Abstract
Pemphigus vulgaris (PV) autoantibodies directly inhibit desmoglein (Dsg) 3-mediated transinteraction. Because cellular signaling also seems to be required for PV pathogenesis, it is important to characterize the role of direct inhibition in pemphigus acantholysis to allow establishment of new therapeutic approaches. Therefore, we modeled the Dsg1 and Dsg3 sequences into resolved cadherin structures and predicted peptides targeting the adhesive interface of both Dsg3 and Dsg1. In atomic force microscopy single molecule experiments, the self-designed cyclic single peptide specifically blocked homophilic Dsg3 and Dsg1 transinteraction, whereas a tandem peptide (TP) consisting of two combined single peptides did not. TP did not directly block binding of pemphigus IgG to their target Dsg antigens but prevented PV-IgG-induced inhibition of Dsg3 transinteraction in cell-free (atomic force microscopy) and cell-based (laser tweezer) experiments, indicating stabilization of Dsg3 bonds. Similarly, PV-IgG-mediated acantholysis and disruption of Dsg3 localization in HaCaT keratinocytes was partially blocked by TP. This is the first evidence that direct inhibition of Dsg3 binding is important for PV pathogenesis and that peptidomimetics stabilizing Dsg transinteraction may provide a novel approach for PV treatment.
Autoantibodies in pemphigus vulgaris (PV)4 are mainly directed against the adhesion molecules desmoglein (Dsg) 3 and Dsg1 (1, 2). Recently, we provided direct evidence that PV autoantibodies directly block Dsg3 transinteraction (3). In contrast, Dsg1 autoantibodies in pemphigus foliaceus (PF) were found to disrupt Dsg1 transinteraction most likely via cellular signaling events rather than by direct inhibition (3, 4). However, it still remained unanswered whether direct inhibition of Dsg3 transinteraction is the sole pathogenic mechanism in mucosal-dominant PV where autoantibodies against Dsg3 but not against Dsg1 are present (1). Alternatively, direct inhibition could act in concert with autoantibody-induced cellular signaling or may just be an epiphenomenon secondary to skin blistering (1, 5–7).
The idea of direct interference with Dsg transinteraction by pemphigus autoantibodies has been proposed when Dsg3 was discovered to be a cadherin-type cell adhesion molecule (8, 9). From structural and mutational analyses, it was concluded that classical cadherins are able to form adhesive dimers through their N-terminal cadherin extracellular domain (EC1) via different interaction schemes (10–13). Investigations of desmosomes using electron tomography from tissue sections also indicated various interaction models for desmosomal cadherins (14). Some of these interaction models resembled the interaction schemes found in the high resolution crystal structure analyses of N- and E-cadherin.
This study was designed to modulate Dsg3 and Dsg1 transinteraction by using peptides fitting the most likely Dsg adhesive interface revealed by three-dimensional modeling. In an earlier study, it was reported that peptides targeting the putative Dsg cell adhesion recognition site blocked desmocadherin-mediated adhesion (15). Homology modeling of Dsg3 and Dsg1 based on the structure of E-cadherin and N-cadherin led to the identification of a peptide sequence expected to block Dsg1 and Dsg3 transinteraction by occupying a predicted binding site. By combining two of these single peptides (SPs) via a flexible linker, a so-called tandem peptide (TP) was generated. This peptide was expected to stabilize Dsg adhesion by cross-linking the binding pockets of two transinteracting Dsg3 or Dsg1 molecules, similar to peptides directed against the adhesive interface of N-cadherin (16). Here we show that TP prevented the effects of PV-IgG on Dsg3 adhesion and attenuated PV-IgG-induced acantholysis in human keratinocytes. These data indicate a significant role of autoantibody-induced direct inhibition of Dsg3 binding in PV pathogenesis and may provide a novel therapeutic approach in PV treatment.
EXPERIMENTAL PROCEDURES
Modeling of the Dsg3 and Dsg1 EC1 and EC2 Domain—The high resolution crystal structure of E-cadherin (Protein Data Bank (PDB) entry 1EDH) was used as a structural template to model the EC1 and EC2 of human Dsg1 and Dsg3. Amino acid sequence alignments were produced using CLUSTALW. Amino acid residues of E-cadherin were exchanged for the corresponding amino acids of Dsg1 and Dsg3 using the ProteinDesign tool of Quanta2006 (Accelrys Inc., San Diego, CA). Insertions and deletions were modeled manually. Structure refinement was performed initially by removing steric clashes using side chain rotamer search tools in Xbuilt, Quanta2006. Backbone and side chain conformations were refined subsequently by energy minimization (500 steps steepest gradient energy minimization) and short molecular dynamics simulation (10 ps in vacuo simulation) employing only geometrical energy terms and stepwise removing conformational harmonic potentials for the backbone atoms (first round, energy force constant 50 kcal mol-1Å-2; second round, 5 kcal mol-1Å-2; third round, no harmonic potentials). The final models for Dsg1 and Dsg3 exhibited good backbone and side chain geometry; Ramachandran plot analysis showed that no residues exhibited backbone torsion angles in the disallowed area.
Design and Application of Dsg Antagonistic (SP) and Agonistic (TP) Peptides—To design peptides that possibly antagonize the transinteraction of Dsg1 and Dsg3, we structurally aligned our Dsg models onto the N-cadherin crystal structure, mimicking the putative transinteraction between two cadherin molecules (PDB entry 1NCH). This arrangement was used to form a Dsg1 (or Dsg3) transinteracting pair. One peptide was designed that comprised a fragment of the main binding interface. The amino acid sequence of this SP was LNSMGQD, corresponding to Leu-81–Asp-87 of Dsg1. Because of the homology of this sequence to the corresponding Dsg3 sequence (LNAQGLD), SP was also supposed to modulate Dsg3 transinteraction. SP was then cyclized by adding cysteine residues at the N- and C-terminal peptide end. The degree of flexibility of the peptide in its linear and cyclic form was simulated by running molecular dynamics simulation in an explicit water shell (250 ps, 8 Å water layer, CHARMm force field 22) using CHARMm, Quanta2006. The so-called TP was generated by combining two SPs linked via a flexible aminohexan linker coupled to one cysteine of the SP. The sequences of the control peptides CP-1 and CP-2 were RVDAE and N-Ac-CRVDAE-Aminohexan-RVDAEC-NH2, respectively, and derived from a study targeting VE-cadherin transinteraction with single (CP-1) and tandem (CP-2) peptides. The underlined peptide sequence denotes circularization via a disulfide bond between the given cysteine residues. SP, TP, and CP-2 peptides were obtained from a commercial supplier (PSL GmbH, Heidelberg, Germany). Peptide CP-1 was synthesized according to the Bachem practice of solid-phase peptide synthesis (all chemicals were supplied by Novabiochem (Darmstadt, Germany)). For most experiments, SP and CP-1 were used at 200 μm and TP and CP-2 was used at 20 μm.
Cell Culture and Test Reagents—The immortalized human keratinocyte cell line HaCaT was cultivated as described (3). PV antibody AK 23 was purchased from Biozol (Eching, Germany) and used at 75 μg/ml for experiments.
Purification and Preparation of Pemphigus IgG and Recombinant Dsg-Fc—Purification of the IgG fractions of a patient with a mucocutaneous PV (PV-IgG 1, both Dsg1 and Dsg3 antibodies) and one with mucous PV (PV-IgG 2, only Dsg3 antibodies), was performed as described previously (3). Final IgG concentrations were adjusted to 150–500 μg/ml for experiments. Recombinant human Dsg3 and Dsg1 were expressed and purified as described (3).
Atomic Force Microscopy (AFM) and Laser Tweezer Measurements—Effects of pemphigus IgG, AK 23, and peptides on cell-free homophilic Dsg3 and Dsg1 binding activities in AFM force distance cycles or Dsg3 and Dsg1 bead binding in laser tweezer experiments on HaCaT cells were conducted and analyzed as described recently (3).
Dsg1 and Dsg3 ELISA—For quantification of antibody binding to Dsg1 and Dsg3, ELISA with recombinant Dsg1 and Dsg3 was performed according to the manufacturer's instructions (Medical and Biological Laboratories, Nagoya, Japan). Peptides, AK 23, and pemphigus IgG were incubated with recombinant Dsgs on the ELISA plates in concentrations as described above. For detection of AK 23 binding to Dsg3, peroxidase-coupled mouse anti-human antibody was exchanged by peroxidase-coupled goat anti-mouse antibody. All conditions were run at least in triplicates.
Cytochemistry—HaCaT cells were stained with mouse monoclonal Dsg3 antibody (Zytomed Systems GmbH) as described previously (3).
Dispase-based Keratinocyte Dissociation Assay—The assay was performed as described in the literature with the following modifications (17, 18). HaCaT cells were seeded on 12-well plates and grown to confluence. After incubation for 24 h under various conditions, cells were washed with Hanks' buffered salt solution and treated for 30 min with 0.3 ml of dispase II (2.4 units/ml, Sigma) at 37 °C. Afterward, dispase solution was carefully removed, and cells were dissolved in 0.5 ml of Hanks' buffered salt solution. Mechanical stress was then applied by pipetting 10 times with a 1-ml pipette. Finally, dissociation was quantified by counting and averaging cell fragments in three defined areas of each condition under a binocular microscope. Every condition was repeated at least four times.
RESULTS
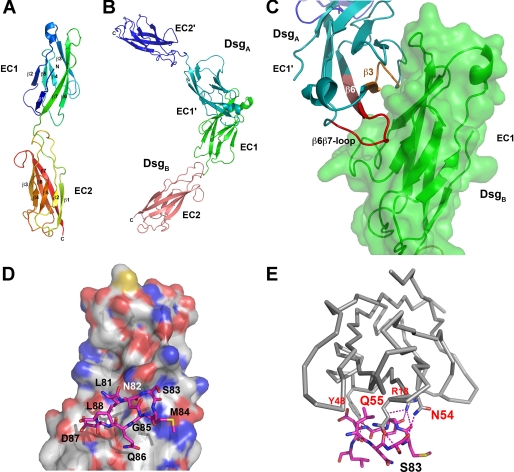
Conformational Epitope Modeling for Peptide Design and Function—A model for the EC1-EC2 domain pair of Dsg1 and Dsg3 was built based on the homology to known structures of other cadherins (Fig. 1) (10, 11). First we used the structure of the EC1-EC2 domain of E-cadherin (PDB entry 1EDH, (11)) as a template. Amino acid sequences of Dsg1 and Dsg3 of various species were aligned to that of N- and E-cadherin, and residues that differed between Dsg1 (or Dsg3) and E-cadherin EC1-EC2 were exchanged to finally present the amino acid sequence of human Dsg1 (or human Dsg3). Insertions and deletions between the Dsg1 or Dsg3 and E-cadherin were located only in the loop region within the EC2 domain (β2β3, β3β4, and β6β7 loop) and thus had no effect on the overall fold of the N-terminal EC1 domain of our Dsg1 or Dsg3 model. After refinement by removing steric clashes between side chains, backbone conformation was refined by short molecular dynamics simulations in vacuo using only geometrical terms. Analysis of the backbone hydrogen bond network revealed that the secondary structure was well conserved to the structural template, indicating that the amino acid differences between E-cadherin and Dsg1 or Dsg3 did not lead to major structural rearrangements. The obtained model for Dsg1 (Fig. 1A) was compared with the recently determined NMR structure of Dsg2 EC1 (PDB entry 2YQG), revealing a root mean square deviation of only 1.3 Å over 80 Cα atoms, thereby confirming that cadherins are structurally well conserved and that our models of Dsg1 or Dsg3 can be used for modeling the transinteraction between Dsgs.
FIGURE 1.
Dsg1 EC1 and EC2 modeling and peptide design. A, secondary structure elements as well as the N and C termini of the Dsg1 EC1 and EC2 model are indicated. B, model of the Dsg1 transinteraction as shaped on the basis of the N-cadherin crystal structure. The two EC1 domains interact via a large interface suggesting an in vivo interaction. C, blow-up of the interface region with peptide fragments possibly interfering with transinteraction shown in orange and red. A similar model was obtained for Dsg3 (data not shown). D, an SP designed to block Dsg transinteraction corresponded to residues Leu-81 to Leu-88 located in the putative binding site on Dsg1, the latter being illustrated by a van der Waals surface representation in atom color code. E, in comparison with D, the putative polar interactions (hydrogen bonds) are shown as stippled lines in magenta. Note that SP formed numerous H-bonds with residues in the Dsg1 binding site.
The final model of Dsg1 (or Dsg3) EC1-EC2 was subsequently structurally aligned to a dimer assembly of N-cadherin and its second molecule in the asymmetric unit representing a putative transinteracting cadherin molecule pair (Fig. 1, B and C) (10). This arrangement showed that β-strand 3 (residues Gln-33 to Ser-39 of Dsg1; residues Lys-33 to Ser-39 of Dsg3), the β3β4 loop and part of β-strand 4 (residues Asp-44 to Gln-55 of Dsg1; residues Asp-44 to Lys-55 of Dsg3), and β-strand 6 (residues Tyr-77 to Gln-86 of Dsg1; residues Thr-77 to Leu-86 of Dsg3) would constitute possible peptide fragments that mimic the transinteracting cadherin molecules (Fig. 1C). Out of these fragments, the so-called SP (amino acid sequence LNSMGQD, Fig. 1, D and E) was used, which should target both Dsg1 and Dsg3 because of sequence homologies. Recent structural analyses demonstrated that this sequence is located within an important immunological region of the extracellular Dsg1 and Dsg3 domains (19).
Because linear peptides may exhibit low binding affinity due to conformational flexibility that leads to entropic penalties upon binding, we designed cyclic peptides that should exist in a preformed conformation similar to those also observed in the transinteraction model of Dsg1 or Dsg3. Molecular simulation runs in explicit solvent confirmed that the backbone flexibility of the cyclized peptide was greatly limited when compared with the linear peptides but would not constrain the cyclic peptide into a conformer that deviates from the modeled transinteracting peptide. In this study, the designed SP was used to inhibit the interaction of Dsg1 or Dsg3. To stabilize interactions between two Dsg molecules, the so-called TP was generated by dimerizing two SPs.
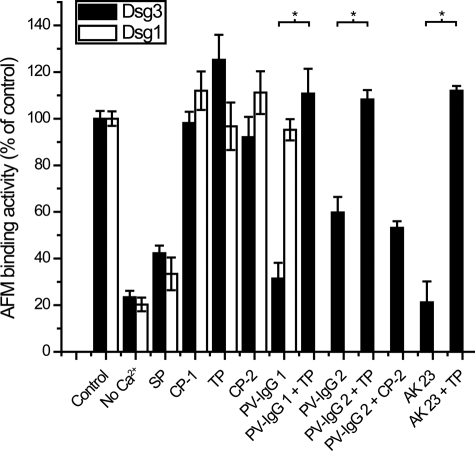
SP Blocked Homophilic Dsg3 and Dsg1 Transinteraction, whereas TP Prevented PV-IgG- and AK 23-induced Inhibition of Dsg3 Binding in Cell-free AFM Studies—As a proof of principle, we first studied the effect of the designed peptides in cell-free AFM experiments (Fig. 2). Recombinant Dsg molecules were covalently coupled to the tip and plate of the AFM setup to probe homophilic Dsg transinteraction in force distance cycles. The resulting binding activities revealed the amount and extent of Dsg transinteraction events. As expected, Dsg3 and Dsg1 binding activities (Fig. 2) were strongly reduced after Ca2+ depletion to 23% ± 3% and 20% ± 3% of controls, respectively, demonstrating Ca2+-dependent desmocadherin interaction. For the antagonistic SP, a concentration of 200 μm was found to efficiently reduce Dsg3 and Dsg1 transinteraction to 42% ± 3% and 33% ± 7% of control levels. Higher SP concentrations were also tested but did not yield different results (data not shown). Specificity of Dsg transinteraction for SP action was demonstrated by the VE-cadherin-targeting control peptide CP-1 (200 μm), which did not block Dsg3 or Dsg1 transinteraction. In contrast, the TP did not significantly alter Dsg3 and Dsg1 binding activities when applied at 20 μm and 0.5 h of preincubation, although a slight increase in binding activity was noted for Dsg3 (125% ± 11%). Higher concentrations were not used because of negative effects on Dsg transinteraction, possibly because occupation of binding sites by single TPs without cross-linking of two Dsg molecules resulted in decreased binding activities. At concentrations below 20 μm, both SP and TP had diminished effects in inhibiting Dsg transinteraction or preventing PV-IgG-mediated blocking of Dsg3 transinteraction as stated below, respectively (data not shown). Although direct binding of SP to Dsg3 or Dsg1 could not be demonstrated by this approach, our experiments indicate that SP directly inhibited Dsg transinteraction, whereas TP in a concentration of 20 μm did not. We hypothesized that TP did not increase Dsg binding activities because the measured binding activities already ranged at maximum under control conditions, and unbinding forces of single Dsg bonds were not increased by TP. A control VE-cadherin tandem peptide (CP-2, 20 μm) did not alter Dsg transinteraction in AFM studies.
FIGURE 2.
SP blocked homophilic Dsg3 or Dsg1 transinteraction and TP prevented PV-IgG- and AK 23-induced blocking of Dsg3 transinteraction in cell-free AFM studies. Binding activities of Dsg3 (black bars) and Dsg1 (white bars) fusion proteins coupled to the tip and plate of the AFM setup were strongly reduced after Ca2+ depletion. SP (200 μm) also blocked Dsg3 and Dsg1 transinteraction, whereas TP (20 μm) and the corresponding control peptides CP-1 and CP-2 did not. PV-IgG 1 and PV-IgG 2 interfered with Dsg3 transinteraction. Preincubation with TP (20 μm, p = 0.00360 and p = 0.00036, respectively) but not CP-2 (20 μm, p = 0.4139) prevented PV-IgG 1- and PV-IgG 2-induced loss of Dsg3 adhesion. Similar results were obtained with mouse PV antibody AK 23 (p = 0.00275). PV-IgG 1 did not directly inhibit homophilic Dsg1 transinteraction (n = 3–4 for each condition).
Next, we investigated whether TP was effective to block PV-IgG-induced loss of Dsg3 binding. As reported previously, PV-IgG directly interfered with Dsg3 transinteraction (3). This blocking effect was confirmed for two additional PV-IgG fractions, PV-IgG 1 (31% ± 7%) and PV-IgG 2 (60% ± 7%), in this study. Interestingly, preincubation with TP (20 μm) completely prevented PV-IgG-induced loss of transinteraction (binding activities were 111% ± 11% and 108% ± 4%, respectively). The control tandem peptide CP-2 (20 μm), however, did not prevent PV-IgG-mediated reduction in Dsg3 transinteraction (53% ± 3% Dsg3 binding activity), demonstrating specificity of Dsg3 transinteraction for TP. We performed similar experiments with mouse PV antibody AK 23, which targets the Dsg3 adhesive interface and blocked Dsg3 transinteraction (3, 20). AK 23-induced inhibition of Dsg3 transinteraction (21% ± 9%) was also prevented by TP (112% ± 2%). As we have already demonstrated previously (3), PV-IgG did not directly inhibit Dsg1 transinteraction (Fig. 2). Therefore, AFM experiments to study Dsg1 transinteraction with TP preincubation prior to pemphigus IgG treatment were not required. Taken together, AFM experiments demonstrated that TP efficiently prevented PV-IgG-induced loss of Dsg3 transinteraction.
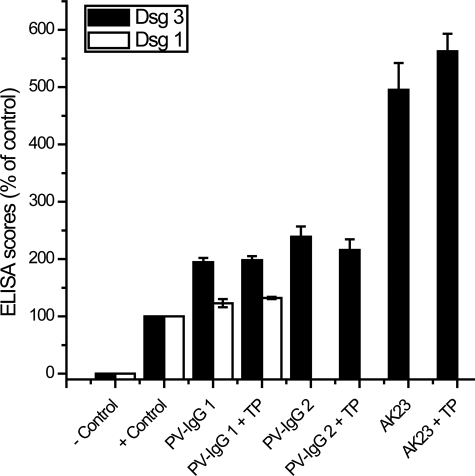
TP Did Not Interfere with Binding of Pemphigus IgG to Dsg1 and Dsg3—It is possible that TP-induced blocking of PV-IgG-mediated loss of Dsg3 transinteraction resulted from interference with binding of pemphigus IgG to Dsg1 and Dsg3. To address this important issue, we analyzed the binding of pemphigus IgG to recombinant Dsg1 and Dsg3 using commercial Dsg ELISA (Fig. 3). ELISA scores for PV-IgG and AK 23 assessed in the absence or presence of TP were not significantly different, indicating that TP did not block binding of pemphigus IgG to Dsg1 and Dsg3. Similarly, Dsg3 binding of AK23, which is directed against a putative adhesive domain of Dsg3, was not prevented by simultaneous TP incubation.
FIGURE 3.
TP did not block binding of pemphigus IgG to Dsg1 and Dsg3. ELISA experiments with recombinant Dsg3 (black bars) and Dsg1 (white bars) revealed that autoantibody scores of PV-IgG 1, PV-IgG 2, and AK 23 in the absence or presence of TP (20 μm) were not significantly different (p = 0.4040, p = 0.751, p = 0.2624, respectively), indicating that TP did not interfere with binding of pemphigus IgG to Dsg1 and Dsg3, respectively. (n = 3 for each condition).
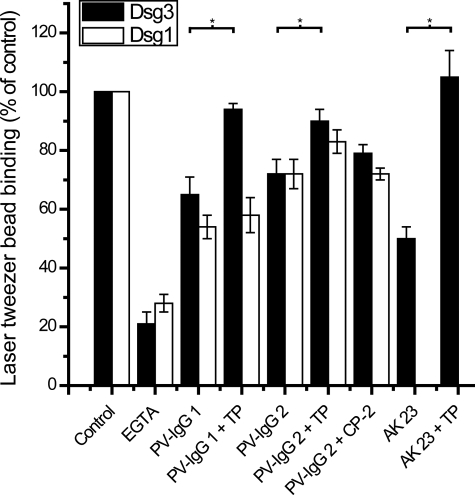
TP Prevented PV-IgG- and AK 23-induced Loss of Dsg3 Bead Binding on Keratinocytes—In the following, TP experiments were extended to laser tweezer experiments on cultured human keratinocytes (HaCaT) where binding of microbeads coated with recombinant Dsg3 (black bars) or Dsg1 (white bars) was probed by laser displacement (Fig. 4). The number of surface-bound Dsg3 and Dsg1 beads resisting laser beam displacement was reduced by simultaneous incubation with EGTA (5 mm, 30 min) to 21% ± 4% and 28% ± 3%, which demonstrated Ca2+-dependent bead binding. Similar to preceding studies (3, 4, 21, 22), incubation of monolayers with attached beads for 30 min with PV-IgG 1 and PV-IgG 2 caused significant loss of both Dsg3-coated and Dsg1-coated beads (Dsg3 bead values, 65% ± 6% and 72% ± 5%; Dsg1 bead values, 54% ± 4% and 72% ± 5%, respectively). PV-IgG-induced reduction in Dsg3 bead binding was significantly prevented by a 30-min preincubation of cells with TP at a concentration of 20 μm (94% ± 2% and 90% ± 4% for PV-IgG 1 and PV-IgG 2, respectively), consistent with cell-free Dsg3 AFM experiments. PV-IgG-mediated effects on Dsg bead binding was unaltered by preincubation with 20 μm CP-2, demonstrating the specificity of Dsg3 bead binding for TP. AK 23 resulted in reduced Dsg3 bead binding (50% ± 4%), an effect that was completely blocked by preincubation with TP as well (105% ± 9%). On the other hand, TP treatment was unable to prevent PV-IgG-mediated reduction of Dsg1 bead binding (values were 58% ± 6% and 83% ± 4% for PV-IgG 1 and PV-IgG 2). Because PV-IgG 1 did not directly inhibit Dsg1 transinteraction (Fig. 2), although containing Dsg1 autoantibodies, loss of Dsg1 binding in keratinocytes was likely caused by more indirect mechanisms such as induction of cellular signaling pathways that were not blocked by TP-mediated stabilization of Dsg transinteraction.
FIGURE 4.
In cell-based laser tweezer experiments, TP prevented PV-IgG- or AK 23-induced reduction of Dsg3 bead binding but not PV-IgG-mediated loss of Dsg1 bead binding. The numbers of HaCaT surface-bound Dsg3 (black bars) and Dsg1 beads (white bars) resisting laser beam displacement were reduced by incubation with EGTA (5 mm, 30 min). Incubation of monolayers with attached beads for 30 min with IgG fractions from PV patients significantly reduced the number of bound Dsg3 and Dsg1 beads. However, preincubation with TP (20 μm) for 30 min prevented AK 23-, PV-IgG 1-, and PV-IgG 2-induced reduction of Dsg3 (p = 0.00021, p = 0.01728, p = 0.03142, respectively) but not reduction of Dsg1 bead binding (p = 0.5512, p = 0.1311, for PV-IgG 1 and PV-IgG 2, respectively). Preincubation with CP-2 (20 μm) did not alter PV-IgG-mediated loss in Dsg3 (p = 0.3522) and Dsg1 bead binding (p = 0.9688) (n = 6 for each condition).
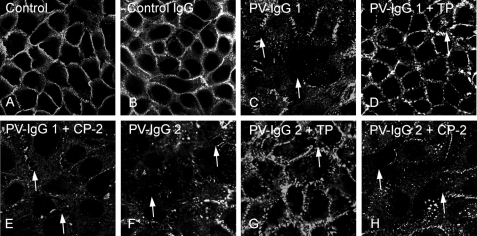
PV-mediated Acantholysis Was Partially Prevented by TP—Next, we evaluated whether TP would be protective against pemphigus IgG-mediated acantholysis in human keratinocytes using Dsg3 immunostaining to label desmosomes (Fig. 5). Following incubation with medium only (Fig. 5A) or control IgG (Fig. 5B), Dsg3 was detected at cell borders. Incubation with PV-IgG 1 and PV-IgG 2 resulted in gross disruption of Dsg3 staining, often accompanied by intercellular gap formation (Fig. 5, C and F, arrows). Intercellular gap formation was further substantiated by F-actin staining (data not shown) (22). PV-IgG-mediated Dsg3 disorganization was largely prevented by simultaneous incubation of HaCaT cells with TP at a concentration of 20 μm (Fig. 5, D and G). Under these conditions, Dsg3 was present at all segments of the cell membrane, but localization was discontinuous. Nevertheless, this protocol did not completely suppress intercellular gap formation (Fig. 5, D and G, arrows). Simultaneous incubation with CP-2 did not prevent PV-IgG-induced effects on keratinocytes (Fig. 5, E and H).
FIGURE 5.
TP treatment partially prevented PV-IgG-mediated disruption of Dsg3 localization in human keratinocytes. Dsg3 immunostaining of HaCaT keratinocytes revealed Dsg3 distribution along cell borders following incubation with medium (A) or control IgG (B). Incubation with PV-IgG 1 (C) and IgG 2 (F) resulted in disruption of Dsg3 staining and intercellular gap formation (arrows). PV-IgG-mediated Dsg3 disorganization was largely prevented by incubation of HaCaT cells with TP (20 μm, D and G) but not with CP-2 (20 μm, E and H). Intercellular gap formation was overall reduced but not completely abolished (arrows). Scale bar:20 μm for all panels. (n = 5).
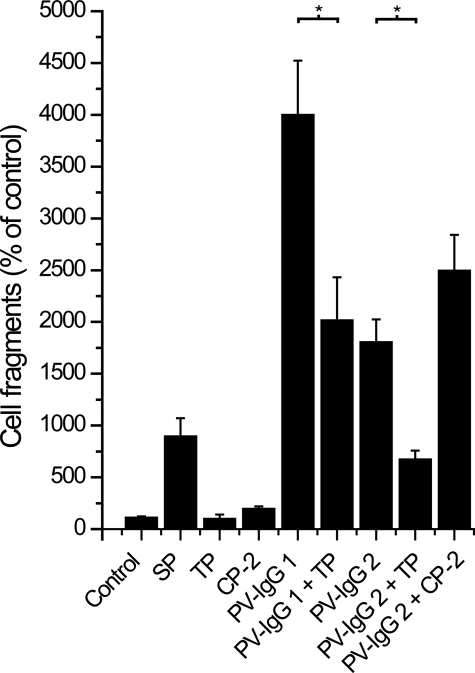
We quantified acantholytic effects of pemphigus IgG and peptide treatment using a dispase-based cell dissociation assay (Fig. 6). In these experiments, treatment of HaCaT cells for 24 h with 200 μm SP resulted in a significantly increased number of cell fragments when compared with controls (895% ± 175%), demonstrating that Dsg binding is crucial for keratinocyte adhesion. TP at 20 μm did not lead to enhanced cell dissociation (100% ± 40%), whereas 20 μm CP-2 treatment resulted in a slight but significant increase (195% ± 24%). PV-IgG 1 caused a strong increase in cell fragments (4000% ± 522%) that was partially blocked by cotreatment with 20 μm of TP (2017% ± 414%). Similar results were obtained with PV-IgG 2; PV-IgG 2 alone dramatically increased cell dissociation (1805% ± 219%), whereas incubation with TP but not CP-2 partially blocked these effects (673% ± 84% and 2494% ± 348%, respectively).
FIGURE 6.
TP treatment partially prevented PV-IgG-mediated acantholysis. In dispase-based keratinocyte dissociation assays, SP treatment caused significant cell dissociation. TP (20 μm) had no effect, whereas treatment with CP-2 (20 μm) slightly increased the number of cell fragments. PV-IgG 1 and PV-IgG 2 incubation alone caused strong acantholysis, which was partially blocked by cotreatment with 20 μm TP (p = 0.01515 and p = 0.00018, respectively). CP-2 had no effect on acantholysis caused by PV-IgG 2 (p = 0.1184). (n = 4).
DISCUSSION
The results of the present study demonstrate for the first time that direct inhibition of Dsg3 transinteraction significantly contributes to PV pathogenesis. This was shown by the use of peptides targeting the adhesive Dsg interface, which were effective to block both PV-IgG-induced direct inhibition of Dsg3 transinteraction and PV-IgG-induced acantholysis in human keratinocytes in vitro. This approach is the first to inhibit pathogenic effects of PV-IgG independent of cellular signaling pathways. Similar approaches using peptidomimetics may be applicable for treatment of pemphigus as well as of other autoantibody-mediated diseases where cell adhesion molecule function is impaired.
Modeling of Dsg sequences into structures of resolved classical cadherins identified a peptide sequence that was expected to block Dsg1 and Dsg3 transinteraction by occupying a critical and conserved Dsg binding site. AFM experiments demonstrated that the single peptide reduced homophilic transinteraction of both Dsg1 and Dsg3. This effect was sequence-specific because a comparable peptide from the equivalent region of VE-cadherin had no effect. In a dispase-based cell dissociation assay, SP caused significant acantholysis, indicating that the peptide reduced desmosomal adhesion in vitro. In the next step, a tandem peptide was generated that, when applied alone, did not interfere with Dsg1 and Dsg3 transinteraction. Nevertheless, preincubation with TP abolished PV-IgG-induced loss of Dsg3 transinteraction under cell-free conditions as probed by AFM, reduced loss of Dsg3-mediated bead binding measured with laser tweezers, and ameliorated autoantibody-induced acantholysis in keratinocytes. Again, specificity was demonstrated in parallel experiments by the lack of effects of a tandem VE-cadherin peptide. Moreover, although we cannot exclude the possibility that the peptides might bind to other (desmo-) cadherins, SP and TP did not have an effect on another cadherin family member, vascular endothelial cadherin, in comparable experiments.5 Taken together, these data provide a first hint that peptidomimetics with stabilizing effects on Dsg3 transinteraction similar to TP could be a novel therapeutic approach for PV treatment.
It has to be noted that direct interaction of the peptides with the adhesive interface of Dsg1 and Dsg3 was not shown in our study. Biacore studies using surface plasmon resonance were not successful because homophilic binding of Dsg-Fc constructs was too weak to be detected by this approach and specific binding of SP and TP to recombinant desmogleins could not be unequivocally demonstrated, most likely because affinity of peptides to recombinant Dsg molecules was rather low (not shown). This is in line with previous studies from the literature, which used recombinant proteins containing the first two extracellular domains of Dsg2 and Dsc2 only to study homophilic and heterophilic binding (23).
Because TP effectively abolished autoantibody-induced loss of Dsg3 transinteraction but did not significantly increase binding activity under cell-free conditions, these data raised the possibility that TP interfered with autoantibody binding to its target Dsg antigen, especially because recent structural analyses indicate that the SP sequence is located in an important immunological region of the Dsg1 and Dsg3 EC1 domain (19). However, ELISA measurements similar to those used to characterize the autoantibody profiles of the sera of patients demonstrated that TP did not affect binding of autoantibodies to Dsg1 and Dsg3. Similarly, binding of AK 23, which is directed against a putative adhesive domain of Dsg3, also was not prevented by simultaneous TP incubation in ELISA experiments. Therefore, a likely interpretation for the AFM experiments with TP alone is that the binding activities of Dsg1 and Dsg3 seem to be maximal under resting conditions and thus cannot be further enhanced by the peptide.
The second important outcome of the study is that direct inhibition of desmoglein transinteraction significantly contributes to pemphigus vulgaris acantholysis. This can be concluded because blocking of PV-IgG-mediated loss of Dsg3 transinteraction by TP was effective to reduce autoantibody-induced acantholysis in vitro. The hypothesis of autoantibody-mediated direct interference with Dsg binding was possible to believe because Dsg3 was discovered to be a cadherin-type cell adhesion molecule (8, 9). Studies using mouse monoclonal Dsg3 antibodies targeting well characterized parts of the Dsg3 extracellular subdomain supported this concept (3, 20). On the other hand, direct inhibitory effects of PV-IgG and PF-IgG on Dsg1 transinteraction were not detectable (3, 4), which led us to propose that direct interference with Dsg binding is specific for PV and may at least in part explain the more severe clinical phenotype of PV when compared with PF (1). Nevertheless, the recent study indicates that other more indirect mechanisms such as cellular signaling pathways or Dsg depletion also seem to be involved because TP did not completely block PV-IgG-induced acantholysis or PV-IgG-induced loss of Dsg1 bead binding. Indeed, accumulating evidence points to a crucial role of signaling mechanisms in pemphigus acantholysis including p38MAPK (24), RhoA (22), plakoglobin (18), c-Myc (25), cdk-2 (26), and tyrosine kinases such as c-Src (27, 28), which seem to be involved in Dsg3 endocytosis and depletion (29, 30). It is possible that some of these cellular pathways are activated as a consequence of direct Dsg inhibition (5). Moreover, we cannot rule out the possibility that TP also has different effects on heterophilic binding of Dsg1 and Dsg3 to other desmosomal cadherins such as desmocollin 3.
Acknowledgments
We thank Lisa Bergauer for excellent technical assistance.
This study was supported by grants from the Deutsche Forschungsgemeinschaft (Grants SFB 487, TP B5, and B2) and the IZKF Würzburg (Grant TP A-51). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PV, pemphigus vulgaris; PF, pemphigus foliaceus; AFM, atomic force microscopy; Dsg, desmoglein; ELISA, enzyme-linked immunosorbent assay; EC, extracellular domain; HaCaT, immortalized human keratinocyte cell line; IgG, immunoglobulins; SP, single peptide; TP, tandem peptide; CP, control peptide; VE-cadherin, vascular endothelial cadherin.
W.-M. Heupel, T. Müller, A. Efthymiadis, E. Schmidt, D. Drenckhahn, and J. Waschke, unpublished observations.
References
- 1.Waschke, J. (2008) Histochem. Cell Biol. 130 21-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanley, J. R., and Amagai, M. (2006) N. Engl. J. Med. 355 1800-1810 [DOI] [PubMed] [Google Scholar]
- 3.Heupel, W. M., Zillikens, D., Drenckhahn, D., and Waschke, J. (2008) J. Immunol. 181 1825-1834 [DOI] [PubMed] [Google Scholar]
- 4.Waschke, J., Bruggeman, P., Baumgartner, W., Zillikens, D., and Drenckhahn, D. (2005) J. Clin. Investig. 115 3157-3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma, P., Mao, X., and Payne, A. S. (2007) J. Dermatol. Sci. 48 1-14 [DOI] [PubMed] [Google Scholar]
- 6.Muller, E. J., Williamson, L., Kolly, C., and Suter, M. M. (2008) J. Investig. Dermatol. 128 501-516 [DOI] [PubMed] [Google Scholar]
- 7.Grando, S. A. (2006) Autoimmunity 39 521-530 [DOI] [PubMed] [Google Scholar]
- 8.Amagai, M., Klaus-Kovtun, V., and Stanley, J. R. (1991) Cell 67 869-877 [DOI] [PubMed] [Google Scholar]
- 9.Jones, J. C. R., Arnn, J., Staehelin, L. A., and Goldman, R. D. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 2781-2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro, L., Fannon, A. M., Kwong, P. D., Thompson, A., Lehmann, M. S., Grubel, G., Legrand, J. F., Als-Nielsen, J., Colman, D. R., and Hendrickson, W. A. (1995) Nature 374 327-337 [DOI] [PubMed] [Google Scholar]
- 11.Nagar, B., Overduin, M., Ikura, M., and Rini, J. M. (1996) Nature 380 360-364 [DOI] [PubMed] [Google Scholar]
- 12.Posy, S., Shapiro, L., and Honig, B. (2008) J. Mol. Biol. 378 952-966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsukasaki, Y., Kitamura, K., Shimizu, K., Iwane, A. H., Takai, Y., and Yanagida, T. (2007) J. Mol. Biol. 367 996-1006 [DOI] [PubMed] [Google Scholar]
- 14.He, W., Cowin, P., and Stokes, D. L. (2003) Science 302 109-113 [DOI] [PubMed] [Google Scholar]
- 15.Tselepis, C., Chidgey, M., North, A., and Garrod, D. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 8064-8069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams, G., Williams, E. J., and Doherty, P. (2002) J. Biol. Chem. 277 4361-4367 [DOI] [PubMed] [Google Scholar]
- 17.Ishii, K., Harada, R., Matsuo, I., Shirakata, Y., Hashimoto, K., and Amagai, M. (2005) J. Investig. Dermatol. 124 939-946 [DOI] [PubMed] [Google Scholar]
- 18.Caldelari, R., de Bruin, A., Baumann, D., Suter, M. M., Bierkamp, C., Balmer, V., and Muller, E. (2001) J. Cell Biol. 153 823-834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong, J. C., and Sinha, A. A. (2008) BMC Immunol. 9 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsunoda, K., Ota, T., Aoki, M., Yamada, T., Nagai, T., Nakagawa, T., Koyasu, S., Nishikawa, T., and Amagai, M. (2003) J. Immunol. 170 2170-2178 [DOI] [PubMed] [Google Scholar]
- 21.Spindler, V., Drenckhahn, D., Zillikens, D., and Waschke, J. (2007) Am. J. Pathol. 171 906-916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waschke, J., Spindler, V., Bruggeman, P., Zillikens, D., Schmidt, G., and Drenckhahn, D. (2006) J. Cell Biol. 175 721-727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Syed, S. E., Trinnaman, B., Martin, S., Major, S., Hutchinson, J., and Magee, A. I. (2002) Biochem. J. 362 317-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berkowitz, P., Hu, P., Warren, S., Liu, Z., Diaz, L. A., and Rubenstein, D. S. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 12855-12860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williamson, L., Raess, N. A., Caldelari, R., Zakher, A., de Bruin, A., Posthaus, H., Bolli, R., Hunziker, T., Suter, M. M., and Mller, E. J. (2006) EMBO J. 25 3298-3309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanza, A., Cirillo, N., Rossiello, R., Rienzo, M., Cutillo, L., Casamassimi, A., de Nigris, F., Schiano, C., Rossiello, L., Femiano, F., Gombos, F., and Napoli, C. (2008) J. Biol. Chem. 283 8736-8745 [DOI] [PubMed] [Google Scholar]
- 27.Chernyavsky, A. I., Arredondo, J., Kitajima, Y., Sato-Nagai, M., and Grando, S. A. (2007) J. Biol. Chem. 282 13804-13812 [DOI] [PubMed] [Google Scholar]
- 28.Delva, E., Jennings, J. M., Calkins, C. C., Kottke, M. D., Faundez, V., and Kowalczyk, A. P. (2008) J. Biol. Chem. 283 18303-18313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calkins, C. C., Setzer, S. V., Jennings, J. M., Summers, S., Tsunoda, K., Amagai, M., and Kowalczyk, A. P. (2006) J. Biol. Chem. 281 7623-7634 [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto, Y., Aoyama, Y., Shu, E., Tsunoda, K., Amagai, M., and Kitajima, Y. (2007) J. Biol. Chem. 282 17866-17876 [DOI] [PubMed] [Google Scholar]