Abstract
We have determined the kinetics of ilicicolin binding and dissociation at center N of the yeast bc1 complex and its effect on the reduction of cytochrome b with center P blocked. The addition of ilicicolin to the oxidized complex resulted in a non-linear inhibition of the extent of cytochrome b reduction by quinol together with a shift of the reduced bH heme spectrum, indicating electron transfer between monomers. The possibility of a fast exchange of ilicicolin between center N sites was excluded in two ways. First, kinetic modeling showed that fast movement of an inhibitor between monomers would result in a linear inhibition of the extent of cytochrome b reduction through center N. Second, we determined a very slow dissociation rate for ilicicolin (k = 1.2 × 10-3 s-1) as calculated from its displacement by antimycin. Ilicicolin binding to the reduced bc1 complex occurred in a single phase (kon = 1.5–1.7 × 105 m-1 s-1) except in the presence of stigmatellin, where a second slower binding phase comprising ∼50% of the spectral change was observed. This second kinetic event was weakly dependent on ilicicolin concentration, which suggests that binding of ilicicolin to one center N in the dimer transmits a slow (k = 2–3 s-1) conformational change that allows binding of the inhibitor in the other monomer. These results, together with the evidence for intermonomeric electron transfer, provide further support for a dimeric model of regulatory interactions between center P and center N sites in the bc1 complex.
The cytochrome bc1 complex is a multisubunit enzyme that generates an electrochemical gradient across the inner mitochondrial or bacterial membrane by transferring electrons from QH22 to cytochrome c. Structurally, the bc1 complex is a dimer of 9–11 subunits, with redox groups in cytochrome b, the Rieske iron-sulfur protein, and cytochrome c1 (1–4). The binding sites for QH2 and Q, termed center P (or Qo site) and center N (or Qi site), are present at opposite sites of each cytochrome b subunit, close to the bL and bH hemes, respectively (Fig. 1). The two bL hemes in the dimer are, depending on the organism, within 13–14 Å of each other, a distance that should theoretically allow electron transfer rates of at least 104 s-1 between the two redox groups (5). The Rieske protein interacts with both monomers by traversing the membrane in a tilted angle from the vicinity of center N of one monomer to center P of the other monomer, where its movable extrinsic domain shuttles one electron at a time from QH2 to cytochrome c1.
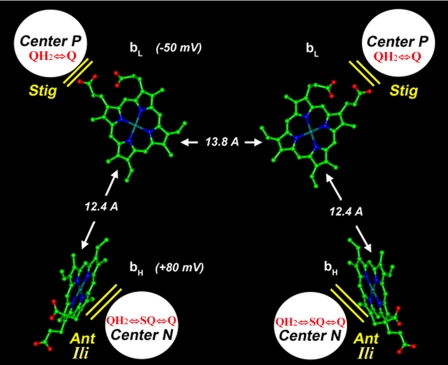
FIGURE 1.
Location of QH2/Q binding sites and b hemes in the yeast bc1 complex dimer. Edge-to-edge distances between heme tetra-pyrrole rings are indicated by arrows along with the heme redox midpoint potentials as measured in the isolated yeast bc1 complex. The approximate locations of center P and center N are also shown along with the redox reactions occurring in each site and their respective inhibitors: stigmatellin (Stig), antimycin (Ant), and ilicicolin (Ili). The structure was constructed from coordinates of the yeast bc1 complex, Protein Data Bank (PDB) code 1EZV (3).
We have previously provided experimental evidence of the functional importance of the dimeric structure of the bc1 complex (reviewed in Ref. 6). Our results have indicated that only one center P in the dimer is able to oxidize QH2 when both center N sites are occupied by the tightly bound inhibitor antimycin but that electrons are able to reduce both bH hemes, implying electron crossover at the level of the bL hemes (7). We obtained more direct evidence for intermonomeric electron equilibration by blocking both center P sites and titrating cytochrome b reduction by QH2 through center N with antimycin (8). In these experiments, non-linear inhibition by antimycin was obtained, and electrons were observed reaching the bH heme where antimycin had been bound before the addition of QH2. Assuming that antimycin did not dissociate from center N during the time scale of the experiments (<1 s), we interpreted these results as proof of electron equilibration between center N sites via the bL hemes. However, determination of dissociation constants for center N ligands is required to definitively discard alternative models that attempt to explain nonlinear titration curves in terms of fast exchange of inhibitor molecules between center N sites (9, 10).
In the present work, we have analyzed the binding and dissociation kinetics of ilicicolin, a center N inhibitor with a lower affinity than antimycin but that binds almost stoichiometrically to the yeast bc1 complex in the μm range (11). We show that the non-linear inhibition of cytochrome b reduction and the bH heme spectral shift induced by ilicicolin cannot be attributed to a fast exchange of the inhibitor between center N sites but instead reveals intermonomeric electron equilibration. Furthermore, we show that binding of ilicicolin is also sensitive to the center P occupants in a manner that indicates a dimeric regulation of the bc1 complex based on the position of the extrinsic domain of the Rieske protein. These results support a dimeric mechanism of half-of-the-sites regulation of the bc1 complex that involves non-rate-limiting electron movement between cytochrome b subunits (12).
EXPERIMENTAL PROCEDURES
Materials—Dodecylmaltoside was obtained from Anatrace. DEAE-Bio Gel A was from Bio-Rad Laboratories. Stigmatellin, myxothiazol, antimycin, and decylubiquinone (2,3-dimethoxy-5-methyl-6-decyl-1,4-benzoquinone) were purchased from Sigma. Ilicicolin was obtained from the Merck sample repository. DBH2 was prepared from decylubiquinone as described before (13) and quantified by UV spectroscopy using an extinction coefficient of 4.14 mm-1 cm-1 at 290 nm (14). Inhibitors were dissolved in ethanol and quantified by UV spectroscopy (15) using extinction coefficients of 4.8 mm-1 cm-1 at 320 nm for antimycin, 65.5 mm-1 cm-1 at 267 nm for stigmatellin, 10.5 mm-1 cm-1 at 313 nm for myxothiazol, and 23.2 mm-1 cm-1 at 248 nm for ilicicolin (11, 16).
Purification of Cytochrome bc1 Complex—Wild-type cytochrome bc1 complex was isolated from Red Star cake yeast as described previously (17), except that dodecylmaltoside concentration was increased to 0.05% in the elution buffers and the volume of DEAE-Bio Gel A was reduced to 25 ml to increase the yield of active enzyme. Quantification of the bc1 complex was performed as described before (18) using extinction coefficients of 17.5 mm-1 cm-1 at 553–539 nm for cytochrome c1 (19) and 25.6 mm-1 cm-1 at 563–579 nm for the average absorbance of the bH and bL hemes in cytochrome b (20).
Inhibition of the Pre-steady State Reduction of the bH Heme and Determination of Spectral Shift by Ilicicolin—Pre-steady state reduction of cytochrome b was followed at room temperature by stopped flow rapid scanning spectroscopy using the OLIS rapid scanning monochromator as described before (18). For these experiments, 3 μm bc1 complex was incubated with 3.6 μm stigmatellin and the indicated concentration of ilicicolin for 5 min in assay buffer containing 50 mm potassium phosphate, pH 7.0, 2 mm sodium azide, 0.2 mm EDTA, and 0.05% Tween 20. The reaction was started by rapid mixing against an equal volume of the same buffer containing 30 μm DBH2. For each experiment, 8–10 data sets were averaged after subtracting the oxidized spectrum. The time course of absorbance changes at 563–579 nm were extracted using software from OLIS and exported to the Origin 5.0 program (OriginLab Corp.).
Spectra from each kinetic trace collected between 0.2 and 0.3 s were averaged using the OLIS software and exported to Origin. To determine spectral shift displacements induced by ilicicolin bound before the addition of DBH2, the average spectrum collected in the absence of ilicicolin was normalized to create a reference spectrum that was subtracted from those collected in the presence of different ilicicolin concentrations. The normalization procedure has been described previously (8) and involves converting to zero the absorbance of each spectrum at two reference wavelengths and then equalizing the maximum absorbance for the bH heme in the reference spectrum to that of each spectrum collected with ilicicolin. In the present work, we decided to use the isosbestic points at 516 and 569 nm as reference wavelengths instead of 539 and 579 nm, which are minimum absorbance values for cytochrome c1 and b, respectively, resulting in more symmetrical ilicicolin-induced spectral shifts than those reported before in the presence of antimycin (8).
Kinetic Modeling—Cytochrome b reduction was simulated using the Dynafit program (Biokin Ltd.), which allows the generation of time-dependent data according to different reaction mechanisms described as a series of kinetic steps (21). The script files describing the mechanisms used for simulations are provided as supplemental data. In all models, an extinction coefficient of 36 mm-1 cm-1 was assumed for bH reduction based on a 70% contribution of this heme to the total absorbance of cytochrome b (22).
All models assumed equilibration of the oxidized dimer (E) with ilicicolin (I) to form three possible complexes (E·I, I·E, and I·E·I). In the model that allowed intermonomeric electron transfer, association (konI) and dissociation rate constants (koffI) of 0.15 × 106 m-1 s-1 and 1 × 10-3 s-1, respectively, were used for ilicicolin based on experimental values we report in this work. QH2 binding and oxidation at center N resulted in reduction of the bH heme and formation of SQ either at half of the dimer (symbolized as SQ·bHE, EbH·SQ) or at both monomers (SQ·bHEbH·SQ), where the bH symbol at either side of the dimer (E) stands for a reduced bH heme. In complexes with an oxidized bH (SQ·bHE and EbH·SQ), the electron was allowed to equilibrate with the other monomer (to form SQ·EbH or bHE·SQ) with a rate that was varied in the simulations. SQ was then allowed to reduce the bH heme that was left oxidized due to the intermonomeric electron transfer to form complexes in which the two bH hemes were in the reduced state and Q was bound at either center N site (Q·bHEbH or bHEbH·Q). Rates for the individual reaction steps are included in the supplemental data file scripts.
A second model used the same values for konI and koffI as above but assumed different rates of inhibitor exchange between monomers, resulting in interconversion between E·I and I·E. Because no intermonomeric electron transfer was assumed in this model, the only pathway for reduction of a bH heme was by QH2 oxidation to form SQ (SQ·bHE, EbH·SQ and SQ·bHEbH·SQ). A final model did not allow electron transfer or inhibitor exchange between monomers but assumed different higher values for the dissociation rate of the inhibitor (koffI) while maintaining the koffI/konI ratio to preserve the same overall binding affinity.
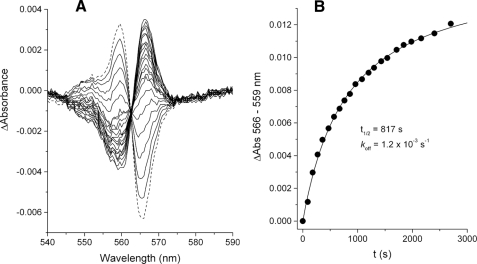
Determination of the Dissociation Rate of Ilicicolin from Center N—The rate of displacement of ilicicolin from center N by antimycin was measured spectroscopically in an Aminco DW-2a spectrophotometer. The dithionite-reduced spectrum of 4 μm bc1 complex was collected between 520 and 640 nm and stored as a baseline before the addition of 10 μm ilicicolin and incubation for 5 min. The spectrum with ilicicolin was collected and stored and was considered as representing time 0. After the addition of 10 μm antimycin, spectra were collected every 2 min. The difference in absorbance between 566 and 559 nm, corresponding to the antimycin-induced spectral shift of the bH heme, was plotted relative to the time 0 spectrum and fitted to a monophasic exponential function in Origin.
Kinetics of Inhibitor Binding to the bc1 Complex—Purified bc1 complex was diluted to 4 μm in the same buffer used for pre-steady state experiments, reduced with dithionite, and mixed rapidly against an equal volume of buffer containing different concentrations of ilicicolin. Where indicated, 5 μm myxothiazol or stigmatellin or different concentrations of DBH2 were added to the enzyme 2 min before rapid mixing. The fully reduced spectrum before mixing was stored as a baseline and subtracted from all subsequent scans, 8–10 data sets were averaged, and the time course of absorbance change at selected wavelengths was exported to the Origin program. The difference in absorbance between the peak (560 nm) and trough (566 nm) of the spectral shift induced by ilicicolin binding was plotted and fitted to monophasic or biphasic exponential functions.
RESULTS
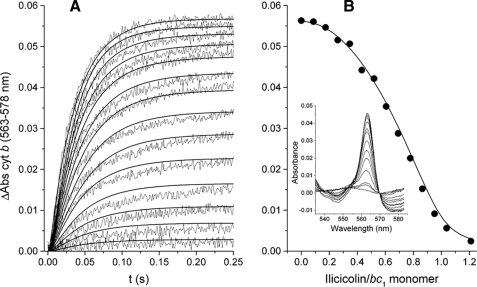
Non-linear Inhibition of Cytochrome b Reduction by Ilicicolin—The pre-steady state reduction of the stigmatellin-bound bc1 complex by DBH2 at different ilicicolin concentrations is shown in Fig. 2. Cytochrome b reduction kinetics were compared with a model that assumed non-rate-limiting intermonomeric electron equilibration (see supplemental data for details), which was able to reproduce the different increase in inhibition at low and high ilicicolin concentrations (Fig. 2A, solid curves). This non-linear inhibition in the extent of cytochrome b reduction (Fig. 2B, circles) was simulated by assuming that inhibition of one center N site per dimer by ilicicolin still allowed both monomers to be reduced to the same extent as dimers with no inhibitor (Fig. 2B, solid curve). The lack of contribution of center P to cytochrome b reduction in the presence of stigmatellin during the time scale of the assay was indicated by the lack of cytochrome c1 reduction (Fig. 2B, inset).
FIGURE 2.
Inhibition of the pre-steady state reduction of cytochrome b (cyt b) through center N by ilicicolin. A shows the reduction traces of 1.5 μm stigmatellin-inhibited cytochrome bc1 complex by 15 μm DBH2 preincubated in the presence of increasing ilicicolin concentrations (from 0 to 1.2 ilicicolin/bc1 monomer in 0.085 intervals). Solid curves correspond to the simulated kinetics at each ilicicolin concentration assuming intermonomeric electron transfer between center N sites at a rate of 500 s-1 (see supplemental data for details). The maximum extent of reduction at each ilicicolin concentration is shown in B (solid circles) taken from the spectra collected at 0.25 s (inset). The solid curve represents the expected extent of cytochrome b reduction assuming that dimers with only one ilicicolin bound contribute to the total absorbance as much as dimers with no ilicicolin. Enzyme-inhibitor complexes were calculated using the equilibration model described in the supplemental data assuming association and dissociation rates for ilicicolin of konI = 1.5 × 105 m-1 s-1 and koffI = 1 × 10-3 s-1, respectively.
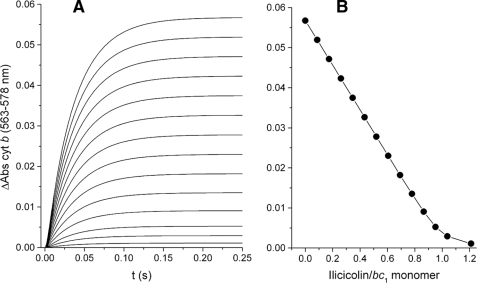
Unimpeded electron equilibration of both cytochrome b subunits through only one center N site per dimer is consistent with fast electron transfer between monomers. However, an alternative model (9) that is still invoked to explain non-linear inhibition curves in the bc1 complex (10) proposes the existence of fast intermonomeric movement of tightly bound inhibitors between center N sites. As shown by the simulation in Fig. 3, such a mechanism would result in linear inhibition of cytochrome b reduction by a center N inhibitor. Interestingly, the same kinetic pattern was obtained irrespectively of the value assigned to the rate of the hypothetical intermonomeric inhibitor exchange (not shown), as long as both center N sites are assumed to be simultaneously active. Because movement of an inhibitor between monomers would still result in the same fraction of inhibited center N sites, linear inhibition curves would always be expected in such a mechanism. A variant of this model in which the dissociation rate (koffI) for the center N inhibitor was increased while maintaining the koff/kon ratio constant also yielded a linear decrease in the extent of cytochrome b reduction together with a loss of stoichiometric binding (see supplemental Fig. S1).
FIGURE 3.
Simulation of the inhibition of pre-steady state reduction by an inhibitor that moves between center N sites. Simulated kinetic traces at the same concentrations of ilicicolin as in Fig. 2 were obtained assuming a rate of inhibitor movement of 500 s-1 between the two active sites in the dimer and no intermonomeric electron transfer (A). Identical curves were obtained with rates ranging from 0 to 50,000 s-1 (see supplemental data for details on the model). cyt b, cytochrome b. B shows the extent of cytochrome b reduction obtained at 0.25 s for each of the simulated reduction kinetic curves, resulting in a linear inhibition pattern.
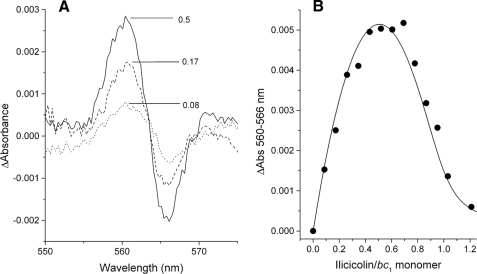
Reduction of the bH Heme in Ilicicolin-blocked Center N Sites—Ilicicolin bound at center N induces a blue shift in the absorbance maximum of the reduced bH heme (11). However, if substoichiometric concentrations of ilicicolin are bound to the oxidized and center P-blocked bc1 complex before the addition of DBH2, no spectral shift is expected to occur unless electrons can equilibrate into the bH heme of the ilicicolin-bound center N from the uninhibited monomer. As shown in Fig. 4, such a shift was observed within the first 300 ms after the addition of DBH2 (Fig. 4A). The amplitude of the ilicicolin-induced shift corresponded to the expected proportion of dimers with one inhibitor bound per dimer (Fig. 4B). This observation indicates that electrons entering the dimer through the uninhibited center N can reach the bH heme in the opposite monomer in a few hundred ms, unless ilicicolin is assumed to move within this time scale from an oxidized to a reduced center N site. This last possibility is in conflict with the kinetic analysis of Figs. 2 and 3 (see above) and also requires the doubtful assumption that the stable SQ formed in the vicinity of the reduced bH heme can dissociate rapidly to allow ilicicolin binding (8).
FIGURE 4.
Ilicicolin-induced spectral shift of the reduced bH heme absorbance upon reduction through the uninhibited center N site in the dimer. A shows the difference between the averaged spectra collected between 0.2 and 0.3 s at the indicated ilicicolin/bc1 monomer ratios and the average and normalized spectra collected in the absence of ilicicolin. B shows the maximal amplitude of the spectral shift induced by ilicicolin at different inhibitor/bc1 monomer ratios. The solid curve represents the relative change in concentration of dimers with only one ilicicolin bound at a center N site obtained at different inhibitor/enzyme ratios. Enzyme-inhibitor complexes were calculated using the equilibration model described in the supplemental data assuming association and dissociation rates for ilicicolin of konI = 1.5 × 105 m-1 s-1 and koffI = 1 × 10-3 s-1, respectively.
Slow Dissociation of Ilicicolin from Center N—To examine the possibility of fast ilicicolin dissociation from center N and reequilibration in the dimer, the dissociation rate of this inhibitor was determined by measuring the rate of its displacement by antimycin, which binds more tightly to center N and generates a distinct red shift in the spectrum of reduced bH heme (16). Antimycin binds to available center N sites within a few hundred ms when added at μm concentrations (12). As shown in Fig. 5A, when ilicicolin was already bound to center N, the antimycin-induced red shift occurred simultaneously with the disappearance of the blue shift caused by bound ilicicolin. The time scale of the displacement by antimycin was in the order of tens of minutes (Fig. 5B), yielding a dissociation rate of (koffI = 1.2 × 10-3 s) for ilicicolin. This value was unmodified by the presence of center P inhibitors during the displacement assay or by varying the concentrations of ilicicolin and antimycin (data not shown), indicating that antimycin and ilicicolin are competing only for center N and not for some unspecific hydrophobic site in the enzyme or in the detergent micelles to which the inhibitors might need to bind before gaining access to center N. These results conclusively exclude the possibility of fast movement of ilicicolin between center N sites because this would require a fast dissociation rate from center N that should have resulted in a much faster replacement by antimycin than was experimentally observed.
FIGURE 5.
Rate of ilicicolin displacement from center N by antimycin. A shows the spectral shift generated after the addition of 10 μm ilicicolin to 4 μm fully reduced bc1 complex and incubation for 5 min (dashed spectrum). 10 μm antimycin was then added, and subsequent spectra (solid traces) were collected every 2 min, showing the simultaneous loss of the blue shift and appearance of a red shift, which indicate ilicicolin dissociation and antimycin binding, respectively. The time course of the spectral red shift induced by the binding of antimycin is shown in B. The solid curve corresponds to the fitting of the data to a one-exponential function, which yielded the indicated rate constant for ilicicolin dissociation.
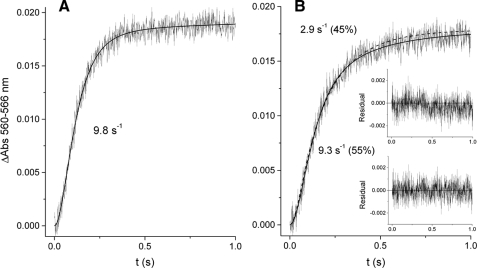
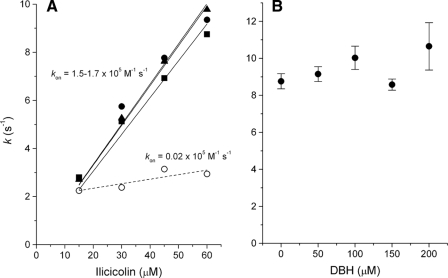
Binding Kinetics of Ilicicolin to Center N—The time course of ilicicolin binding to the vicinity of the bH heme showed monophasic kinetics in the presence of myxothiazol (Fig. 6A) or in the absence of center P inhibitors (not shown). In contrast, stigmatellin induced a change in the kinetics of ilicicolin binding that resulted in two kinetic phases, each comprising approximately half of the spectral shift amplitude (Fig. 6B). The first phase had a rate similar to that observed when center P was unoccupied or bound with myxothiazol. As shown in Fig. 7A, this fast phase was dependent on ilicicolin concentration, indicating that it was limited by the diffusion of the inhibitor to center N. However, the second slower phase observed only in the presence of stigmatellin was very weakly dependent on the concentration of ilicicolin, suggesting that a relatively slow change in conformation at half of the center N sites was limiting binding of the inhibitor.
FIGURE 6.
Binding kinetics of ilicicolin to center N. The time-dependent increase in the amplitude of the blue shift generated by rapid mixing of 60 μm ilicicolin with 4 μm dithionite-reduced bc1 complex was determined after equilibrating the enzyme with myxothiazol (A) or stigmatellin (B). The best fit to either a single exponential (A) or a double exponential (B) function is shown as solid curves. A single exponential function in B (dashed curve) resulted in a worse fit as determined by comparing the residual plots (inset) resulting from the single (top) and double (bottom) exponential fits. Numbers in parentheses in B indicate the relative absorbance change attributable to each kinetic phase obtained from the double exponential fitting.
FIGURE 7.
Ilicicolin binding rates at different inhibitor or DBH2 concentrations. In A, the rates of ilicicolin binding obtained in the absence of center P inhibitor (squares) with myxothiazol (triangles), as well as the initial rate in the presence of stigmatellin (solid circles), were fitted to a linear regression (solid lines) to obtain the binding rate constant for ilicicolin (kon). The slower ilicicolin binding rates obtained with stigmatellin at center P(open circles) showed a very weak concentration dependence, as shown by the very low apparent kon value obtained by a linear fit (dashed line). B shows the binding rate of 60 μm ilicicolin to center N in the presence of different DBH2 concentrations and in the absence of center P inhibitors.
The fast phase of ilicicolin binding to the reduced bc1 complex was insensitive to the presence of DBH2 (Fig. 7B), whereas the slow phase observed in the presence of stigmatellin disappeared at the lowest concentrations of DBH2 assayed, rendering the kinetics of ilicicolin binding monophasic (data not shown). As we have shown previously for antimycin binding (8), these results suggest a very weak affinity for DBH2 in the reduced complex that prevents competition with binding of center N inhibitors. However, transient binding of DBH2 before mixing with the inhibitor was apparently able to trigger the conformational change that allowed all center N sites to bind ligands in a single phase.
DISCUSSION
Ilicicolin is an antibiotic that exerts an inhibitory effect at center N of the bc1 complex in a manner that differs in certain respects from that of antimycin (11). The most notable difference is the opposite direction of the spectral shift of the bH heme absorbance induced by ilicicolin with respect to antimycin, which indicates a different effect on the electronic environment of the heme macrocycle. Another difference is the affinity of these two ligands, with antimycin (Kd ∼ 10 pm, according to Ref. 16) binding much tighter to most bc1 complexes than ilicicolin, which inhibits the activity of the yeast and bovine bc1 complexes with an IC50 of 3–12 and 200–250 nm, respectively (11, 23) and very poorly in the case of the bacterial enzyme (24). The mutations that confer resistance to both inhibitors are also different (23, 24), indicating that several of the residues that interact with each of these molecules are not the same, and that might possibly contribute to the weaker binding of ilicicolin. Nevertheless, the fact that both inhibitors displace the stable SQ formed at center N (25) and that antimycin displaces ilicicolin (Fig. 5) indicates that their binding sites are overlapping.
In a previous work, we showed that incubation with increasing concentrations of antimycin resulted in cytochrome b reduction kinetics through center N that could not be fitted to a model of fast inhibitor movement (8). In the present work, we have shown that the non-linear inhibition pattern in the extent of cytochrome b reduction upon titration with ilicicolin, which we also observed previously with antimycin, cannot be reproduced by assuming fast exchange of inhibitors (Figs. 2 and 3). Careful examination of the original work where such a model was first proposed (9) reveals that non-linear curves are expected only if active sites in the dimer are assumed to function in an alternating fashion so that a single movable inhibitor molecule always shifts to the site that is already inactive and therefore does not inhibit the overall reaction. Our kinetic modeling shows that if the two sites in the dimer are assumed to be active from the outset, fast or slow inhibitor movement becomes irrelevant in terms of the fraction of inhibited sites in the population of enzyme (Fig. 3). Other authors have claimed that non-linear titration curves can be generated if the inhibitor blocks a non-rate-limiting step in the reaction (10). That argument is not applicable to our present results because the inhibitor used directly blocks the only reaction measured, that is, cytochrome b reduction by QH2 at center N. Therefore, the only mechanism that can explain the kinetic pattern of inhibition by center N inhibitors is intermonomeric electron transfer (Fig. 2).
As we have discussed elsewhere (6), electron equilibration from one center N to the other in the dimer is expected to occur in <20 ms based on the distance between heme groups and electron tunneling calculations. Recently, it has been argued that bL to bL electron transfer does not exist based on the linear inhibition by myxothiazol of cytochrome b reduction through center P, and poorly characterized interference effects were invoked in an attempt to justify how two heme groups at such a close distance from each other do not share electrons across the dimer interface (10). However, it is difficult to conceive a logical reason for natural selection to conserve a close distance between the bL hemes across the phylogenetic scale without taking advantage of the beneficial effects that electron equilibration would have in maintaining cytochrome b maximally oxidized (8). Furthermore, we have already explained why myxothiazol inhibition is in fact expected to be linear based on the reported half-of-the-sites activity of the center P sites in the dimer (6), rendering such titration curves irrelevant in terms of proving or disproving intermonomeric electron transfer.
We previously reported that one antimycin molecule bound to oxidized dimers was able to generate a red shift in the reduced bH heme absorbance upon the addition of QH2 (8), indicating that electrons could rapidly equilibrate from the uninhibited center N to the heme in the blocked monomer. The reasonable assumption was made that antimycin could not dissociate from the oxidized center N to bind to a reduced site. However, we did not experimentally determine the dissociation rate for antimycin at that time. In the present work, we show that ilicicolin also induces a spectral shift in those dimers where one inhibitor was bound before the addition of QH2 (Fig. 4). More importantly, we experimentally demonstrate that this effect is not due to a fast dissociation of ilicicolin from center N (Fig. 5). A very slow dissociation rate was also reported (although no numerical value was provided) for funiculosin (26), another center N inhibitor with an affinity similar to that of ilicicolin (24). Thus, ilicicolin can still be defined as a tightly bound or pseudo-irreversible inhibitor in the case of the yeast bc1 complex, mainly due to its very slow dissociation rate from center N, which was found to be in the range of tens of minutes.
The values we have obtained for the ilicicolin association (konI = 1.5–1.7 × 105 m-1 s-1, Fig. 7A) and dissociation rate constants (koffI = 1.2 × 10-3 s-1, Fig. 5) allow us to calculate a Kd of 7–8 nm, which is within the range of IC50 values reported before from activity assays using yeast mitochondrial membranes or isolated enzyme (11). There is consequently an ∼1000-fold difference in the Kd of antimycin and ilicicolin. Because the kon value we have previously reported for antimycin (12) is only five times higher than the present value for ilicicolin, we conclude that most of the difference in affinity between the two inhibitors is attributable to the dissociation rate, which can thus be estimated to be 200 times lower for antimycin or close to 6 × 10-6 s-1. This means that antimycin dissociates from center N in a time scale of days, and not in ms, as implied by models that have proposed fast movement of inhibitors between center N sites within (or even between) dimers (9). Consequently, our previous and present results regarding the spectral shift induced by antimycin or ilicicolin bound to the oxidized enzyme provide direct evidence of intermonomeric electron equilibration between cytochrome b subunits.
Electron movement between monomers in the bc1 complex is especially relevant under physiological conditions in which the mitochondrial transmembrane potential favors electron occupancy at the bL hemes by decreasing the effective potential difference with respect to the bH hemes (27). As discussed before (8, 28), intradimeric electron equilibration would also aid in maintaining forward electron flow at higher QH2/Q ratios that might exist under pathological conditions, such as ischemia (29). Dimeric functioning of the bc1 complex under these conditions would minimize inhibition of cytochrome b oxidation and subsequent electron leakage to oxygen at center P caused by an excess of QH2 binding at center N.
Further evidence for the functional relevance of the dimeric structure of the bc1 complex comes from the effect that the position of the Rieske protein has on half of the center N sites as evidenced by the biphasic binding of ilicicolin in the presence of stigmatellin (Fig. 6). We have previously discussed similar results with antimycin to propose a model in which the simultaneous location of the two Rieske protein peripheral domains close to the center P sites in the dimer delays or transiently impedes SQ stability at one center N (12). This proposed mechanism was based on the concentration independence of the second phase of antimycin binding, which we interpreted as reflecting a conformational change that was transmitted from one center N site to the other upon initial binding of antimycin to one monomer. Interestingly, the rate of the concentration-independent binding event we have now obtained with ilicicolin (Fig. 7A) is identical to that we determined with antimycin (12), although the diffusion-limited initial phase is different by a factor of ∼5. This supports our interpretation that binding of any tight ligand to the second center N site is limited by the same relatively slow conformational change. The poor binding of DBH2 to center N in the reduced enzyme as evidenced by the lack of its effect on antimycin (12) and ilicicolin (Fig. 7B) binding rates is consistent with other kinetic results that suggest that center N binds QH2 preferentially when the bH heme is oxidized and Q when the heme is reduced, favoring the formation of SQ·bH3+ complexes that maintain cytochrome b favorably poised to accept electrons from center P (25). Therefore, we conclude that the binding properties of tight inhibitors at center N resemble those of the stabilized SQ, evidencing the regulatory interactions between center P and center N sites that impede simultaneous activity of the four QH2/Q binding sites in the bc1 complex dimer to promote optimal electron flow (6).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant GM 20379. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains Dynafit script files, supplemental text, and a supplemental figure.
Footnotes
The abbreviations used are: QH2, quinol; Q, quinone; SQ, semiquinone; DBH2, decylubiquinol (2,3-dimethoxy-5-methyl-6-decyl-1,4-benzoquinol); E, oxidized dimer; I, ilicicolin.
References
- 1.Xia, D., Yu, C. A., Kim, H., Xian, J. Z., Kachurin, A. M., Zhang, L., Yu, L., and Deisenhofer, J. (1997) Science 277 60-66 [DOI] [PubMed] [Google Scholar]
- 2.Zhang, Z. L., Huang, L. S., Shulmeister, V. M., Chi, Y. I., Kim, K. K., Hung, L. W., Crofts, A. R., Berry, E. A., and Kim, S. H. (1998) Nature 392 677-684 [DOI] [PubMed] [Google Scholar]
- 3.Hunte, C., Koepke, J., Lange, C., Rossmanith, T., and Michel, H. (2000) Structure (Camb.) 8 669-684 [DOI] [PubMed] [Google Scholar]
- 4.Berry, E. A., Huang, L. S., Saechao, L. K., Pon, N. G., Valkova-Valchanova, M., and Daldal, F. (2004) Photosynth. Res. 81 251-275 [DOI] [PubMed] [Google Scholar]
- 5.Osyczka, A., Moser, C. C., Daldal, F., and Dutton, P. L. (2004) Nature 427 607-612 [DOI] [PubMed] [Google Scholar]
- 6.Covian, R., and Trumpower, B. L. (2008) Biochim. Biophys. Acta 1777 1079-1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covian, R., Gutierrez-Cirlos, E. B., and Trumpower, B. L. (2004) J. Biol. Chem. 279 15040-15049 [DOI] [PubMed] [Google Scholar]
- 8.Covian, R., and Trumpower, B. L. (2005) J. Biol. Chem. 280 22732-22740 [DOI] [PubMed] [Google Scholar]
- 9.Bechmann, G., Weiss, H., and Rich, P. R. (1992) Eur. J. Biochem. 208 315-325 [DOI] [PubMed] [Google Scholar]
- 10.Crofts, A. R., Holland, J. T., Victoria, D., Kolling, D. R., Dikanov, S. A., Gilbreth, R., Lhee, S., Kuras, R., and Kuras, M. G. (2008) Biochim. Biophys. Acta 1777 1001-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutierrez-Cirlos, E. B., Merbitz-Zahradnik, T., and Trumpower, B. L. (2004) J. Biol. Chem. 279 8708-8714 [DOI] [PubMed] [Google Scholar]
- 12.Covian, R., and Trumpower, B. L. (2006) J. Biol. Chem. 281 30925-30932 [DOI] [PubMed] [Google Scholar]
- 13.Trumpower, B. L., and Edwards, C. A. (1979) J. Biol. Chem. 254 8697-8706 [PubMed] [Google Scholar]
- 14.Rich, P. R. (1984) Biochim. Biophys. Acta 768 53-79 [DOI] [PubMed] [Google Scholar]
- 15.Gutierrez-Cirlos, E. B., Merbitz-Zahradnik, T., and Trumpower, B. L. (2002) J. Biol. Chem. 277 1195-1202 [DOI] [PubMed] [Google Scholar]
- 16.Von Jagow, G., and Link, T. A. (1986) Methods Enzymol. 126 253-271 [DOI] [PubMed] [Google Scholar]
- 17.Ljungdahl, P. O., Pennoyer, J. D., Robertson, D. E., and Trumpower, B. L. (1987) Biochim. Biophys. Acta 891 227-241 [DOI] [PubMed] [Google Scholar]
- 18.Snyder, C. H., and Trumpower, B. L. (1998) Biochim. Biophys. Acta 1365 125-134 [DOI] [PubMed] [Google Scholar]
- 19.Yu, C. A., Yu, L., and King, T. E. (1972) J. Biol. Chem. 247 1012-1019 [PubMed] [Google Scholar]
- 20.Berden, J. A., and Slater, E. C. (1970) Biochim. Biophys. Acta 216 237-249 [DOI] [PubMed] [Google Scholar]
- 21.Kuzmic, P. (1996) Anal. Biochem. 237 260-273 [DOI] [PubMed] [Google Scholar]
- 22.Rich, P. R., Jeal, A. E., Madgwick, S. A., and Moody, J. A. (1990) Biochim. Biophys. Acta 1018 29-40 [DOI] [PubMed] [Google Scholar]
- 23.Ding, M. G., Di Rago, J. P., and Trumpower, B. L. (2006) J. Biol. Chem. 281 36036-36043 [DOI] [PubMed] [Google Scholar]
- 24.Rotsaert, F. A., Ding, M. G., and Trumpower, B. L. (2008) Biochim. Biophys. Acta 1777 211-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Covian, R., Zwicker, K., Rotsaert, F. A., and Trumpower, B. L. (2007) J. Biol. Chem. 282 24198-24208 [DOI] [PubMed] [Google Scholar]
- 26.Kamensky, Y., Konstantinov, A. A., Kunz, W. S., and Surkov, S. (1985) FEBS Lett. 181 95-99 [DOI] [PubMed] [Google Scholar]
- 27.Shinkarev, V. P., and Wraight, C. A. (2007) FEBS Lett. 581 1535-1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Covian, R., and Trumpower, B. L. (2008) Biochim. Biophys. Acta 1777 1044-1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenefsky, E. J., and Hoppel, C. L. (2003) Arch. Biochem. Biophys. 420 287-297 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.