Abstract
The telomeric protein TRF1 negatively regulates telomere length by inhibiting telomerase access at the telomere termini, suggesting that the protein level of TRF1 at telomeres is tightly regulated. Regulation of TRF1 protein abundance is essential for proper telomere function and occurs primarily through post-translational modifications of TRF1. Here we describe RLIM, a RING H2 zinc finger protein with intrinsic ubiquitin ligase activity, as a TRF1-interacting protein. RLIM increases TRF1 turnover by targeting it for degradation by the proteasome in a ubiquitin-dependent manner, independently of Fbx4, which is known to interact with and negatively regulate TRF1. Whereas overexpression of RLIM decreases the level of TRF1 protein, depletion of endogenous RLIM expression by small hairpin RNA increases the level of TRF1 and leads to telomere shortening, thereby impairing cell growth. These results demonstrate that RLIM is involved in the negative regulation of TRF1 function through physical interaction and ubiquitin-mediated proteolysis. Hence, RLIM represents a new pathway for telomere maintenance by modulating the level of TRF1 at telomeres.
Telomeres, the specialized nucleoprotein complexes at the ends of eukaryotic chromosomes, are essential for the maintenance of chromosome integrity, and their deregulation has been implicated in aging and cancer (1). Properly capped telomeres provide protection from nucleolytic degradation and prevent end-to-end fusion between chromosome ends (2, 3). In the absence of functional telomere maintenance pathways, dividing cells show a progressive loss of telomeric DNA during successive rounds of cell division because of a DNA end replication problem (4, 5). In humans, telomerase activity is expressed in a majority of immortalized cells but is undetectable in most normal somatic cells, suggesting that activation of telomerase is necessary for the proliferation of primary and transformed cells (6–8).
Telomere maintenance relies on associations between the telomeric DNA repeats and specific binding proteins. The six major telomeric proteins (TRF1, TRF2, RAP1, TIN2, POT1, and TPP1) have been shown to form a large complex, referred to as the mammalian telosome/shelterin, and participate in telomere regulation (9–11). Among the telomeric proteins, TRF1 and TRF2 directly bind to the double-stranded telomeric repeats and interact with a number of proteins to maintain telomere structure and length (12). Both proteins contain a C-terminal DNA binding motif that is closely related to the Myb domain and an internal conserved TRF2 homology domain that mediates dimerization (13). TRF2 has an essential role in end protection (14) and stabilizes a terminal loop structure called the t-loop, thereby concealing telomere termini from the action of telomerase and other enzymatic activities (15). TRF2 also works closely with its associated protein RAP1 (16). In comparison, TRF1 negatively regulates telomere length by inhibiting access of telomerase at telomere termini. Overexpression of TRF1 in telomerase-positive cells results in a gradual shortening of telomeres, whereas a dominant negative mutant induces inappropriate telomere elongation (12, 17).
Post-translational modifications of TRF1 play important roles in modulating telomere length homeostasis by determining the abundance of TRF1 at telomeres (19–21). We have previously identified casein kinase 2 (CK2) as a TRF1-interacting protein (22). CK2 interacts with and phosphorylates TRF1 in vitro and in cells. CK2-mediated phosphorylation is required for the efficient telomere binding of TRF1, suggesting a novel role of CK2 as a positive regulator for determining the level of TRF1 at telomeres. Furthermore, CK2 phosphorylation appears to be critical for TRF1-mediated telomere length control. Recently, it was reported that Polo-like kinase 1 phosphorylates TRF1 and that its phosphorylation is involved in both TRF1 overexpression-induced apoptosis and the telomere binding ability of TRF1 (23). In addition, it has been reported that ATM interacts with and phosphorylates TRF1 in response to ionizing DNA damage (24).
Telomere length is also regulated by tankyrase 1 through its interaction with TRF1 (25, 26). Tankyrase 1 poly(ADP-ribosyl)ates TRF1 and releases it from telomeres, allowing access of telomerase to telomeres and subsequently telomere elongation (27). Thus, tankyrase 1 is a positive regulator of telomere length. The inhibition of TRF1 by tankyrase 1 is in turn controlled by TIN2 (28). TIN2 forms a ternary complex with TRF1 and tankyrase 1 and appears to protect TRF1 from being modified by tankyrase 1. Partial knockdown of TIN2 by small interfering RNA results in loss of TRF1 from telomeres, leading to subsequent telomere elongation (29).
TRF1 can be dissociated from telomeres by either activation of tankyrase 1 (25) or inhibition of CK2 (22). The dissociated telomere-unbound form of TRF1 is subsequently degraded via ubiquitin-mediated proteolysis (19). It has been reported previously that Fbx4, a member of the F-box family of proteins, interacts with TRF1 and promotes its ubiquitination in vitro and in vivo (21). Thus, sequential post-translational modification of TRF1, including poly(ADP-ribosyl)ation by tankyrase 1 (25), phosphorylation by CK2 (22), and ubiquitination by Fbx4 (21), may modulate telomere length homeostasis by determining the level of TRF1 at telomeres.
In a search for proteins capable of interacting with TRF1, we identified RLIM using the yeast two-hybrid screening assay. RLIM was previously identified as an E3 ubiquitin ligase able to target CLIM for proteasome-dependent degradation, thereby inhibiting developmental LIM homeodomain activity (30–32). RLIM increases TRF1 turnover by targeting it for degradation by the proteasome in a ubiquitin-dependent manner, independently of Fbx4. Whereas overexpression of RLIM promotes degradation of TRF1, depletion of endogenous RLIM expression by small hairpin RNA (shRNA) stabilizes TRF1 and leads to telomere shortening, thereby impairing cell growth. Overall, these results demonstrate that RLIM, like Fbx4, is a critical negative regulator of TRF1 protein abundance and represents a new pathway for telomere maintenance by regulating the level of TRF1 at telomeres.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screening—Yeast two-hybrid screening was performed as described previously (33). Briefly, the full-length TRF1 cDNA was fused to the LexA DNA binding domain and transformed by the lithium acetate method into the EGY48 yeast strain. Expression of the LexA-TRF1 fusion protein was verified by Western blotting using anti-LexA antibody. The stable strain was transformed again with a HeLa cDNA library fused to the activation domain vector pB42AD (Clontech).
Recombinant Protein Expression and Antibody Production—The RLIM-V5 expression vector was constructed by inserting the BamHI and XhoI fragment from the full-length Rnf12 cDNA encoding RLIM (generated by PCR with the appropriate synthetic primers) into pcDNA3.1A/V5 (Invitrogen) (31). The expression vectors for GST-RLIM were constructed by cloning the full-length and truncated fragments from the Rnf12 cDNA into pGEX-5X-1, and the GST fusion proteins were expressed and purified by using a glutathione-Sepharose column according to the manufacturer's instructions (Amersham Biosciences). To raise antibodies against RLIM, the fragment containing amino acid residues 1–310 was isolated by cleaving GST-RLIM fusion protein with factor Xa and used to immunized rabbits. Anti-RLIM antibodies were affinity-purified using a GST-RLIM column.
GST Pulldown, Immunoprecipitation, and Immunoblot—GST pulldown, immunoprecipitation, and immunoblot analyses were performed as described previously (33). Briefly, the expression vectors were transfected into H1299 cells using Lipofectamine 2000 (Invitrogen) for 30 h followed by lysis. For the GST pulldown assay, the cellular supernatants were precleared with glutathione-Sepharose 4B (Amersham Biosciences) and incubated with glutathione-Sepharose beads containing GST fusion proteins. For immunoprecipitation, the supernatants were preincubated with protein A-Sepharose (Amersham Biosciences) and incubated with primary antibodies precoupled with protein A-Sepharose beads. The immunoprecipitated proteins were washed extensively and subjected to immunoblot analysis. Immunoprecipitation and immunoblotting were performed using anti-TRF1 (Santa Cruz Biotechnology), anti-FLAG (Sigma), anti-V5 (Invitrogen), and anti-HA (Santa Cruz Biotechnology) antibodies as specified. All of the immunoblots are representatives of at least three experiments that demonstrated similar results.
In Vitro Ubiquitination Assay—GST-TRF1 expressed in bacteria was used as substrate for in vitro ubiquitination assay. RLIM-V5 was immunoprecipitated by anti-V5 antibody from lysates of transfected H1299 cells and used as E3 enzyme. The ubiquitination assay was carried out in ubiquitination buffer (50 mm Tris, pH 7.5, 2 mm ATP, 2.5 mm MgCl2, 0.5 mm dithiothreitol, and 0.05% Nonidet P-40) containing E1 (50 ng), UbcH5a (200 ng), and His-tagged ubiquitin (0.8 μg). The reactions were incubated at 30 °C for 1 h and terminated with 2× SDS loading buffer. The proteins were separated on a 10% SDS-PAGE. In some experiments, GST-RLIM was expressed in bacteria and used as the E3 enzyme for in vitro ubiquitination assay. Ubiquitinated proteins were visualized by immunoblotting using an anti-ubiquitin antibody (Zymed Laboratories Inc.) or anti-TRF1 antibody (Santa Cruz Biotechnology).
In Vivo Ubiquitination Assay—H1299 cells were transfected with HA-ubiquitin, FLAG-TRF1, and RLIM-V5 expression vectors, either alone or in combination, followed by MG132 treatment to inhibit proteasome function. Cell lysates were subjected to immunoprecipitation with anti-FLAG antibody followed by immunoblotting analysis with anti-HA antibody to illuminate ubiquitin-modified TRF1.
Establishment of Stable Cell Lines—To establish cell lines stably expressing RLIM, the full-length Rnf12 cDNA was subcloned into a pLentiM1.4 expression vector (Macrogen) and transfected into 293T cells with VSV-G (vesicular stomatitis virus glycoprotein) expression vector and gag-pol expression vector. The culture supernatant containing viral vector particles was harvested 48 h after transfection and filtered through a 0.45-μm membrane filter (Nalgene). Green fluorescent protein expression of transduced cells was observed and photographed under fluorescence microscope. The virus titers were in the range of 107 transduction units/ml medium. HT1080 cells were transduced with the RLIM-expressing lentivirus containing 6 μg/ml Polybrene (Sigma) and selected with 0.5 μg/ml puromycin (Sigma). Multiple independent single clones were isolated and checked for protein expression by immunoblotting with anti-RLIM antibody.
RNA Interference—Two shRNA expression lentiviral vectors for targeting RLIM gene were generated by inserting double-stranded oligonucleotides (targeting nucleotides: 5′-GGCAACAAACTTCGTAAACTA-3′ for shRNA1 and 5′-GGTCTCAGACACCAAACAACA-3′ for shRNA2) into the shLenti2.4G lentiviral vector (Macrogen), which is designed to produce shRNA from the U6 promoter and to express enhanced green fluorescent protein from the hCMV (human cytomegalovirus) promoter (34, 35). The scrambled sequence (5′-AATCGCATAGCGTATGCCGTT-3′) was used as a control and did not correspond to any known gene in the data bases. The nucleotide sequence of the constructs was verified by sequencing. Lentiviral vector particles were produced and transduced as described above. Of the two different shRNA viral constructs used, one (shRNA2) substantially reduced RLIM level as determined by quantitative immunoblotting. The RNAi-resistant RLIM was silently mutated in the shRNA2 target sequence (underlined nucleotide) without altering the amino acid sequence with the following primer: 5′-AACTCGGTCTAGATCTCAAACTCCAAACAACACT-3′. To exclude the problems associated with the off-target effects of shRNA, two different siRNA duplexes targeting RLIM (targeting nucleotides: 5′-GUGAGAACCUAUGUCAGUA-3′ for siRNA1 and 5′-GACCAACCUAGAGGACUCA-3′ for siRNA2) were transiently transfected into HT1080 cells using Lipofectamine 2000 (Invitrogen) for 3 days.
Immunofluorescence Staining and Fluorescence-activated Cell Sorter (FACS) Analysis—Cells were fixed with 2% paraformaldehyde and permeabilized with 0.5% Triton X-100 in phosphate-buffered saline. Cells were blocked in phosphate-buffered saline containing 2% bovine serum albumin and incubated with mouse anti-TRF1 antibody (Sigma), and rabbit anti-RLIM antibody. After washing, cells were incubated with Alexa Fluor 568 goat anti-mouse immunoglobulin and Alexa Fluor 488 goat anti-rabbit immunoglobulin (Molecular Probes). DNA was stained with 4,6-diamino-2-phenylindole (Vectashield, Vector Laboratories). For FACS analysis, cells were washed with phosphate-buffered saline and fixed 30 min in ice-cold 70% ethanol. The fixed cells were resuspended in phosphate-buffered saline containing RNase A (200 μg/ml) and propidium iodide (50 μg/ml) and incubated in dark for 15 min at room temperature. Cell cycle distribution was examined by flow cytometry using a FACScan flow cytometer (BD Biosciences).
Terminal Restriction Fragment Analysis—To measure the telomere length, genomic DNA was digested with RsaI and HinfI and separated on a 0.7% agarose gel. DNA samples were transferred to a nylon membrane (Hybond N+; Amersham Biosciences) and hybridized with a 32P-labeled probe (TTAGGG)20. Signals were detected by a phosphoimage analyzer (Fuji Photo Film).
RESULTS
Identification of RLIM as a TRF1-interacting Factor—To identify TRF1-interacting factors, we screened a HeLa cell cDNA library using a yeast two-hybrid system. With the full-length TRF1 as bait, 26 positive clones were obtained and sequenced. One of the isolated clones contained the Rnf12 cDNA encoding RLIM (Fig. 1A). RLIM was identified previously as an E3 ubiquitin ligase able to target CLIM for proteasome-dependent degradation, thereby inhibiting developmental LIM homeodomain activity (32). Rnf12-specific mRNA is widely expressed in many tissues and encodes the RLIM protein, which contains a RING H2 zinc finger domain found in a diverse group of ubiquitin ligases (36). Rnf12 orthologs have been identified in a wide spectrum of species including Xenopus, chick, mouse, and human, suggesting an evolutionarily conserved function of RLIM (31). Interestingly, RLIM has been recognized by autologous antibodies in patients with renal cell carcinoma (37). This suggests that altered RLIM protein expression may be relevant to cancer.
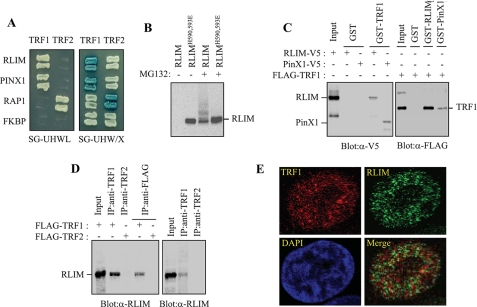
FIGURE 1.
Physical interaction between TRF1 and LRIM. A, analysis of the physical interaction between TRF1 and RLIM using the yeast two-hybrid assay. PinX1, RAP1, and unrelated FKBP52 were used as the TRF1-binding, TRF2-binding, and negative control, respectively. The growth on the SG-HWUL plate and the blue signal on the SG-HWU/X plate indicate activation of the reporter genes, LacZ and LEU2, respectively. S, synthetic; G, galactose; H, histidine (-); W, tryptophan (-); U, uracil, (-); L, leucine (-); X, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal). B, H1299 cells were transfected with V5-RLIM or V5-RLIMH590,593E and either untreated or treated with 10 μm MG132 for 4 h. Cell lysates were analyzed by immunoblotting with anti-V5 antibody probe. C, interaction between TRF1 and RLIM in vitro. Left, GST and GST-TRF1 were immobilized on glutathione-Sepharose and incubated with exogenously expressed RLIM-V5 and PinX1-V5. After washing and SDS-PAGE, bound RLIM and PinX1 were detected by immunoblotting with anti-V5 antibody. Right, GST, GST-RLIM, and GST-PinX1 were incubated with exogenously expressed FLAG-TRF1 followed by immunoblotting with anti-FLAG antibody. D, coimmunoprecipitation of TRF1 and RLIM. Left, H1299 cells were transfected with FLAG-TRF1 or FLAG-TRF2 and then subjected to immunoprecipitation as indicated followed by immunoblotting with anti-RLIM antibody. Right, H1299 cells were subjected to immunoprecipitation with either anti-TRF1 or -TRF2 antibodies followed by immunoblotting with anti-RLIM antibody. E, H1299 cells were treated with 10 μm MG132 for 4 h and analyzed by indirect immunofluorescence. Paraformaldehyde-fixed cells were stained with anti-TRF1 (red) and anti-RLIM antibodies (green). DNA was stained with 4,6-diamidino-2-phenylindole (blue).
To confirm the direct interaction between TRF1 and RLIM, we performed GST pulldown experiments. When the RLIM-V5 expression vector was transfected into H1299 cells, RLIM was not detected in lysates directly probed with anti-V5 antibody (Fig. 1B). This suggests that RLIM may be rapidly degraded through autoubiquitination, like other RING finger proteins that promote their own ubiquitination/degradation (38, 39). In the presence of MG132, however, there was a marked increase in both unmodified and ubiquitinated RLIM species. RLIMH590,593E, with mutations in the RING finger domain, was not autoubiquitinated (Fig. 1B). Therefore, RLIM-V5 in cells pretreated with MG132 was tested for its ability to bind to GST-TRF1 that was immobilized on glutathione-Sepharose beads. GST-TRF1, but not the control GST, bound to RLIM, indicating that TRF1 interacts with RLIM in vitro (Fig. 1C). PinX1, which was known to interact with TRF1 (40), was used as a positive control. Likewise, GST-RLIM and GST-PinX1, but not the control GST, precipitated FLAG-TRF1 expressed in H1299 cells. To determine whether TRF1 and RLIM associate in vivo, H1299 cells were transfected with Flag-TRF1 or FLAG-TRF2 and subjected to immunoprecipitation. RLIM was detected in anti-TRF1 and anti-FLAG immunoprecipitates when FLAG-TRF1 was expressed but not in anti-TRF2 and anti-FLAG immunoprecipitates when FLAG-TRF2 was expressed (Fig. 1D). Endogenous TRF1 also immunoprecipitated with endogenous RLIM in H1299 cells, indicating that TRF1 interacts with RLIM in mammalian cells (Fig. 1D). When subcellular distribution of endogenous TRF1 and RLIM was examined by immunofluorescence, both proteins were clearly localized in the nucleus of the cell. In addition, the RLIM protein, appears to colocalize with TRF1 in distinct subnuclear pockets of fluorescence (Fig. 1E). Thus, RLIM and TRF1 coimmunoprecipitate and colocalize in cells.
To map the region in RLIM that interacts with TRF1, different RLIM fragments were expressed in H1299 cells (Fig. 2A) and subjected to GST pulldown experiments. GST-TRF1 bound to RLIM and its N-terminal 310 fragment but not its C-terminal 311–624 fragment, indicating that N-terminal region of RLIM is required for in vitro association with TRF1 (Fig. 2B). Likewise, GST-RLIM and GST-RLIM-(1–310), but not GST-RLIM-(311–624), precipitated FLAG-TRF1 expressed in H1299 cells (Fig. 2C). We next determined the domain in TRF1 that is important for RLIM binding. A series of TRF1 fragments were fused to GST and used in the in vitro binding assay (Fig. 2D). GST-TRF1 fragment encompassing residues 265–378 bound to RLIM (Fig. 2E). In contrast, GST-TRF1 fragments encompassing residues 1–66, 67–155, 156–316, or 379–439 all failed to associate with RLIM, indicating that RLIM binds a region between the homodimerization domain and the Myb-like motif of TRF1.
FIGURE 2.
Mapping the interaction domains on TRF1 and RLIM. A, schematic representation of the region of RLIM involved in binding to TRF1. B, the region of RLIM involved in binding to TRF1 was analyzed in a GST pulldown assay. H1299 cells were transfected with various RLIM-V5 mutants and subjected to immunoblotting analysis with anti-V5 antibody directly (left) or were first precipitated by GST or GST-TRF1 (right). C, H1299 cells transfected with FLAG-TRF1 were precipitated by GST or the various truncated GST-RLIM fusion proteins followed by immunoblotting with anti-FLAG antibody. D, schematic representation of TRF1 truncations fused to GST. E, GST or the various truncated GST-TRF1 fusion proteins were affinity-purified and incubated with H1299 cell extracts followed by detection of endogenous RLIM by immunoblotting with anti-RLIM.
RLIM Promotes Ubiquitination of TRF1 in Vitro and in Vivo—The finding that TRF1 interacts physically with RLIM led us to hypothesize that RLIM may target TRF1 for ubiquitination and thereby promote its proteasome-dependent degradation. To examine whether RLIM ubiquitinates TRF1 directly, we performed in vitro ubiquitination assay. Recombinant GST-TRF1 was incubated with E1, UbcH5a, and His-ubiquitin in the presence of purified RLIM proteins. RLIM-V5 was expressed in H1299 cells and affinity-purified using anti-V5 antibody-coupled beads from cells pretreated with MG132 because of autoubiquitination. Anti-ubiquitin antibody detected a smear of slower migrating bands that represent polyubiquitinated TRF1 (Fig. 3A). This modification was substantially enhanced by adding increasing amounts of RLIM. The smear was undetectable when GST-TRF1, E1, and UbcH5a, or His-ubiquitin, were omitted. When the same blot was probed with anti-V5 antibody, unmodified and self-ubiquitinated products of RLIM were visualized (Fig. 3A). To exclude the possibility that other cellular proteins co-precipitated with RLIM from the transfected H1299 cell lysates may influence TRF1 ubiquitination, we used bacterially expressed GST-RLIM as an E3 ligase for in vitro ubiquitination assay. As shown in Fig. 3B, TRF1 was efficiently ubiquitinated by adding increasing amounts of GST-RLIM.
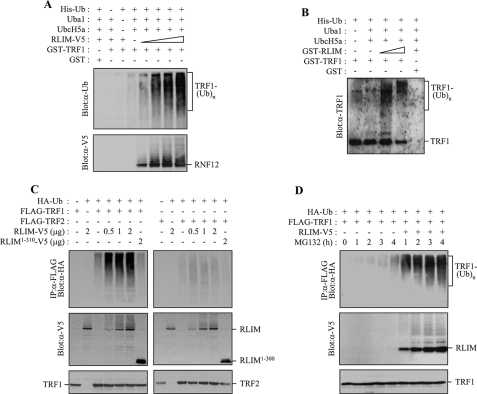
FIGURE 3.
RLIM serves as an E3 ubiquitin ligase for TRF1. A, RLIM-V5 was expressed in H1299 cells and affinity-purified from cells pretreated with 10 μm MG132 for 4 h using anti-V5 antibody-coupled with protein A-Sepharose beads. GST-TRF1 was incubated with increasing amounts of RLIM-V5 in the presence of E1, UbcH5a (E2), and His-ubiquitin (Ub) as indicated. After the ubiquitination reaction, the samples were analyzed by SDS-PAGE and immunoblotting with anti-ubiquitin antibody to reveal ubiquitinated products. B, GST-TRF1 was incubated with increasing amounts of GST-RLIM expressed in bacteria as indicated. Ubiquitinated products were detected by immunoblotting with anti-TRF1 antibody. C, H1299 cells were co-transfected with HA-ubiquitin, FLAG-TRF1 (or FLAG-TRF2), and together with either increasing amounts of RLIM-V5 or RLIM-(1–310)-V5 as specified and treated with 10 μm MG132 for 4 h. Immunoprecipitation was performed with anti-FLAG antibody before probing with anti-HA antibody. D, H1299 cells were co-transfected with HA-ubiquitin, FLAG-TRF1, and together with or without RLIM-V5 as indicated and treated with 10 μm MG132 for the indicated times. Anti-FLAG immunoprecipitates and cell lysates were analyzed by immunoblotting with anti-HA antibody and anti-V5 antibody probes, respectively.
To examine whether RLIM contributes to the ubiquitination of TRF1 in vivo, H1299 cells were co-transfected with HA-ubiquitin, FLAG-TRF1 (or FLAG-TRF2), and RLIM-V5 (or RLIM-(1–310)-V5) and then subjected to immunoprecipitation. Anti-FLAG immunoprecipitates were probed with anti-HA antibody to illuminate ubiquitin-modified TRF1 (Fig. 3C). Ubiquitin conjugates of TRF1 were readily detected when RLIM was expressed, and the levels of ubiquitinated TRF1 were correlated with the amounts of ectopically expressed RLIM. In contrast, RLIM-(1–310), which does not contain a RING finger motif, failed to ubiquitinate TRF1, indicating that the RING finger domain is essential for E3 activity of RLIM. Interestingly, ubiquitination of TRF2 was not observed even when RLIM was highly expressed. To further examine the in vivo role of RLIM in the ubiquitination of TRF1, H1299 cells were co-transfected with HA-ubiquitin, FLAG-TRF1, and together with or without RLIM-V5 and immunoprecipitated with anti-FLAG antibody followed by immunoblotting with anti-HA antibody (Fig. 3D). Although no TRF1-ubiquitin conjugates were observed in untreated cells, a low level was detected after 4 h of MG132 treatment. In marked contrast, co-expression of RLIM caused a dramatic increase in the amounts of ubiquitinated TRF1 species. When the expression levels of RLIM were examined by immunoblotting with anti-V5 antibody, both unmodified and self-ubiquitinated RLIM species increased in a time-dependent manner in response to MG132 treatment (Fig. 3D). These data indicate that RLIM is an E3 ligase for TRF1 ubiquitination in mammalian cells.
RLIM Expression Reduces the Steady-state Level of TRF1—Because RLIM promotes ubiquitination of TRF1, we next examined its ability to regulate the level of TRF1. H1299 cells were co-transfected with FLAG-TRF1 (or FLAG-TRF2) and increasing amounts of RLIM-V5, followed by immunoblotting with anti-FLAG antibody. Overexpression of RLIM resulted in a clear reduction in the level of TRF1 in a dose-dependent manner but overexpression of RLIM-(1–310) did not affect the level of TRF1 (Fig. 4A). In contrast, the expression level of TRF2 was not affected by overexpression of RLIM. On the basis of these data, we concluded that RLIM acts as a negative regulator of TRF1 function through ubiquitin-mediated proteolysis in mammalian cells. To examine whether reduction in TRF1 level is due to a RLIM-related decrease in the transcription level, the impact of RLIM on gene expression of TRF1 was evaluated using reverse transcription-PCR analysis. Whereas RLIM mRNA levels were much higher in cells expressing the gene (relative to control cells), we observed no significant difference in the steady-state levels of TRF1 mRNA in cells expressing RLIM and the empty vector (Fig. 4B).
FIGURE 4.
Overexpression of RLIM reduces the steady-state level of TRF1 protein. A, H1299 cells were co-transfected with FLAG-TRF1 (or FLAG-TRF2) and either increasing amounts of RLIM-V5 or RLIM-(1–310)-V5 as specified. The levels of ectopically expressed TRF1 (or TRF2) and RLIM were determined by immunoblotting with anti-FLAG antibody and anti-V5 antibody, respectively. An antibody against β-actin was used as a loading control. B, representative results of reverse transcription-PCR analysis for the expression of RLIM and TRF1 genes in H1299 cells transfected with RLIM. Reverse transcription-PCR products from each sample were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) signal. C, H1299 cells were transfected with RLIM-V5, HA-Fbx4, or both and treated with 100 μg/ml cycloheximide together with, or without, 10 μm MG132 for the indicated times. The nuclear fractions were analyzed on immunoblots with anti-TRF1 antibody or anti-TATA-binding protein (TBP) antibody. D, graphical representation of relative TRF1 levels normalized against the TATA-binding protein loading control. The graph represents an average of three independent experiments.
Although we showed that RLIM associates with and efficiently ubiquitinates TRF1, our data did not exclude the existence of other E3 ligases that target TRF1. Recently, it was reported that Fbx4, a member of the F-box family of proteins, targets TRF1 for ubiquitin-mediated degradation (21). To investigate whether RLIM promotes the turnover of TRF1 protein independently of Fbx4, we examined the effects of RLIM or FBX4 on the half-life of TRF1. H1299 cells were transfected with RLIM, FBX4, or both, incubated with cycloheximide to block new protein synthesis, and then analyzed by immunoblotting with anti-TRF1 antibody. As shown in Fig. 4C, and graphically in Fig. 4D, TRF1 was turned over with a half-life of ∼4.5 h. In contrast, overexpression of either RLIM or Fbx4 caused a clear reduction in the half-life of TRF1, which was further decreased in cells co-transfected with both RLIM and Fbx4. Interestingly, expression of RLIM, but not Fbx4, had little effect on cellular levels of TRF1 for the first 3 h. Turnover of TRF1 was blocked by the MG132 treatment, indicating that TRF1 degradation is mediated by the proteasome. Taken together, these data indicate that RLIM, like Fbx4, can negatively regulate the steady-state level of TRF1 protein.
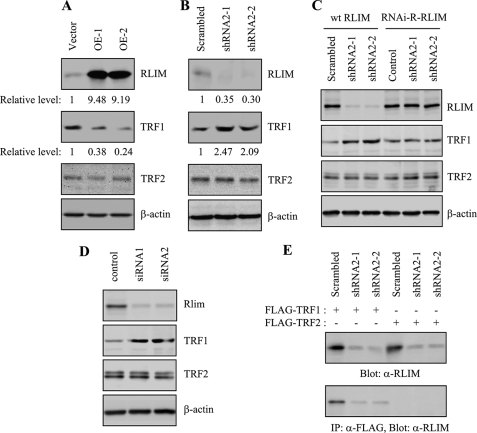
RLIM Regulates the Level of Endogenous TRF1—To examine the direct effect of RLIM on the level of endogenous TRF1, we established HT1080 cell lines stably expressing RLIM or control vector. Two independent clones were isolated to rule out the effect of clonal variation. Level of RLIM was examined by immunoblotting with anti-RLIM antibody. The amount of ectopically expressed RLIM was ∼9.3-fold greater than that of endogenous protein (Fig. 5A). Cells expressing RLIM and control vector grew normally and exhibited no detectable differences in growth rates or morphology over 75 population doublings. Whereas endogenous TRF1 was readily detectable in vector control cells, the amount of TRF1 protein was significantly reduced in all RLIM-expressing clones examined (Fig. 5A). In contrast, the level of endogenous TRF2 was not affected by overexpression of RLIM.
FIGURE 5.
RLIM regulates the levels of endogenous TRF1 protein. A, HT1080 clones stably expressing RLIM-V5 (OE-1 and OE-2) or the control empty vector were harvested at 75 population doublings and subjected to immunoblotting with anti-RLIM, anti-TRF1, anti-TRF2, or β-actin antibodies. The relative levels of RLIM and TRF1 were determined from three independent experiments using β-actin as a loading control. B, HT1080 clones stably expressing RLIM shRNA2 (shRNA2-1 and shRNA2-2) or scrambled shRNA were harvested at 75 population doublings and subjected to immunoblotting with anti-RLIM, anti-TRF1, anti-TRF2, or β-actin antibodies. The relative levels of RLIM and TRF1 were determined from three independent experiments using β-actin as a loading control. C, HT1080 cells stably expressing RLIM shRNA2 (shRNA2-1 and shRNA2-2) or scrambled shRNA were transfected with wild type RLIM (wt RLIM) or RNAi-R-RLIM and examined for expression of endogenous RLIM, TRF1, and TRF2 by immunoblotting. D, HT1080 cells were transiently transfected with siRNA duplexes, either nontargeting control or targeting RLIM (siRNA1 and siRNA2) for 3 days and examined for the expression of endogenous RLIM, TRF1, and TRF2 by immunoblotting. E, HT1080 cells stably expressing RLIM shRNA2 (shRNA2-1 and shRNA2-2) or scrambled shRNA were transfected with FLAG-TRF1 or FLAG-TRF2 and subjected to immunoprecipitation (IP) with anti-FLAG antibody, followed by immunoblotting with anti-RLIM antibody.
To confirm the role of RLIM in a more physiological setting, the expression of endogenous RLIM was inhibited using shRNA produced from a lentiviral vector. Of the two different shRNA viral constructs used, one (shRNA2) substantially reduced RLIM level as determined by quantitative immunoblotting (Fig. 5B). Depletion of endogenous RLIM by shRNA2 in HT1080 cells caused a pronounced accumulation of endogenous TRF1 protein, indicating that RLIM negatively regulates TRF1 expression. These results were reproducible with two independently isolated clones (shRNA2-1 and shRNA2-2). Endogenous TRF2 level was not altered by depletion of RLIM. To further examine the specificity of RLIM shRNA2, we performed the rescue assay with RNAi-resistant RLIM (RNAi-R-RLIM). Three nucleotides of wild type RLIM were mutated to create resistance to shRNA2, and the encoding protein was not altered. Cells stably expressing shRNA2-1 or shRNA2-2 were transfected with wild type RLIM or RNAi-R-RLIM and examined for the expression of endogenous RLIM by immunoblotting (Fig. 5C). The expression of RLIM was inhibited in cells transfected with wild type RLIM but not in cells transfected with RNAi-R-RLIM. When wild type RLIM was transfected, the TRF1 level in the shRNA2 cells was significantly higher than that in the scrambled shRNA cells. However, the expression of RNAi-R-RLIM in the shRNA2 cells attenuated the accumulation of endogenous TRF1 to a similar level of the scrambled shRNA cells. To exclude the problems associated with the off-target effects of shRNA, HT1080 cells were transiently transfected with two different siRNA duplexes targeting RLIM and subjected to immunoblotting (Fig. 5D). Both siRNA1 and siRNA2 decreased the level of RLIM protein. TRF1 level in cells transfected with siRNA duplexes targeting RLIM was significantly higher than that in cells transfected with control nontargeting siRNA. RLIM siRNA did not alter the quantities of TRF2 protein.
Because the expression of RLIM was inhibited by RLIM shRNA2, we further validated in vivo association of TRF1 and RLIM in the shRNA2 cells. Cells stably expressing shRNA2-1 or shRNA2-2 were transfected with FLAG-TRF1 or FLAG-TRF2 and subjected to immunoprecipitation (Fig. 5E). When FLAG-TRF1 was expressed, the signal of immunoprecipitated RLIM was significantly reduced in the RLIM shRNA2 cells compared with the scrambled shRNA cells. However, RLIM was not detected in anti-FLAG immunoprecipitates when FLAG-TRF2 was expressed. These results clearly indicate that the protein immunoprecipitated with TRF1 that is detected with anti-RLIM antibody is RLIM.
We further confirmed that the amount of endogenous RLIM is involved in regulating the stability of endogenous TRF1. The stability of TRF1 protein was monitored by immunoblotting with anti-TRF1 antibody in cell lines stably expressing RLIM or shRNA2-1 or the scrambled vector after cycloheximide treatment to inhibit new protein synthesis. Overexpression of RLIM reduced the half-life of TRF1 when compared with the scrambled vector cells (Fig. 6). In contrast, depletion of RLIM by shRNA2-1 significantly extended the half-life of TRF1. In all cell lines, TRF1 was stabilized upon treatment with MG132. Taken together, these results indicate that RLIM regulates the half-life of endogenous TRF1 through ubiquitin-mediated proteolysis.
FIGURE 6.
RLIM regulates the half-life of endogenous TRF1. A, stable clones expressing the control vector, RLIM-V5 (OE-1), or shRNA (shRNA2-1) were treated with 100 μg/ml cycloheximide together with, or without, 10 μm MG132 for the indicated times followed by immunoblotting with anti-TRF1 antibody or anti-actin antibodies. B, graphical representation of relative TRF1 levels normalized against the β-actin loading control. The graph represents an average of three independent experiments.
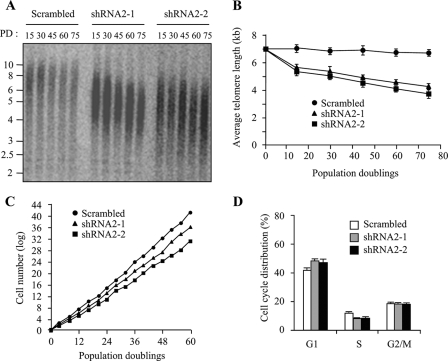
Depletion of RLIM Leads to Progressive Telomere Shortening and Impaired Cell Growth—Given that shRNA-mediated depletion of RLIM increases the amount of endogenous TRF1 protein, we wished to determine whether inhibition of RLIM is attended by change in telomere length. We performed a terminal restriction fragment length analysis in HT1080 cells stably expressing RLIM shRNA2 at various population doublings. Cells stably expressing RLIM shRNA2-1 or shRNA2-2 maintained the decreased level of RLIM and the increased level of TRF1 over 75 population doublings (see Fig. 5B). Control cells maintained a stable terminal restriction fragment length with an average telomere length of ∼7 kb up to 75 population doublings (Fig. 7, A and B). However, inhibition of RLIM led to a clear reduction in telomere length in two independent stable cell lines. These findings suggest that depletion of endogenous RLIM resulted in an increase of TRF1 expression and subsequent progressive telomere shortening. It is noteworthy that stable overexpression of RLIM did not cause a marked increase in telomere length over 75 population doublings (data not shown). This might be because telomeres in RLIM-overexpressing cells apparently do not elongate as much as observed in the terminal restriction fragment length analysis.
FIGURE 7.
Depletion of RLIM leads to progressive telomere shortening and impaired cell growth. A, genomic blot of telomere restriction fragments in stable HT1080 clones expressing RLIM shRNA2 (shRNA2-1 or shRNA2-2) or the control scrambled shRNA. Genomic DNA was isolated at the indicated population doublings (PD), digested with RsaI and HinfI, and analyzed by Southern blotting using a telomere repeat probe. B, graphical representation of average terminal restriction fragment length versus population doubling number from two independent experiments. C, growth curves of stable HT1080 clones expressing RLIM shRNA2 (shRNA2-1 or shRNA2-2) or the control scrambled shRNA. Stable clones were replated every 3–4 days to maintain log-phase growth and to calculate the growth rate. D, cell cycle profiles of stable clones expressing RLIM shRNA2. Stable clones were harvested at 60 population doublings, and cell cycle profiles were determined by propidium iodide staining and flow cytometry. The results represent the average of three independent experiments.
Finally, we examined the effects of RLIM inhibition on growth suppression. As shown in Fig. 7C, the growth rates of RLIM-depleted cells were significantly reduced when compared with control cells. To determine whether this growth inhibition correlates with an altered cell cycle distribution, RLIM-depleted cells were subjected to flow cytometric analysis by propidium iodide staining. RLIM-depleted cells exhibited an increase in the proportion of cells in G1 and a decrease in the proportion of cells in S phase compared with control cells (Fig. 7D). This was reflected by an increase in the G1/S ratio from 3.5 to 5.9, which is consistent with growth arrest in the G1 phase of the cell cycle. As inhibition of RLIM expression resulted in an increase in TRF1 protein, these data suggest that RLIM plays an important role in determining the level of TRF1 in cells and is involved in telomere length-mediated growth suppression and cell cycle control.
DISCUSSION
TRF1 negatively regulates the telomere length by inhibiting the interaction between telomeres and telomerase (12, 17). This suggests that the abundance of TRF1 protein at telomeres is tightly regulated. Sequential post-translational modifications of TRF1, including phosphorylation, poly(ADP-ribosyl)ation, and ubiquitination, play a critical role in modulating telomere length homeostasis by determining the level of TRF1 at telomeres (21, 22, 25). Here we describe a novel function of RLIM for regulating the abundance of TRF1 in mammalian cells. RLIM interacts with TRF1 and promotes its ubiquitination in vitro and in vivo, thus acting as a negative regulator of TRF1. Whereas overexpression of RLIM reduces the half-life of endogenous TRF1 protein, depletion of RLIM results in an increase of endogenous TRF1 expression, subsequently leading to progressive telomere shortening and impaired cell growth. These results support the hypothesis that RLIM plays a critical role in regulating the level of TRF1 and participates in telomere maintenance.
It has been reported previously that TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres (19). Recently, Fbx4 was shown to interact with and target TRF1 for ubiquitin-mediated degradation by acting as a substrate-specific adaptor of a Cul1-based ubiquitin ligase complex (21). Like Fbx4, RLIM has ubiquitin ligase activity and can promote ubiquitination and degradation of TRF1. Overexpression of either RLIM or Fbx4 in cells causes a clear reduction in the half-life of TRF1, which is further decreased in cells co-transfected with both genes. These data suggest that RLIM may function independently of Fbx4. Moreover, RLIM binds to the internal region between the dimerization and the Myb domain of TRF1 (residues 265–378), whereas Fbx4 associates with the N-terminal region in the dimerization domain of TRF1 (residue 48–155). Thus, it will be informative to determine whether both proteins can bind TRF1 simultaneously. Because RLIM and Fbx4 are each capable of promoting ubiquitination of TRF1, RLIM appears to target TRF1 for ubiquitin-mediated degradation through an Fbx4-independent mechanism. However, this does not exclude the possibility that the two proteins may function cooperatively under physiological conditions.
The functional similarity between RLIM and Fbx4 raises important questions about the physiological significance of multiple pathways that exert negative control over TRF1. TRF1 binds to telomeric DNA as a homodimer and can be covalently modified for determining its abundance at telomeres (18, 19). These findings suggest that different forms of TRF1 can coexist within a cell. The different forms of TRF1 may be targeted for ubiquitination by different pathways, depending on cell type or physiological condition. For example, tankyrase 1 poly(ADP-ribosyl)ates TRF1 and releases it from telomeres, allowing access of telomerase to telomeres (19, 25). The fact that the tankyrase 1-dependent turnover of TRF1 requires Fbx4 indicates that FBx4 is downstream of tankyrase 1 in the TRF1 degradation pathway (21). Recently, it was reported that CK2-mediated phosphorylation enhances the ability of TRF1 to bind telomeric DNA in vitro and in vivo (22). On the contrary, inhibition of CK2 results in the nonphosphorylated form of TRF1, which in turn leads to a decrease in its DNA binding activity. It will be of interest to determine whether the dissociated telomere-unbound form of TRF1 is degraded by different ubiquitination mechanism.
When TRF1 is dissociated from telomeres either by activation of tankyrase 1 (19) or inhibition of CK2 (22), telomerase can gain access to the TRF1-free telomeres to add the telomeric repeats. The resulting telomere-unbound TRF1 could be ubiquitinated by RLIM and/or Fbx4, and subsequent degradation of TRF1 by the proteasome prevents its rapid reassociation with telomeres. The ability of RLIM to promote ubiquitin-mediated degradation of TRF1 was efficiently inhibited by the MG132 treatment, suggesting that TRF1 degradation occurs in a separate step from its dissociation from telomeres. This is consistent with previous findings that telomere-bound TRF1 is protected from ubiquitination (19). Based on this hypothesis, RLIM could preferentially interact with and ubiquitinate the telomere-dissociated form of TRF1 to rapidly degrade this protein. Thus, telomeres seem to be assembled with newly synthesized TRF1 rather than the dissociated telomere-unbound TRF1 to establish telomerase-inaccessible telomeres.
Overall, our results provide an insight into the new cellular function of RLIM in addition to its function to inhibit developmental LIM homeodomain activity (31, 32). Although important questions about redundancy and the physiological significance of multiple E3 ligases that control the abundance of TRF1 at telomeres remain to be resolved, our results indicate that RLIM represents a new pathway for modulating telomere length homeostasis by acting as a negative regulator of TRF1.
This work was supported by grants from the Molecular Aging Research Center, Korean Ministry of Health and Welfare; the Research Program on Dual Regulation Mechanisms of Aging and Cancer, Korea Science and Engineering Foundation (M1075604000107N560400110); Korea Research Foundation (KRF-2005-C00097); and the Seoul Development Institute. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TRF, telomeric repeat binding factor; CK2, casein kinase 2; E3, ubiquitin-protein isopeptide ligase; GST, glutathione S-transferase; shRNA, small hairpin RNA; siRNA, small interfering RNA; RNAi, RNA interference; RNAi-R-RLIM, RNAi-resistant RLIM; FACS, fluorescence-activated cell sorter; HA, hemagglutinin; E1, ubiquitin-activating enzyme.
References
- 1.Smogorzewska, A., and de Lange, T. (2004) Annu. Rev. Biochem. 73 177-208 [DOI] [PubMed] [Google Scholar]
- 2.Blasco, M. A., Lee, H. W., Hande, M. P., Samper, E., Lansdorp, P. M., DePinho, R. A., and Greider, C. W. (1997) Cell 91 25-34 [DOI] [PubMed] [Google Scholar]
- 3.Harrington, L. (2005) Chromosome Res. 13 493-504 [DOI] [PubMed] [Google Scholar]
- 4.Lingner, J., Cooper, J. P., and Cech, T. R. (1995) Science 269 1533-1534 [DOI] [PubMed] [Google Scholar]
- 5.Cerone, M. A., Autexier, C., Londoño-Vallejo, J. A., and Bacchetti, S. (2005) Oncogene 24 7893-7901 [DOI] [PubMed] [Google Scholar]
- 6.Kim, N. W., Piatyszek, M. A., Prowse, K. R., Harley, C. B., West, M. D., Ho, P. L. C., Coviello, G. M., Wright, W. E., Weinrich, S. L., and Shay, J. W. (1994) Science 266 2011-2015 [DOI] [PubMed] [Google Scholar]
- 7.Bodnar, A. G., Ouellette, M., Frolkis, M., Holt, S. E., Chiu, C. P., Morin, G. B., Harley, C. B., Shay, J. W., Lichtsteiner, S., and Wright, W. E. (1998) Science 279 349-352 [DOI] [PubMed] [Google Scholar]
- 8.Hahn, W. C., Counter, C. M., Lundberg, A. S., Beijersbergen, R. L., Brooks, M. W., and Weinberg, R. A. (1999) Nature 400 464-468 [DOI] [PubMed] [Google Scholar]
- 9.de Lange, T. (2005) Genes Dev. 19 2100-2110 [DOI] [PubMed] [Google Scholar]
- 10.O'Connor, M. S., Safari, A., Xin, H., Liu, D., and Songyang, Z. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 11874-11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu, D., O'Connor, M. S., Qin, J., and Songyang, Z. (2004) J. Biol. Chem. 279 51338-51342 [DOI] [PubMed] [Google Scholar]
- 12.Smogorzewska, A., van Steensel, B., Bianchi, A., Oelmann, S., Schaefer, M. R., Schnapp, G., and de Lange, T. (2000) Mol. Cell. Biol. 20 1659-1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fairall, L., Chapman, L., Moss, H., de Lange, T., and Rhodes, D. (2001) Mol. Cell 8 351-361 [DOI] [PubMed] [Google Scholar]
- 14.van Steensel, B., Smogorzewska, A., and de Lange, T. (1998) Cell 92 401-413 [DOI] [PubMed] [Google Scholar]
- 15.Griffith, J. D., Comeau, L., Rosenfield, S., Stansel, R. M., Bianchi, A., Moss, H., and de Lange, T. (1999) Cell 97 503-514 [DOI] [PubMed] [Google Scholar]
- 16.O'Connor, M. S., Safari, A., Liu, D., Qin, J., and Songyang, Z. (2004) J. Biol. Chem. 279 28585-28591 [DOI] [PubMed] [Google Scholar]
- 17.van Steensel, B., and de Lange, T. (1997) Nature 385 740-743 [DOI] [PubMed] [Google Scholar]
- 18.Bianchi, A., Smith, S., Chong, L., Elias, P., and de Lange, T. (1997) EMBO J. 16 1785-1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang, W., Dynek, J. N., and Smith, S. (2003) Genes Dev. 17 1328-1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishiyama, A., Muraki, K., Saito, M., Ohsumi, K., Kishimoto, T., and Ishikawa, F. (2006) EMBO J. 25 575-584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, T. H., Perrem, K., Harper, J. W., Lu, K. P., and Zhou, X. Z. (2006) J. Biol. Chem. 281 759-768 [DOI] [PubMed] [Google Scholar]
- 22.Kim, M. K., Kang, M. R., Nam, H. W., Bae, Y. S., Kim, Y. S., and Chung, I. K. (2008) J. Biol. Chem. 283 14144-14152 [DOI] [PubMed] [Google Scholar]
- 23.Wu, Z. Q., Yang, X., and Liu, X. (2008) J. Biol. Chem. 283 25503-25513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kishi, S., Zhou, X. Z., Ziv, Y., Khoo, C., Hill, D. E., Shiloh, Y., and Lu, K. P. (2001) J. Biol. Chem. 276 29282-29291 [DOI] [PubMed] [Google Scholar]
- 25.Smith, S., and de Lange, T. (2000) Curr. Biol. 10 1299-1302 [DOI] [PubMed] [Google Scholar]
- 26.Cook, B. D., Dynek, J. N., Chang, W., Shostak, G., and Smith, S. (2002) Mol. Cell. Biol. 22 332-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, S., Giriat, I., Schmitt, A., and de Lange, T. (1998) Science 282 1484-1487 [DOI] [PubMed] [Google Scholar]
- 28.Kim, S. H., Kaminker, P., and Campisi, J. (1999) Nat. Genet. 23 405-412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye, J. Z., and de Lange, T. (2004) Nat. Genet. 36 618-623 [DOI] [PubMed] [Google Scholar]
- 30.Bach, I., Rodriguez-Esteban, C., Carrière, C., Bhushan, A., Krones, A., Rose, D. W., Glass, C. K., Andersen, B., Izpisúa Belmonte, J. C., and Rosenfeld, M. G. (1999) Nat. Genet. 22 394-399 [DOI] [PubMed] [Google Scholar]
- 31.Ostendorff, H. P., Bossenz, M., Mincheva, A., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Lichter, P., and Bach, I. (2000) Genomics 69 120-130 [DOI] [PubMed] [Google Scholar]
- 32.Ostendorff, H. P., Peirano, R. I., Peters, M. A., Schlüter, A., Bossenz, M., Scheffner, M., and Bach, I. (2002) Nature 416 99-103 [DOI] [PubMed] [Google Scholar]
- 33.Lee, G. E., Yu, E. Y., Cho, C. H., Lee, J., Muller, M. T., and Chung, I. K. (2004) J. Biol. Chem. 279 34750-34755 [DOI] [PubMed] [Google Scholar]
- 34.Follenzi, A., Ailles, L. E., Bakovic, S., Geuna, M., and Naldini, L. (2000) Nat. Genet. 25 217-222 [DOI] [PubMed] [Google Scholar]
- 35.Dull, T., Zufferey, R., Kelly, M., Mandel, R. J., Nguyen, M., Trono, D., and Naldini, L. (1998) J. Virol. 72 8463-8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joazeiro, C. A., Wing, S. S., Huang, H., Leverson, J. D., Hunter, T., and Liu, Y. C. (1999) Science 286 309-312 [DOI] [PubMed] [Google Scholar]
- 37.Scanlan, M. J., Gordan, J. D., Williamson, B., Stockert, E., Bander, N. H., Jongeneel, V., Gure, A. O., Jäger, D., Jäger, E., Knuth, A., Chen, Y. T., and Old, L. J. (1999) Int. J. Cancer 83 456-464 [DOI] [PubMed] [Google Scholar]
- 38.Joazeiro, C. A., and Weissman, A. M. (2000) Cell 102 549-552 [DOI] [PubMed] [Google Scholar]
- 39.Kim, J. H., Park, S. M., Kang, M. R., Oh, S. Y., Lee, T. H., Muller, M. T., and Chung, I. K. (2005) Genes Dev. 19 776-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou, X. Z., and Lu, K. P. (2001) Cell 107 347-359 [DOI] [PubMed] [Google Scholar]