TAR DNA-binding protein 43) is the major disease protein of sporadic and familial frontotemporal lobar degeneration (FTLD)4 with ubiquitin-positive, tau-negative inclusions (FTLD-U) with or without motor neuron disease as well as sporadic and the majority of familial amyotrophic lateral sclerosis (ALS) cases (1, 2). Human TDP-43 is encoded by the TARDBP gene on chromosome 1. It is a 414-amino acid nuclear protein with two highly conserved RNA recognition motifs (RRM1 and RRM2) and a C-terminal tail with a typical glycine-rich region that mediates protein-protein interactions, including interactions with other heterogeneous ribonucleoprotein (hnRNP) family members such as hnRNP A1, A2/B1, and A3 (3). Thus, TDP-43 is a ubiquitously expressed RNA/DNA-binding protein that also interacts with other nuclear proteins such as splicing factors. As such, TDP-43 is implicated in repression of gene transcription, regulation of exon splicing, and the functions of nuclear bodies (4–9).
Pathological TDP-43 accumulates as insoluble aggregates in the central nervous system neurons and glia of patients with FTLD-U and ALS (1). Moreover, FTLD-U patients can develop ALS, and ALS patients often suffer from a dementia consistent with FTLD-U (10). We therefore proposed that these diseases are part of a clinicopathological spectrum of the same neurodegenerative process collectively referred to as TDP-43 proteinopathy (1, 2). TDP-43 inclusions are present as cytoplasmic, neuritic, or nuclear inclusions, and affected neurons show a dramatic depletion of normal nuclear TDP-43 (1, 11, 12). To mimic this nuclear clearance and to model the sequestration of endogenous TDP-43 into cytoplasmic aggregates, we overexpressed TDP-43 with mutated nuclear localization signals (ΔNLS-TDP-43) in cultured cells that showed a reduction in endogenous nuclear TDP-43 and accumulations of insoluble cytoplasmic aggregates (13). Moreover, overexpression of TDP-43 with a mutated nuclear export signal (ΔNES-TDP-43) resulted in the formation of insoluble nuclear TDP-43 aggregates (13).
Pathological TDP-43 is hyperphosphorylated, ubiquitinated, and abnormally cleaved so that C-terminal fragments (CTFs) of TDP-43 accumulate in cells of affected CNS areas (1). Indeed, we recently showed that insoluble TDP-43 CTFs are selectively enriched in affected cortical regions compared with the spinal cord of both FTLD-U and ALS cases (14). These observations suggest that TDP-43 is differentially processed in brain versus spinal cord and that TDP-43 CTFs might seed inclusion formation and aggregation in cortical neurons. Because CTFs contain the TDP-43 glycine-rich region, the accumulation of CTF-rich aggregates may result in abnormal interactions with proteins involved in the splicing machinery of affected cells. However, little is known about how pathological TDP-43 is cleaved to generate TDP-43 CTFs or the biochemical properties of disease-related TDP-43 CTFs.
Here we address these questions by identifying the cleavage site of an endogenous TDP-43 CTF purified from FTLD-U brains. We also model the aggregation of this and several other TDP-43 CTFs in the cytoplasm of cultured cells and show that expression of TDP-43 CTFs is sufficient to generate cytoplasmic aggregates. Moreover, these insoluble CTFs are ubiquitinated and abnormally phosphorylated at sites similar to those in human FTLD-U and ALS CNS samples. Finally, we demonstrate that cells expressing TDP-43 CTFs display abnormal splicing of the cystic fibrosis transmembrane conductance regulator (CFTR), which is relevant to TDP-43 function because alternative splicing of this gene is known to be regulated by TDP-43 (6). Our results recapitulate unique features of pathological TDP-43 that are hallmarks of FTLD-U and ALS and implicate the generation of TDP-43 CTFs as a key event in the pathogenesis of TDP-43 proteinopathies.
EXPERIMENTAL PROCEDURES
Constructs—N-terminal truncations (see below) were generated by PCR of human TDP-43 using the following primers: 177-TDP-43, 5′GGATCCATGCTTCCTAATTCTAAGCAAAGCC-3′; 187-TDP-43, 5′-GGATCCATGCCTTTGAGAAGCAGAAAAGTGT-3′; 197-TDP-43, 5′-GGATCCATGCGCTGTACAGAGGACATGACTG-3′; and 208-TDP-43, 5′-GGATCCCGGGGTTCTTCTCTCAGTACGGGGAT-3′ and 5′-TCTAGAGCTACATTCCCCAGCCAGAAGACTTAGA-3′. The addition of a Myc epitope tag to the 5′-end of the TDP-43 N-terminal truncations was achieved by PCR, using the following primers: 5-Myc-177-TDP-43, 5′-GGATCCATGGAACAAAAACTCATCTCGGAAGAGGATCTGCTTCCTAATTCTAAGCAAAGCC-3′; Myc-187-TDP-43, 5′-GGATCCATGGAACAAAAACTCATCTCGGAAGAGGATCTGCCTTTGAGAAGCAGAAAAGTGT-3′; Myc-197-TDP-43, 5′-GGATCCATGGAACAAAAACTCATCTCGGAAGAGGATCTGCGCTGTACAGAGGACATGACTG-3′; and Myc-208-TDP-43, 5′-GGATCCATGGAACAAAAACTCATCTCGGAAGAGGATCTGCGGGAGTTCTTCTCTCAGTACGGGGAT-3′ and 5′-TCTAGAGCTACATTCCCCAGCCAGAAGACTTAGA-3′. All of the PCR products were cloned into the pGEM-T vector (Promega, Madison, WI). Following sequence analysis, the PCR products were subcloned into pcDNA 5/To plasmid (Invitrogen) using restriction sites BamHI and XbaI. A diagram of each TDP-43 CTF is shown in Fig. 1B. Myc-WT-TDP-43, Myc-ΔNLS-TDP-43, and Myc-ΔNES-TDP-43 were previously described (13). Human TDP-43 short hairpin RNA (targeted to the 3′-untranslated region) was obtained from OriGene (Rockville, MD; catalog number TR308946). pcDNA 3.1 α-synuclein was previously described (15).
FIGURE 1.
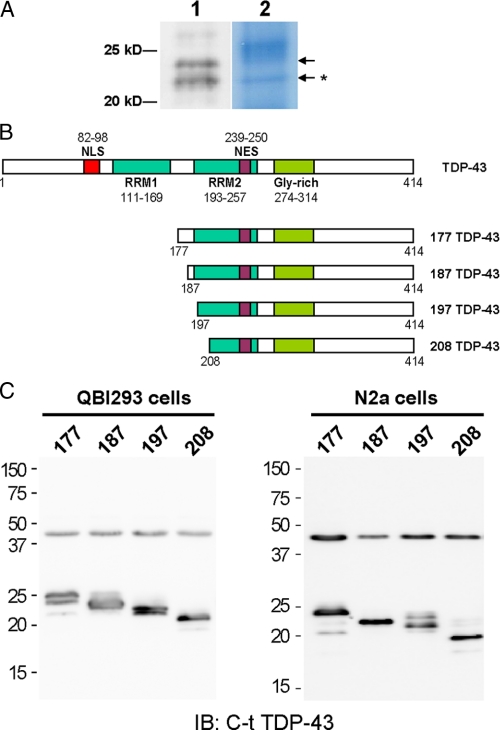
N-terminal cleavage site identification and generation of TDP-43 CTFs. A, CTFs used for N-terminal cleavage site determination after immunoprecipitation proteins from urea extract of FTLD-U brain were resolved on Bis-Tris SDS-PAGE and immunoblotted with anti-C-t-TDP-43 pAb (lane 1) or stained with Amido Black (lane 2). N-terminal sequencing was done on stained protein bands corresponding to the immunoreactive bands (arrows). The primary sequence of the ∼22-kDa band was identified to be TDP-43 with Arg208 as the N terminus (arrow with asterisk). B, schematic representation of TDP-43 protein highlighting the most prominent features (domains and localization signals) of the full-length protein, and the four untagged TDP-43 CTFs generated for mammalian cell expression. Gly-rich, glycine-rich domain. C, QBI-293 and N2a cells were transfected with the indicated TDP-43 CTF constructs, and the total cell extracts were subsequently analyzed by immunoblotting (IB) with C-t TDP-43 pAb. Endogenous TDP-43 migrates as a 43-kDa band in all transfections.
Antibodies—Commercial antibodies used in this study were: rabbit anti-TDP-43 polyclonal antibody (pAb) raised to amino acids 1–260 (Protein Tech Group, Chicago, IL), a human specific mouse monoclonal (mAb) raised to the same TDP-43 sequence (2E2-D3) (Abnova, Taipei, Taiwan), anti-Myc mAb (9E10; Santa Cruz Biotechnology, Santa Cruz, CA), anti-HA (12CA5 mouse mAb; Roche Applied Science), and anti-α-glyceraldehyde-3-phosphate dehydrogenase (6C5 mouse mAb; Advanced ImmunoChemical Inc, Long Beach, CA). A rabbit anti-TDP-43 pAb raised to amino acids 394–414 (C-t TDP-43 pAb) was described previously (14). A rat phospho-specific mAb that recognizes TDP-43 phosphorylated at Ser409/Ser410 (p409/410 TDP-43) was developed and characterized elsewhere (16).
Immunoprecipitation (IP) and N-terminal Cleavage Site Analysis—Sarkosyl-insoluble urea soluble extracts from FTLD-U brains with abundant TDP-43 CTFs were used for IP. Because N-terminal cleaved TDP-43 fragments migrate similarly to IgG light chains, anti-TDP-43 specific mAbs were cross-linked to protein A/G-agarose (Santa Cruz Biotechnology, Santa Cruz, CA) with the homobifunctional imidoester cross-linker dimethyl pimelimidate for IP use. Dialyzed urea fractions in RIPA buffer (0.1% SDS, 1% Nonidet P-40, 0.5% sodium dexoycholate, 5 mm EDTA, 150 mm NaCl, 50 mm Tris-HCl, pH 8.0) or diluted SDS fractions from CNS were precleared with protein A/G-agarose and subjected to IP with antibody-protein A/G-agarose beads. Bound proteins and fragments were then eluted from the beads with SDS sample buffer without dithiothreitol (10 mm Tris-HCl, pH 6.8, 1 mm EDTA, 1% SDS, 10% sucrose) at 80 °C to minimize the dissociation of IgG light chain from the bead complex. Eluted proteins were reduced with dithiothreitol at elevated temperature before resolution by 12% Bis-Tris NuPage® (Invitrogen) SDS-PAGE using MOPS buffer system and transferred to sequence grade polyvinylidene difluoride membranes (Bio-Rad). Typically, two gels were run: one for immunoblotting and the other for N-terminal sequencing. For N-terminal sequencing, the membrane was stained with 0.1% Amido Black, and protein bands that correspond to the TDP-43 immuno-positive bands (∼20–25 kDa) on the companion immunoblots were excised. Cleavage sites of TDP-43 fragments were determined by N-terminal automated Edman sequencing on an Applied Biosystems 494 protein sequencer at the Wistar Institute Proteomics Facility. At minimum, eight cycles of sequencing are conducted for amino acid sequence.
Cell Culture and Transfection—QBI-293, Neuro2a, and COS-7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% l-glutamate. The cells were transfected using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. In some experiments, transfected cells were treated with 10 μm MG132 (Sigma-Aldrich) for 16 h.
Immunofluorescence Studies—Cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS), permeabilized with 0.2% Triton X-100 (Sigma) in PBS for 10 min, blocked with 5% powdered milk in PBS for 2 h, and incubated overnight with primary antibody at 4 °C. Primary antibodies were visualized with secondary antibodies conjugated with Alexa Fluor 488 and Alexa Fluor 594 (Vector Laboratories, Burlingame, CA), and the nuclei were detected using 4′,6′-diamino-2-phenylindole. All of the cells were analyzed using a Nikon TE-2000-E (Nikon, Tokyo, Japan), and images were captured using a CoolSnap-HQ camera (Photometrics, Tuscon, AZ). All of the micrographs show images representative of the total cell population. For quantification of aggregates, several random fields/sample were analyzed, and the percentage of transfected cells that displayed TDP-43 positive accumulations was calculated.
Solubility and Biochemical Analysis—To examine the solubility profile of TDP-43, sequential extractions were performed. The cells were washed twice with PBS, lysed in cold RIPA buffer containing 1 mm phenylmethylsulfonyl fluoride, a mixture of protease inhibitors (1 mg/ml pepstatin, leuptin, N-p-Tosyl-l-phenylalanine chloromethyl ketone, Nα-Tosyl-l-lysine chloromethyl ketone hydrochloride, trypsin inhibitor; Sigma), and a mixture of phosphatase inhibitors (2 mm imidazole, 1 mm NaF, 1 mm sodium orthovanadate; Sigma). The cell lysates were sonicated and then cleared by centrifugation at 100,000 × g for 30 min at 4 °C to generate the RIPA soluble samples. To prevent carry-overs, the resulting pellets were washed with RIPA buffer (i.e. resonicated and recentrifuged). Only the supernatants from the first centrifugation were analyzed. RIPA insoluble pellets were then extracted with urea buffer (7 m urea, 2 m thiourea, 4% CHAPS, 30 mm Tris, pH 8.5), sonicated, and centrifuged at 70,000 × g for 30 min at 22 °C. Protease and phosphatase inhibitors were added to all buffers prior to use (1 mm phenylmethylsulfonyl fluoride and a mixture of protease and phosphatase inhibitors). Protein concentration was determined by bicinchoninic acid method (Pierce), and proteins were resolved by 10 or 15% SDS-PAGE and transferred to nitrocellulose membranes. Following transfer, nitrocellulose membranes were blocked in 5% powdered milk and incubated in the primary antibody overnight at 4 °C. Primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, Wegate, PA), and the blots were developed with Renaissance Enhanced Luminal Reagents (PerkinElmer Life Sciences). The digital images were acquired using a Fuji Film Intelligent Darkbox II (Fuji Systems, Stamford, CT). For quantification of TDP-43 CTFs insolubility, densitometric analysis of RIPA-soluble and insoluble fractions of at least three different experiments was performed using Image Quant 5.0 software (Molecular Dynamics Inc, Sunnyvale, CA). Where indicated, cell lysates or postmortem brain tissue from FTLD-U cases sequentially extracted as previously described (1) were dephosphorylated by dialysis (20 mmol/liter Tris and 0.2 mmol/liter EDTA, pH 8.0) and treated with Escherichia coli alkaline phosphatase (Sigma) for 2 h at 56 °C.
Splicing Analysis—TDP-43 functional activity was assayed through evaluation of CFTR splicing. First, various TDP-43 constructs were transiently transfected into QBI-293 cells using Lipofectamine 2000 reagent (Invitrogen) following standard manufacturer protocols. Forty-eight hours later, a hybrid minigene construct (a generous gift from Dr. F. Baralle, International Centre for Genetic Engineering and Biotechnology, Trieste, Italy) designed to evaluate CFTR exon 9 splicing was transiently transfected into the same cells (6, 17). Supplemental Fig. S3 depicts the structure of the minigene construct, which consists of CFTR exon 9 with portions of the CFTR flanking introns inserted between exons from a hybrid fibronectin-α-globin gene (18). The relative exclusion of exon 9 in the presence of various TDP-43 constructs was then evaluated by primer extension from the flanking exons of exon 9. Total RNA was prepared from cells 72 h after transfection of TDP-43 constructs and 24 h after transfection of the TG(13)T(5) CFTR minigene reporter construct, and RT-PCR was performed using 3 μg of total RNA and 2 μl of the resulting cDNA as described previously (17). The primers used were: Bra2, TAGGATCCGGTCACCAGGAAGTTGGTTAAATCA; a2-3, CAACTTCAAGCTCCTAAGCCACTGC. PCR conditions were as follows: 95 °C for 10 min (hot start), followed by 30 cycles of denaturing at 95 °C for 30 s, annealing at 57 °C for 30 s, and elongation at 72 °C for 60 s. The PCR products were visualized on a 1.5% agarose gel; relative amounts of different splice products were quantified and visualized using the Agilent 2100 Bioanalyzer on a DNA 1000 chip. The experiments were performed in duplicate and repeated at least three times.
RESULTS
N-terminally Cleaved Sites of TDP-43 CTFs—Previously, we have shown that cortical TDP-43 inclusions in FTLD-U and ALS brains are composed predominantly of CTFs (14). To better understand the biological significance of the CTFs, we determined their cleavage sites by N-terminal sequencing. Cortical urea extracts of FTLD-U brains containing high levels of CTFs were immunoprecipitated with anti-TDP-43 mAb, and the resultant proteins were resolved on SDS-PAGE gels and immunoblotted with a pAb raised to the extreme C terminus (C-t) of TDP-43. Two protein bands with apparent molecular masses of ∼24 and ∼22 kDa were recognized by the anti-C-t TDP-43 pAb (Fig. 1A). The same bands were identified on Amido Black-stained duplicate polyvinylidene difluoride blots and were excised for N-terminal sequencing (Fig. 1A, arrows). Results from the ∼22-kDa band gave a primary sequence beginning at Arg208 in TDP-43 (Fig. 1A, arrow with asterisk), but no sequence was obtained from the ∼24-kDa band. Similar results were obtained from four separate experiments using two different FTLD-U cases. The identification of the N terminus of a CTF together with our previous liquid chromatography/tandem mass spectrometry studies on TDP-43 CTFs showing the presence of residues at the extreme C terminus (1) allow us to conclude that we have identified a pathological TDP-43 fragment spanning amino acid residues 208–414 (designated as 208 TDP-43).
To study the biochemical properties of TDP-43 CTFs and to determine whether the expression of these fragments recapitulated pathological features of authentic CTFs isolated from FTLD-U and ALS brains, we developed a series of vectors for expression in cultured cells. Plasmids containing the 208 TDP-43 CTF as well as slightly longer CTFs containing residues 177–414, 187–414, or 197–414 of TDP-43 (designated as 177 TDP-43, 187 TDP-43, and 197 TDP-43, respectively) were generated (Fig. 1B). Each TDP-43 CTF cDNA was expressed in QBI-293 cells, a human embryonic kidney cell line, as well as N2a, a mouse neuroblastoma cell line, and the electrophoretic mobility of the fragments migrated between 20 and 25 kDa (Fig. 1C).
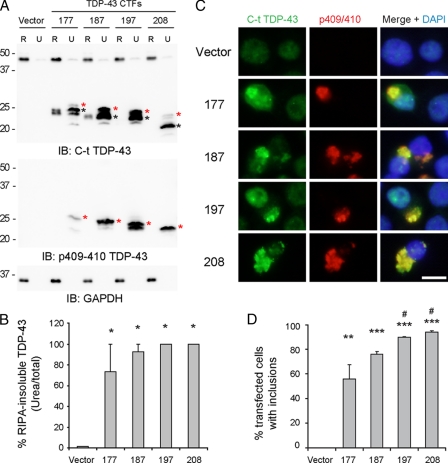
TDP-43 CTFs Expressed in Cultured Cells Are Insoluble and Hyperphosphorylated—Previous studies have shown that TDP-43 CTFs isolated from FTLD-U and ALS brains are insoluble and hyperphosphorylated (1). To determine whether these pathological properties can be recapitulated in cultured cells, we expressed all four TDP-43 CTFs constructs (i.e. 177 TDP-43, 187 TDP-43, 197 TDP-43, and 208 TDP-43) in N2a (Fig. 2) and QBI-293 (supplemental Fig. S1) cells. Sequential extraction of cells overexpressing each of the four CTFs with RIPA and urea buffers showed a progressive increase in the degree of RIPA insolubility going from the largest to the smallest CTF with 177 TDP-43 CTFs being the most soluble and 197 and 208 TDP-43 CTFs being the most insoluble (Fig. 2, A and B, and supplemental Fig. S1). Quantitative immunoblotting showed that almost 100% of the two smaller TDP-43 CTFs are insoluble in RIPA and can only be extracted by urea (Fig. 2B). However, expression of TDP-43 CTFs tagged with the Myc epitope in QBI-293 cells (supplemental Fig. S2) increased their solubility when compared with their untagged counterparts (compare supplemental Figs. S1 and S2).
FIGURE 2.
TDP-43 CTFs expressed in N2a neuroblastoma cells are insoluble and hyperphosphorylated. A, N2a cells 48 h post-transfection with vector alone or the different TDP-43 CTFs were sequentially extracted with RIPA (R) and urea buffer (U). Immunoblotting (IB) was conducted with C-t TDP-43 pAb or p409/410 phospho-specific TDP-43 mAb. Note the slower migrating p409/410-positive immunobands detected with both antibodies (highlighted with the asterisks) that were only recovered in the insoluble fraction of all TDP-43 CTF extracts. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. B, densitometric analysis of C-t TDP-43 pAb immunoblots shown in A. Immunobands corresponding to endogenous TDP-43 (vector condition) or TDP-43 CTFs were quantified and expressed as percentages of RIPA-insoluble/total. The error bars represent S.E. of three different experiments. One-way analysis of variance (p = 0.0107) shows significant differences between vector condition and all CTFs (*, p < 0.05, Tukey's multiple comparison test). C, double labeling using C-t TDP-43 pAb (green), p409/410 TDP-43 mAb (red) and counterstained with 4′,6′-diamino-2-phenylindole (DAPI, blue) for nuclei. N2a cells transfected with vector or TDP-43 CTFs show that CTF-expressing cells form cytoplasmic aggregates. Phosphorylated TDP-43 was only observed in C-t-positive aggregates, but not in endogenous TDP-43 from control or untransfected cells. Scale bar, 10 μm. D, quantification of the percentage of inclusion-bearing cells in TDP-43 CTF-transfected Neuro2a cells. Note the correlation between TDP-43 CTF solubility shown in B and aggregate formation. The error bars represent S.E. (n = 3). One-way analysis of variance (p = 0.0003) followed by Tukey's multiple comparison test. #, p < 0.05 compared with 177 CTF; **, p < 0.01 and *, p < 0.001 compared with Vector.
We also detected multiple immunobands (particularly those recovered in the urea fractions) upon expression of TDP-43 CTFs in cultured cells (Fig. 2A and supplemental Fig. S1). Because pathological TDP-43 CTFs recovered from FTLD-U and ALS brains are hyperphosphorylated at multiple sites including hyperphosphorylation at Ser409 and Ser410 (p409/410), we asked whether the CTFs expressed in transfected cells are also hyperphosphorylated (19). Using a rat mAb specific for p409/410 (16), we found that all four TDP-43 CTFs displayed robust phospho-specific signals and that labeled phospho-immunobands showed slower apparent electrophoretic mobility than the main protein band recognized by the anti-C-t TDP-43 pAb (Fig. 2A and supplemental Fig. S1, red asterisks highlight the same immunobands detected by C-t TDP-43 and p409/410 antibodies, and the black asterisk identifies the main protein band recognized only by the anti-C-t TDP-43 pAb). Significantly, although the electrophoretic mobility of nonphosphorylated 208 TDP-43 CTFs was close to 20 kDa, the phosphorylated counterpart migrated at ∼22 kDa. Moreover, phosphorylated TDP-43 CTFs were detected only in the urea fraction, and phosphorylation at Ser409 and Ser410 was not seen in endogenous TDP-43 recovered from RIPA extractions (Fig. 2A and supplemental Fig. S1). Thus, like pathological TDP-43 CTFs recovered from diseased brains, TDP-43 CTFs in our in vitro models are phosphorylated, and the phosphorylated TDP-43 CTFs are insoluble.
To characterize the morphology of these phosphorylated, insoluble TDP-43 CTFs, we conducted double label immunofluorescence analysis of transfected cells using anti-C-t TDP-43 pAb and the rat anti-p409/410 mAb. Phosphorylated Ser409/Ser410 was detected within C-t TDP-43-positive aggregates when each of the four TDP-43 CTFs was expressed (Fig. 2C and supplemental Fig. S1). Phosphorylated endogenous nuclear TDP-43 was not seen in control or untransfected cells, and the foci of aggregated phosphorylated TDP-43 CTFs were observed within diffuse cytoplasmic TDP-43 immunoreactivity when longer, more soluble CTFs (e.g. 177 TDP-43) were expressed (Fig. 2C and supplemental Fig. S1). Occasionally, we observed CTFs in the nucleus, and we attribute this to the overexpression and small size of the CTFs. Finally, we also observed a direct correlation between the solubility of the TDP-43 CTFs and aggregate formation such that the more insoluble CTFs, i.e. 208 TDP-43, consistently showed increased percentage of inclusion-bearing cells with larger aggregates (Fig. 2, B and D). Similar results were obtained using a third cell line, COS-7 cells (data not shown).
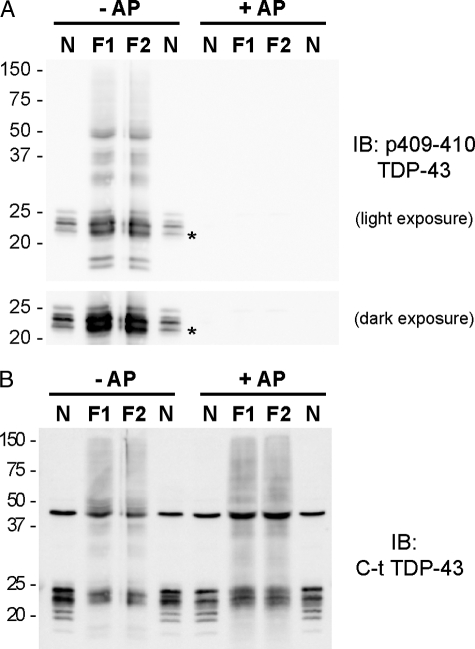
To determine whether the size of our phosphorylated TDP-43 CTFs expressed in cultured cells correspond to endogenous CTFs from FTLD-U brains, we compared the electrophoretic mobility of TDP-43 fragments expressed in N2a cells and showed that they resembled the pathological FTLD-U fragments (Fig. 3A). Indeed, because the phosphorylated 208 TDP-43 fragment expressed in N2a cells co-migrated with an immunoband at ∼22 kDa of TDP-43 CTFs from FTLD-U brains, we suggest that the origin of our N-terminal sequenced fragment beginning with Arg208 came from a phosphorylated FTLD-U CTF (Fig. 3A). Dephosphorylated TDP-43 CTFs from N2a cells also co-migrated with FTLD-U CTFs (Fig. 3B). Therefore, although the exact cleavage sites of the larger TDP-43 CTFs from FTLD-U brains remain to be determined, the close resemblance of the electrophoretic migration of these fragments to those expressed in N2a cells suggests that the biochemical properties of the expressed fragments we observed here likely reflect that of pathological TDP-43 CTFs in disease brains.
FIGURE 3.
Comparison of TDP-43 CTFs from N2a cells and from FTLD-U brains. The electrophoretic mobility of a mixture of CTFs TDP-43 from N2a cell lysates (N) was compared with sarkosyl-insoluble fraction from FTLD-U cases (F1 and F2) before (–) and after (+) dephosphorylation with alkaline phosphatase (AP). A and B, immunoblot (IB) probed with p409/410 TDP-43 mAb (A) and C-t TDP-43 pAb (B). Note that the immunobands from TDP-43 CTFs mixture of the transfected N2a extracts shown in A (lanes N) display similar electrophoretic mobility with CTFs extracted from FTLD-U brain samples (F1 and F2). The asterisk in A denotes the phosphorylated 208 TDP-43 CTF co-migrating with the ∼22-kDa band from FTLD-U brains.
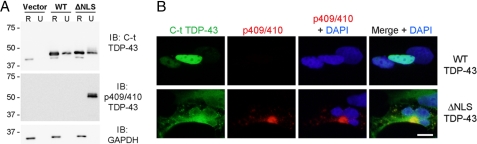
To determine whether full-length TDP-43 expressed in the cytoplasm is also phosphorylated, we transfected mutant TDP-43 with defective nuclear localization signals (ΔNLS-TDP-43) that we characterized in a previous study (13) and found robust phosphorylation at Ser409/410 of ΔNLS-TDP-43 in the insoluble fraction (Fig. 4). Interestingly, phosphorylation of Ser409/410 was not detected in RIPA-soluble ΔNLS-TDP-43 or overexpressed WT-TDP-43 in both the RIPA and urea fractions. Similar results were obtained using N2a cells (data not shown). Thus, phosphorylation of both full-length TDP-43 and CTFs of TDP-43 at Ser409/410 is detected in the insoluble cytoplasmic aggregates. Taken together, these observations suggest a correlation between phosphorylation and insolubility, regardless of whether TDP-43 accumulates as the full-length protein or as CTFs in the cytoplasm.
FIGURE 4.
Cytoplasmic full-length TDP-43 expression results in phosphorylated aggregates. A, QBI-293 cells 48 h post-transfection with empty vector, Myc-TDP-43-WT (WT), or Myc-TDP-43-ΔNLS (ΔNLS) were sequentially extracted with RIPA (R) and urea buffer (U). Immunoblotting (IB) was conducted with anti-C-t TDP-43 pAb or anti-p409/410 phospho-TDP-43 mAb. Myc-TDP-43 (top band) migrates slower than endogenous TDP-43 (lower band). Note that although both WT (nuclear) and ΔNLS (cytoplasmic) displayed considerable amounts of RIPA-insoluble material, only the latter was p409/410-positive. glyceraldehyde-3-phosphate dehydrogenase was used as a loading control. B, double label immunofluorescence of vector, WT, and ΔNLS TDP-43 transfected QBI-293 cells immunostained with C-t TDP-43 pAb (green) and p409/410 TDP-43 mAb (red) antibodies. Only aggregated cytoplasmic TDP-43 is hyperphosphorylated. Scale bar,10 μm.
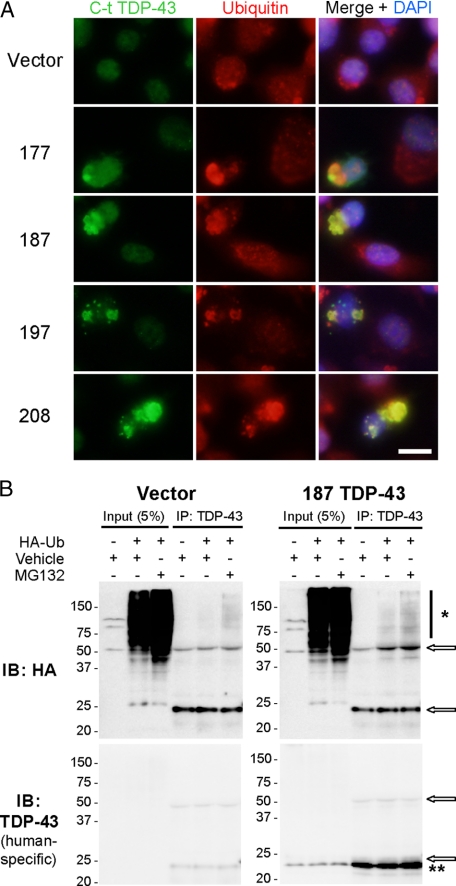
TDP-43 CTF Aggregates Are Ubiquitinated—We recently showed that ubiquitin-positive TDP-43 inclusions in FTLD-U and ALS brains are enriched in TDP-43 CTFs (14). Therefore, we investigated whether aggregated TDP-43 CTFs in transfected cells were also ubiquitinated. Immunofluorescence analysis of transfected N2a or QBI-293 cells displayed strong colocalization of aggregated TDP-43 with ubiquitin; this was not seen in nontransfected neighboring cells or vector-only transfected cells (Fig. 5A and data not shown). To directly demonstrate that TDP-43 CTFs are ubiquitinated, 187 TDP-43 was cotransfected with HA-tagged ubiquitin and incubated with or without MG132, a proteasome inhibitor. Immunoprecipitation of the cell lysates with TDP-43 antibodies followed by immunoblotting with anti-HA antibody (12CA5 mAb) revealed a high molecular mass smear of ubiquitinated TDP-43 species, enhanced in the presence of 187 TDP-43 expression (Fig. 5B). Control immunoprecipitation reactions carried out using an unrelated antibody (anti-tau T46 mAb) showed little or no high molecular mass smear, irrespective of TDP-43 CTF expression (data not shown). MG132 treatment after transfection greatly increased the abundance of ubiquitinated TDP-43 concomitant with TDP-43 CTF expression (Fig. 5B). Lack of TDP-43-positive high molecular mass smear in the TDP-43 immunoblot is likely due to relatively low levels of ubiquitinated CTFs and as such below the sensitivity for detection with this antibody. These results are consistent with the notion that exogenous expression of TDP-43 CTFs leading to the formation of cytoplasmic aggregates also results in increased ubiquitination.
FIGURE 5.
TDP-43 CTFs are ubiquitinated. A, double labeling of N2a cells after transfection with vector or TDP-43 CTFs. The cells were stained 48 h after transfection using anti-C-t TDP-43 pAb (green) and anti-ubiquitin (FK2 mAb, red) antibodies. The majority of TDP-43 aggregates strongly colocalized with multi-ubiquitin antibody signal. Scale bar, 10 μm. B, HA or human-specific TDP-43 mAb immunoblots (IB) of input or immunoprecipitated N2a cell lysates cotransfected with empty vector or 187 TDP-43 and HA-tagged ubiquitin (HA-Ub) in the presence (+) or absence (–) of MG132 10 μm (16 h). Note the presence of the MG132-dependent, ubiquitinated HA positive high molecular mass smear (*) when 187 TDP-43 is expressed (**). The arrows indicate IgG bands. DAPI,4′,6′-diamino-2-phenylindole.
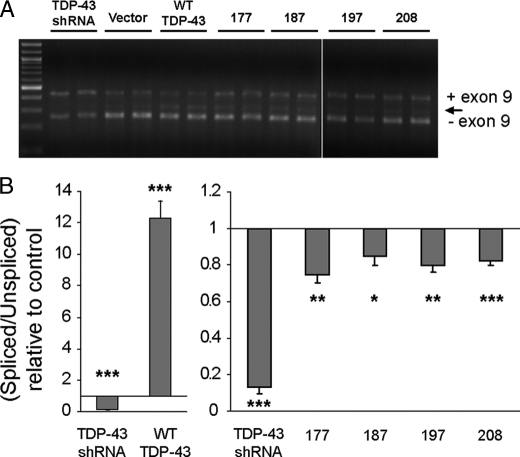
Accumulation of TDP-43 CTFs Compromises RNA Splicing of CFTR—One of the biological functions of TDP-43 described previously is the regulation of alternative splicing. In particular, exon skipping mediated through interactions of TDP-43 with GU repeats in CFTR and ApoAII pre-mRNA transcripts has been reported (6, 7), whereas the enhancement of exon inclusion for the SMN2 gene has been reported upon overexpression of TDP-43 (8). To determine whether expression of TDP-43 CTFs had any impact on this known biological function of TDP-43, we analyzed splicing efficiency using a previously described minigene reporter system (supplemental Fig. S3) (6). In agreement with previous observations, control experiments in cells transfected with WT-TDP-43 or a TDP-43 short hairpin RNA demonstrate that TDP-43 overexpression or depletion (>90%) led to a decrease or an increase in CFTR exon 9 inclusion, respectively (Fig. 6 and supplemental Fig. S3). Surprisingly, expression of all four TDP-43 CTFs tested resulted in a decrease in exon skipping activity relative to control transfected cells, suggesting a loss-of-function consequence of CTF generation (Fig. 6 and supplemental Fig. S3). The expression of WT-TDP-43 protein, the knockdown of endogenous TDP-43, and expression of all four TDP-43 CTFs were confirmed in the same samples employed for RNA isolation using immunoblot analysis of transfected QBI-293 cells (supplemental Fig. S3). This effect was specific to the TDP-43 CTFs because parallel transfection of α-synuclein, a protein not known to affect RNA splicing, into the same cell line showed no impact on CFTR reporter splicing activity (data not shown). It is noteworthy that although these TDP-43 CTFs do not possess the first RNA recognition motif (RRM1) required for efficient RNA and DNA binding, they still encode the entire C-terminal region containing the glycine-rich domain that is critical for interactions of TDP-43 with proteins displaying known splicing inhibitory activity such as hnRNPs (3, 20). Taken together, these results provide the first evidence of impaired splicing regulatory activity in the presence of abnormal TDP-43 species associated with FTLD-U and ALS pathology.
FIGURE 6.
TDP-43 CTF-expression cells display abnormal CFTR mRNA splicing. A, representative agarose gel electrophoresis analysis of RT-PCR products used to assess CFTR exon 9 inclusion in TG(13)T(5) transfected cells. QBI-293 were first transfected with either vector alone, WT TDP-43, a TDP-43 short hairpin RNA, or TDP-43 CTFs, and 48 h later, they were transfected with the TG(13)T(5) CFTR reporter plasmid. After another 24 h, total RNA was isolated, and RT-PCR was performed using the primers indicated in supplemental Fig S3. PCR products were visualized on agarose gels and quantified using the Agilent 2100 Bioanalyzer. Exon 9 included (+) and excluded (–) RT-PCR products are shown. The arrow indicates a previously reported aberrant splicing product (6) generated by a cryptic splicing site. B, spliced versus unspliced ratios were calculated and then normalized to the values of vector alone transfections. Mean values from at least three different experiments performed in duplicate are shown with S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (Student t test versus vector alone).
DISCUSSION
Our study investigated the role of TDP-43 CTFs in the pathogenesis of two neurodegenerative TDP-43 proteinopathies, i.e. FTLD-U and ALS. To that end, we recovered pathological TDP-43 CTFs from FTLD-U brains and provided the identity of one fragment encompassing amino acid residues 208–414. We further showed that expression of either the 208–414 TDP-43 CTF or slightly longer CTFs in cell culture systems resulted in the formation of ubiquitinated and hyperphosphorylated CTF aggregates. Finally, we demonstrated that TDP-43 CTF accumulation alters the splicing pattern of the TDP-43 mRNA target CFTR. These findings show that the generation of CTFs is sufficient to initiate a number of events — cytoplasmic localization, ubiquitination, phosphorylation, and aggregation of TDP-43 CTFs — that mirror TDP-43 proteinopathy and that these events have functional consequences on gene splicing.
Structurally, it is not surprising that the C-terminal region of TDP-43 may be important in disease pathogenesis, because this region harbors several remarkable features. Although the general domain architecture of TDP-43 is similar to that of other RNA-binding proteins of the RRM family such as hnRNPs, in silico analysis of the extreme C-terminal portion of TDP-43 (PSIPRED Server) (21) indicates that this region displays little organized secondary structure. In this regard, although the full-length protein contains typical motifs (i.e. RRMs and glycine-rich domains), CTFs of TDP-43 (such as those found in disease and modeled in this study) may be viewed as functionally different entities that share some commonalities with other proteins linked to neurodegeneration, such as α-synuclein. These comparatively disordered peptides are aggregation-prone if the right conditions or post-translational modifications are present.
Genetically, the C-terminal region of TDP-43 is implicated in pathogenesis as well. The discovery of multiple ALS-associated TARDBP mutations that map almost exclusively to the C-terminal domain constitutes an intriguing reminder of the potential link between abnormal TDP-43 post-translational modification, localization, or conformation and the pathogenesis of TDP-43 proteinopathies (22–25). Interestingly, several of these reported mutations may create novel phosphorylation sites through substitution to serine, which could provide the basis for abnormal properties of mutated TDP-43. Testing of such a hypothesis awaits the availability of more human neuropathological material from TARDBP mutation patients.
In the experiments described here, we found TDP-43 CTFs to be relatively insoluble and aggregation-prone similar to recent results in yeast (26). Specifically, the expression of a green fluorescent protein-tagged TDP-43 construct encoding a CTF (188–414 TDP-43, similar to a CTF used in this study) in yeast led to the formation of cytoplasmic aggregates as well as cellular toxicity. Yeast and mammalian cells appear to show some important differences in their handling of TDP-43, however. For example, N-terminal fragments of TDP-43 used in the same yeast study remained in the nucleus and showed no apparent aggregation or effect on cell survival (26). In contrast, N-terminal fragments of TDP-43 expressed in a mammalian cell culture system have been reported to mislocalize and form inclusions (27). Unlike in yeast cells, epitope-tagged TDP-43 CTFs are more soluble than their untagged counterparts when expressed in mammalian cells (supplemental Fig. S2). Thus, the biochemical profile of TDP-43 truncations appears to be sensitive to the cellular system used for study.
Our data show that expression of predominantly cytoplasmic species of TDP-43 (i.e. CTFs or NLS mutants) results in phosphorylation by an endogenous mechanism at residues that are pathologically hyperphosphorylated in human TDP-43 proteinopathies such as FTLD-U and ALS. Moreover, immunoblot and immunocytochemistry data showed that this abnormal phosphorylation is seen only with insoluble aggregated TDP-43. These data are compatible with a model of pathogenesis whereby cytoplasmic species of TDP-43 generated by proteolysis and/or redistribution become hyperphosphorylated and ubiquitinated. Other less likely possibilities include that hyperphosphorylation decreases solubility and promote aggregation or that more insoluble species are preferential substrates for yet to be determined protein kinases. Overall, our CTFs results suggest that proteolytic cleavage of TDP-43 may constitute an important event for aggregate formation, occurring independently or upstream of hyperphosphorylation.
If cytoplasmically localized, abnormally hyperphosphorylated and insoluble/aggregated CTFs are bona fide features of disease, what are their downstream functional consequences? This question is difficult to answer because the functions of normal TDP-43 remain incompletely understood. One relatively well characterized biological function of TDP-43, however, is in the regulation of alternative splicing - specifically, the splicing of CFTR, Apo IIA, and SMN (6–8). Of these, the most thoroughly studied splicing function of TDP-43 to date has been CFTR exon 9 skipping, which is mediated through (UG)m(U)n regulatory regions to which TDP-43 binds (6). In the present study, we showed that expression of TDP-43 CTFs alters CFTR splicing, arguing that the generation of pathological CTFs affects normal TDP-43 function. We postulate that this effect may result from altered interactions with members of the hnRNP family of splicing factors. In vitro studies have determined that several members of the hnRNP family (A1, A2/B1, A3, and C1/C2) interact with TDP-43 through its C-terminal region (3). Because hnRNP A/B proteins have known inhibitory splicing properties, the effect of TDP-43 on exon skipping could be mediated via recruitment of an hnRNP-rich inhibitory complex through its C-terminal tail (3, 28). Given that several of these interactors have been shown to shuttle between the cytoplasm and the nucleus (29), excess glycine-rich CTFs in the cytoplasm could alter the nucleocytoplasmic equilibrium of available hnRNP proteins, affecting not only TDP-43-mediated targets but also many different splicing and mRNA export pathways involving hnRNPs. Dysregulation of gene expression could then cause downstream disease effects in TDP-43 proteinopathies.
In this study, we investigated the hypothesis that TDP-43 CTFs are central to disease pathogenesis because they are seen in affected CNS regions from FTLD-U and ALS patients and absent in the CNS of normal individuals. By identifying Arg208 as one of the cleavage sites of endogenous CTFs from FTLD-U brains and by demonstrating that this and other larger TDP-43 CTFs expressed in cultured cells co-migrate with pathological CTFs, we have generated TDP-43 CTF constructs to study the biochemical properties of the fragments. We showed that simply by expressing TDP-43 CTFs in a cell culture system, one can recapitulate key biochemical features of TDP-43 in disease, i.e. insolubility, aggregation, ubiquitination, and hyperphosphorylation. We also showed that expression of TDP-43 CTFs is sufficient to provoke a partial loss of one of the only known biological functions of TDP-43: regulation of CFTR exon 9 splicing. These findings support the idea that generation of TDP-43 CTFs is an important event in FTLD-U and ALS pathogenesis. Thus, we believe that the cell culture model described here will be useful in identifying endogenous signaling cascades that lead to TDP-43 phosphorylation, assessing the impact of splicing alteration on cellular metabolism, and elucidating mechanisms of aberrant protein-protein interaction displayed by pathological TDP-43 species. This information in turn will be vital in the development of targeted therapies for these neurodegenerative diseases.
Supplementary Material
Acknowledgments
We thank Drs. F. Baralle and E. Buratti (International Centre for Genetic Engineering and Biotechnology, Trieste, Italy) for their generosity in sharing the CFTR minigene constructs. We thank Dr. Scott Pesiridis for helpful discussion and Chi Li for technical support.
This work was supported, in whole or in part, by National Institutes of Health Grant AG17586. This work was also supported by Deutsche Forschungsgemeinschaft Grant SFB 596 and Federal Ministry of Education and Research Grant 01GI0704. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: FTLD-U, frontotemporal lobar degeneration with ubiquitin-positive inclusions; ALS, amyotrophic lateral sclerosis; CFTR, cystic fibrosis transmembrane conductance regulator; CTF, C-terminal fragment; hnRNP, heterogeneous nuclear ribonucleoprotein; IP, immunoprecipitation; mAb, monoclonal antibody; NES, nuclear export signal; NLS, nuclear localization signal; pAb, polyclonal antibody; RRM, RNA recognition motifs; RIPA, radioimmunoprecipitation assay; CFTR, cystic fibrosis transmembrane conductance regulator; MOPS, 4-morpholinepropanesulfonic acid; PBS, phosphate-buffered saline; CHAPS, 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonic acid; RT, reverse transcription; C-t, C terminus; CNS, central nervous system.
References
- 1.Neumann, M., Sampathu, D. M., Kwong, L. K., Truax, A. C., Micsenyi, M. C., Chou, T. T., Bruce, J., Schuck, T., Grossman, M., Clark, C. M., McCluskey, L. F., Miller, B. L., Masliah, E., Mackenzie, I. R., Feldman, H., Feiden, W., Kretzschmar, H. A., Trojanowski, J. Q., and Lee, V. M. Y. (2006) Science 314 130–133 [DOI] [PubMed] [Google Scholar]
- 2.Forman, M. S., Trojanowski, J. Q., and Lee, V. M. Y. (2007) Curr. Opin. Neurobiol. 17 548–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buratti, E., Brindisi, A., Giombi, M., Tisminetzky, S., Ayala, Y. M., and Baralle, F. E. (2005) J. Biol. Chem. 280 37572–37584 [DOI] [PubMed] [Google Scholar]
- 4.Ou, S. H., Wu, F., Harrich, D., Garcia-Martinez, L. F., and Gaynor, R. B. (1995) J. Virol. 69 3584–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abhyankar, M. M., Urekar, C., and Reddi, P. P. (2007) J. Biol. Chem. 282 36143–36154 [DOI] [PubMed] [Google Scholar]
- 6.Buratti, E., Dork, T., Zuccato, E., Pagani, F., Romano, M., and Baralle, F. E. (2001) EMBO J. 20 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercado, P. A., Ayala, Y. M., Romano, M., Buratti, E., and Baralle, F. E. (2005) Nucleic Acids Res. 33 6000–6010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bose, J. K., Wang, I. F., Hung, L., Tarn, W. Y., and Shen, C. K. (2008) J. Biol. Chem. 283 28852–28859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, I. F., Reddy, N. M., and Shen, C. K. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy, J. M., Henry, R. G., Langmore, S., Kramer, J. H., Miller, B. L., and Lomen-Hoerth, C. (2007) Arch Neurol. 64 530–534 [DOI] [PubMed] [Google Scholar]
- 11.Arai, T., Hasegawa, M., Akiyama, H., Ikeda, K., Nonaka, T., Mori, H., Mann, D., Tsuchiya, K., Yoshida, M., Hashizume, Y., and Oda, T. (2006) Biochem. Biophys. Res. Commun. 351 602–611 [DOI] [PubMed] [Google Scholar]
- 12.Cairns, N. J., Neumann, M., Bigio, E. H., Holm, I. E., Troost, D., Hatanpaa, K. J., Foong, C., White, C. L., 3rd, Schneider, J. A., Kretzschmar, H. A., Carter, D., Taylor-Reinwald, L., Paulsmeyer, K., Strider, J., Gitcho, M., Goate, A. M., Morris, J. C., Mishra, M., Kwong, L. K., Stieber, A., Xu, Y., Forman, M. S., Trojanowski, J. Q., Lee, V. M. Y., and Mackenzie, I. R. (2007) Am. J. Pathol. 171 227–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winton, M. J., Igaz, L. M., Wong, M. M., Kwong, L. K., Trojanowski, J. Q., and Lee, V. M. (2008) J. Biol. Chem. 283 13302–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Igaz, L. M., Kwong, L. K., Xu, Y., Truax, A. C., Uryu, K., Neumann, M., Clark, C. M., Elman, L. B., Miller, B. L., Grossman, M., McCluskey, L. F., Trojanowski, J. Q., and Lee, V. M. (2008) Am. J. Pathol. 173 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paxinou, E., Chen, Q., Weisse, M., Giasson, B. I., Norris, E. H., Rueter, S. M., Trojanowski, J. Q., Lee, V. M., and Ischiropoulos, H. (2001) J. Neurosci. 21 8053–8061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neumann, M., Kwong, L. K., Lee, E. B., Kremmer, E., Flatley, A., Xu, Y., Forman, M. S., Troost, D., Kretzschmar, H. A., Trojanowski, J. Q., and Lee, V. M. (2009) Acta Neuropathol. 117 137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pagani, F., Buratti, E., Stuani, C., Romano, M., Zuccato, E., Niksic, M., Giglio, L., Faraguna, D., and Baralle, F. E. (2000) J. Biol. Chem. 275 21041–21047 [DOI] [PubMed] [Google Scholar]
- 18.Muro, A. F., Iaconcig, A., and Baralle, F. E. (1998) FEBS Lett. 437 137–141 [DOI] [PubMed] [Google Scholar]
- 19.Hasegawa, M., Arai, T., Nonaka, T., Kametani, F., Yoshida, M., Hashizume, Y., Beach, T. G., Buratti, E., Baralle, F., Morita, M., Nakano, I., Oda, T., Tsuchiya, K., and Akiyama, H. (2008) Ann. Neurol. 64 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buratti, E., and Baralle, F. E. (2001) J. Biol. Chem. 276 36337–36343 [DOI] [PubMed] [Google Scholar]
- 21.Bryson, K., McGuffin, L. J., Marsden, R. L., Ward, J. J., Sodhi, J. S., and Jones, D. T. (2005) Nucleic Acids Res. 33 36–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gitcho, M. A., Baloh, R. H., Chakraverty, S., Mayo, K., Norton, J. B., Levitch, D., Hatanpaa, K. J., White, C. L., 3rd, Bigio, E. H., Caselli, R., Baker, M., Al-Lozi, M. T., Morris, J. C., Pestronk, A., Rademakers, R., Goate, A. M., and Cairns, N. J. (2008) Ann. Neurol. 63 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kabashi, E., Valdmanis, P. N., Dion, P., Spiegelman, D., McConkey, B. J., Vande Velde, C., Bouchard, J. P., Lacomblez, L., Pochigaeva, K., Salachas, F., Pradat, P. F., Camu, W., Meininger, V., Dupre, N., and Rouleau, G. A. (2008) Nat. Genet. 40 572–574 [DOI] [PubMed] [Google Scholar]
- 24.Sreedharan, J., Blair, I. P., Tripathi, V. B., Hu, X., Vance, C., Rogelj, B., Ackerley, S., Durnall, J. C., Williams, K. L., Buratti, E., Baralle, F., de Belleroche, J., Mitchell, J. D., Leigh, P. N., Al-Chalabi, A., Miller, C. C., Nicholson, G., and Shaw, C. E. (2008) Science 319 1668–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Deerlin, V. M., Leverenz, J. B., Bekris, L. M., Bird, T. D., Yuan, W., Elman, L. B., Clay, D., Wood, E. M., Chen-Plotkin, A. S., Martinez-Lage, M., Steinbart, E., McCluskey, L., Grossman, M., Neumann, M., Wu, I. L., Yang, W. S., Kalb, R., Galasko, D. R., Montine, T. J., Trojanowski, J. Q., Lee, V. M., Schellenberg, G. D., and Yu, C. E. (2008) Lancet Neurol. 7 409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson, B. S., McCaffery, J. M., Lindquist, S., and Gitler, A. D. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 6439–6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ayala, Y. M., Zago, P., D'Ambrogio, A., Xu, Y. F., Petrucelli, L., Buratti, E., and Baralle, F. E. (2008) J. Cell Sci. 121 3778–3785 [DOI] [PubMed] [Google Scholar]
- 28.Buratti, E., and Baralle, F. E. (2008) Front. Biosci. 13 867–878 [DOI] [PubMed] [Google Scholar]
- 29.Pinol-Roma, S., and Dreyfuss, G. (1992) Nature 355 730–732 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.