Abstract
Hantaviruses are distributed worldwide and can cause a hemorrhagic fever or a cardiopulmonary syndrome in humans. Mature virions consist of RNA genome, nucleocapsid protein, RNA polymerase, and two transmembrane glycoproteins, G1 and G2. The ectodomain of G1 is surface-exposed; however, it has a 142-residue C-terminal cytoplasmic tail that plays important roles in viral assembly and host-pathogen interaction. Here we show by NMR, circular dichroism spectroscopy, and mutagenesis that a highly conserved cysteine/histidine-rich region in the G1 tail of hantaviruses forms two CCHC-type classical zinc fingers. Unlike classical zinc fingers, however, the two G1 zinc fingers are intimately joined together, forming a compact domain with a unique fold. We discuss the implication of the hantaviral G1 zinc fingers in viral assembly and host-pathogen interaction.
Many viruses in the family Bunyaviridae, which consists of five genera (Hantavirus, Orthobunyavirus, Nairovirus, Phlebovirus, and Tospovirus), cause emerging zoonotic infections in humans (1). Examples are the La Crosse encephalitis orthobunyavirus, Rift Valley fever phlebovirus, and the Crimean-Congo hemorrhagic fever nairovirus (tospoviruses are plant pathogens). Hantaviruses use rodents as their primary reservoir, and although some (e.g. Prospect Hill virus) are nonpathogenic to humans, others (e.g. Andes virus) can cause either the hantavirus cardiopulmonary syndrome or the hemorrhagic fever with renal syndrome in humans (1). Annually, over 150,000 cases of hantaviral infections are reported worldwide (2) with mortality rates reaching as high as 40% (3).
Bunyaviridae viruses are enveloped and have three genomic RNA molecules: the small (S), medium (M), and large (L) segments, and four viral proteins: the RNA polymerase, the nucleocapsid (N) protein, and the membrane glycoproteins, G1 and G2 (1). The ectodomains of G1 and G2 are glycosylated, form a heterodimer on the viral surface, and function as the viral spike proteins (1). In G1 and G2, the N termini form the ectodomains, followed by single pass transmembrane helices, then the C termini or cytoplasmic tails project within the virions. Bunyaviridae viruses lack a matrix protein (4), which link the membrane to the ribonucleoprotein among enveloped viruses (5). Based on this observation, it was suggested that the cytoplasmic tails of the viral glycoproteins might bind the viral ribonucleoprotein (6). Indeed, recent results have shown that the G1 tail binds the viral ribonucleoprotein in phlebovirus (7) and is required for packaging the genome in orthobunyavirus (8). These data suggest that the G1 tail plays a critical role in viral assembly.
Other reports suggest that among hantaviruses, the G1 tail is important in host-pathogen interaction. The G1 tail of human pathogenic hantaviruses inhibits the cellular interferon response (9, 10) against viral infection by disrupting protein-protein interactions (10). In nonpathogenic hantaviruses, by contrast, the interferon response is activated (9, 11). The G1 tail contains conserved immunoreceptor tyrosine-based activation motifs, which are involved in protein-protein interactions in the cellular immune response to viral infection (12). Further, the G1 tail of pathogenic hantaviruses is ubiquitinated and proteasomally degraded (13), which is thought to regulate the activity of the G1 tail (13), whereas the nonpathogenic hantavirus G1 tail is stable.
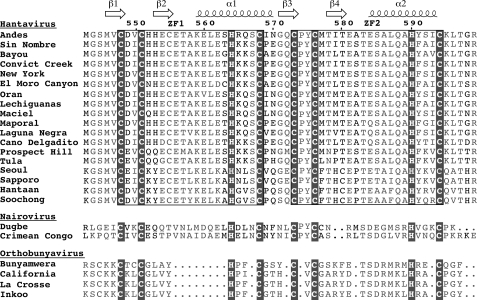
The G1 tail varies in length from 78 residues in orthobunyaviruses to 142 residues among hantaviruses. Sequence alignment shows a region of conserved cysteine and histidine residues in the G1 tail of Bunyaviridae. Further, this region was predicted to form a RING finger motif in the G1 tail of hantavirus (14). Here, we show by NMR that the conserved cysteine/histidine region in the G1 tail of hantaviruses forms two classical ββα-fold zinc fingers (15–18) and not a RING finger structure as suggested earlier (14). We also discuss the implication of our structural findings of the hantavirus G1 tail in the context of viral assembly and host pathogen interaction.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—The cysteine/histidine-rich region (residues 543–599) of the G1 tail of the Andes virus (strain 23) and Prospect Hill virus was subcloned into pET-21a (Novagen) as a C-terminal fusion to a His6-tagged GB1 domain separated by a TEV protease cleavage site. GB1 is the B1 immunoglobulin-binding domain of Streptococcus protein G (19), and a GB1 expression plasmid (obtained from Peter E. Wright, Scripps Research Institute, La Jolla, California) was used in the subcloning. Isotopically (15N or 15N, 13C) labeled protein was overexpressed in bacteria as follows: Freshly transformed Escherichia coli BL21(DE3) was grown in 1 liter M9 minimal media supplemented with 0.1 mm ZnSO4 before and after induction. The cells were grown at 37 °C, induced with 1 mm isopropyl-β-d-thiogalacto-pyranoside at A600 =∼0.8, and protein expression was continued at 15 °C overnight (to a final A600 of ∼2.0). The cells were centrifuged, resuspended in buffer A (20 mm Tris-HCl, pH 8.0, 20 mm NaCl, 1 mm DTT,2 0.1 mm ZnSO4), and lysed by sonication. Cellular debris was removed by centrifugation, and to the supernatant was added one-tenth volume of 1% polyethyleneimine (pH 8) to precipitate the nucleic acids. Following centrifugation, the supernatant was applied to a 5-ml HiTrap Q column (GE Healthcare), and bound protein was eluted with a 100 ml linear gradient of buffer B (20 mm Tris-HCl, pH 8.0, 0.5 m NaCl, 1 mm DTT, 1 mm ZnSO4). Fractions containing the fusion protein were pooled and dialyzed against TEV digestion buffer (50 mm Tris-HCl, pH 8.0, 20 mm NaCl, 1 mm DTT, 1 mm ZnSO4). TEV digestion was carried out at 25 °C for 16 h with 0.16 mg of recombinant TEV protease (20) per 10 ml of fusion protein. The His6-tagged GB1 domain was removed by passing the digest through a 1-ml nickel affinity column (I1408; Sigma); purified G1543–599 was recovered in the flow-through. Recombinant G1543–599 zinc finger retained two extra N-terminal amino acids (Gly-His) resulting from the subcloning.
Site-directed mutagenesis was performed using the QuikChange kit (Stratagene). In total, 7 cysteine and 5 histidine residues (4 native His residues and the cloning artifact, His542) were mutated individually to serine or phenylalanine, respectively, and confirmed by DNA sequencing. Mutants H542F, H553F, H552F, H590F, and C594S were expressed as soluble proteins and were purified by nickel affinity chromatography as previously described (21). Mutants C548S, C551S, C555S, H564F, C568S, C573S, and C576S were expressed as inclusion bodies (despite the presence of the GB1 solubility tag) and were purified as follows. Inclusion bodies were resuspended at room temperature in buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 8 m urea, 1 mm DTT, 0.1 mm ZnSO4, and 0.1 mm phenylmethanesulfonyl fluoride). Solubilized protein was dialyzed into buffer (20 mm Tris-HCl, pH 8.0, 0.5 m NaCl, 6 m urea, 1 mm DTT, and 0.1 mm ZnSO4), loaded into a 5-ml nickel affinity column, and eluted with buffer (20 mm Tris-HCl, pH 8.0, 20 mm NaCl, 6 m urea, 1 mm DTT, 0.1 mm ZnSO4, 1 m imidazole). Eluted protein was refolded using stepwise dialysis to remove urea. None of the mutant fusion proteins was cleaved. The two-dimensional 1H-15N HSQC of GB1 in the fusion protein was used as a marker to determine the refolding of the fusion protein.
RNA Binding Assay—Andes virus (strain 23) was used to inoculate a T175 flask of confluent monolayer of Vero E6 cells at a multiplicity of infection of 0.1 plaque-forming unit/cell and incubated for 14 days in a Biosafety Level 3 environment. The cells were harvested, and total RNA was extracted using TRIzol (15596-018; Invitrogen), ethanol-precipitated, and resuspended in water to a final concentration of 300 ng/μl. The presence of viral and cellular RNA was confirmed by reverse transcription-PCR. Total cellular and viral RNA was incubated at room temperature for 15 min with increasing amounts of Andes G1 zinc finger protein and a known RNA-binding protein, PACT (22) (a gift from Dr. Gaya Amarasinghe, Iowa State University) in binding buffer (10 mm NaPO4, 10 mm NaCl, pH 7.6). Samples were mixed with an equal volume 50% glycerol and loaded in a 0.7% agarose Tris-borate gel for electrophoretic mobility shift assay. The gel was run at 70 V for 50 min in Tris-borate buffer, pH 8.3. The gel was visualized by staining with SYBR Green II RNA specific dye (Invitrogen).
CD Spectroscopy—The GB1 tag was removed in all samples for CD spectroscopy. Each sample contained 5–10 μm protein in buffer (10 mm NaPO4, pH 7.0, 10 mm NaCl, 1 mm DTT, 0.1 mm ZnSO4). CD spectra were acquired in triplicate using a JASCO J-815 Spectropolarimeter at 25 °C and 50 nm/min scan rate. Titration with EDTA and ZnSO4 were applied to the same sample.
NMR Spectroscopy—NMR data were acquired at 25 °C using a Bruker Avance 800 MHz spectrometer equipped with a cryoprobe, processed with NMRPipe (23) and analyzed with NMR-View (24). For NMR structure determination, the G1 zinc-binding domain of the Andes virus was used. Typical NMR samples contained 1 mm 15N- or 15N, 13C-labeled protein in buffer (10 mm NaPO4, pH 7.0, 10 mm NaCl, 1 mm DTT, 0.1 mm ZnSO4) dissolved in 10% D2O or 100% D2O. Backbone assignments were obtained from two-dimensional 1H-15N HSQC (25) and three-dimensional HNCA (26), CBCA(CO)NH (26), HNCACB (27), and HNCO (28). Secondary structures were identified from the Cα, Cβ, C′, and Hα chemical shifts (29). Side chain assignments were obtained from two-dimensional 1H-13C HMQC (30), three-dimensional HBHA(CO)NH (31), and three-dimensional 13C-edited HMQC-NOESY (32) (mixing time (tmix) = 120 ms). The tautomeric ring assignments of Zn2+-coordinated histidines were identified by 15N HMQC (33). NOE cross-peaks were identified from three-dimensional 15N-edited NOESY-HSQC (34) (tmix = 120 ms) and three-dimensional 13C-edited HMQC-NOESY (32) (tmix = 120 ms).
Structure Calculation—NOE distance restraints were classified into upper bounds of 2.7, 3.5, 4.5, and 5.5 Å and a lower bound of 1.8 Å based on peak volumes. Backbone dihedral angles in the α-helical regions were restrained to ϕ (-60 ± 20°) and Ψ (-40 ± 20°). Initial structures were generated by torsion angle dynamics in CYANA (35), followed by molecular dynamics and simulated annealing in AMBER7 (52), first in vacuo and then with the generalized Born potential to account for the effect of solvent during structure calculation. Tight distance restraints that imposed tetrahedral Zn2+ coordination to Cys and His residues were used in the CYANA calculations (36). Structural calculations were also done without Zn2+ restraints to confirm that the domain could fold from NOE-derived restraints only. CYANA and AMBER structure calculation protocols have been described elsewhere (37). Iterative cycles of AMBER calculations followed by refinement of NMR-derived restraints were performed until the structures converged with low restraint violations and good statistics in the Ramachandran plot. A family of 20 lowest energy structures was analyzed using PROCHECK (38), and the graphics were generated using Pymol (39).
RESULTS
Protein Expression—Sequence analysis of the G1 cytoplasmic tail of hantaviruses revealed two highly conserved CX2CX12–13-HX3C (where X is any amino acid) motifs, which suggested the presence of two CCHC-type zinc fingers (Fig. 1). Expression of the Andes virus G1 zinc fingers (residues 543–599) in E. coli resulted in cell death, with cell density reaching only A600 of ∼0.9 after induction at A600 of ∼0.8, suggesting that the zinc finger was toxic to E. coli. Thus, the zinc finger domain was expressed as a GB1 fusion protein. The GB1 tag contained His6 for nickel affinity purification and a TEV protease cleavage site to recover the native G1 zinc finger domain. The fusion protein was overexpressed in soluble form in E. coli, purified under native conditions, and digested with TEV protease to obtain the G1 zinc finger domain.
FIGURE 1.
The G1 tail of Hantaviruses, Nairoviruses, and Orthobunyaviruses (genera of Bunyaviridae) contains a cysteine/histidine-rich region with two CCHC arrays. Structure determination of the Andes virus dual CCHC-region revealed a novel zinc finger domain. Shown are the secondary structures (α-helices and β-strands), zinc-coordinating residues (blocked), the two CCHC motifs (boxed), conserved residues (gray), and residue numbers for the Andes virus G1 sequence. Sequence alignment was generated using CLUSTALW and formatted with ESPript 2.2 (53).
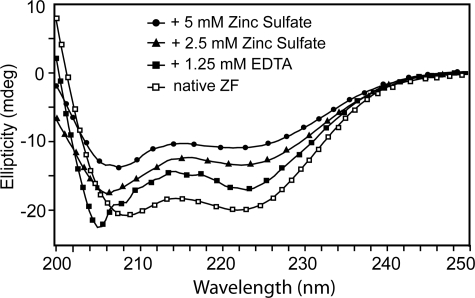
Zn2+ Is Required for Proper Folding—CD spectrum of the Andes virus G1 zinc finger showed a folded α-helical domain with local minima at 209 and 222 nm (Fig. 2). Titration of EDTA to a final concentration of 1.25 mm caused a spectral shift to 205 nm, indicating a partial loss of secondary structure. However, the minimum at 222 nm remained despite EDTA treatment, suggesting that although the global fold is disrupted by removal of zinc ion, some residual helical structure remained. Subsequently, titrating ZnSO4 back into the solution resulted in increased α-helical content, suggesting restoration of the global fold.
FIGURE 2.
CD spectroscopy and titration with EDTA and ZnSO4 of recombinant Andes virus G1 tail CCHC-region (residues 543–599), which was expressed and purified under native conditions, showed that Zn2+-binding is required for the proper folding of this domain. Native G1 tail zinc finger domain showed a folded CD spectrum (open squares). Titration with an excess of EDTA resulted to an unfolded peak (at 204 nm) and reduced the helical peak (at 222 nm) (closed squares). The addition of 2.5 and 5 mm ZnSO4 yielded folded CD spectra (triangles and circles).
NMR data were also used to confirm the requirement for Zn2+ coordination on the proper folding of the zinc finger domain. The Andes virus and the Prospect Hill virus zinc-binding domains purified under native conditions showed well dispersed and sharp peaks in their two-dimensional 1H-15N HSQC spectra (supplemental Fig. S1). After treatment with excess EDTA, peaks in the HSQC of the Andes virus zinc-binding domain deteriorated, showed residual peaks that differ markedly from each other with respect to peak intensities and sharpness, and displayed a collapse of the amide side chains (supplemental Fig. S1). In the presence of excess EDTA, the HSQC spectrum of the Prospect Hill virus showed a collapse of the backbone and side chain amide peaks (supplemental Fig. S1), which indicated that the protein was unfolded.
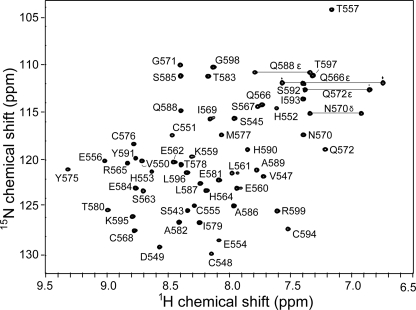
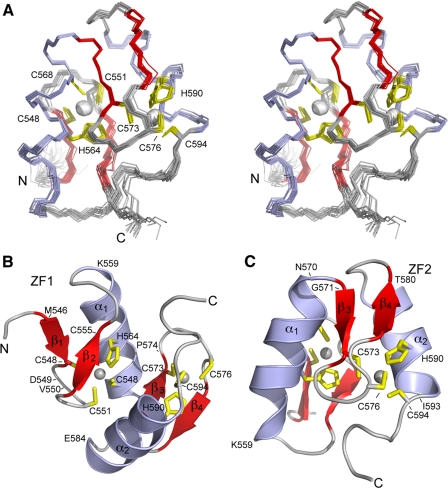
NMR Structure Determination—We determined the NMR structure of the Andes virus zinc finger domain. The Andes virus G1 zinc finger domain showed an excellent well dispersed two-dimensional 1H-15N HSQC (Fig. 3). Nearly complete backbone assignments were obtained from three-dimensional HNCA, CBCA(CO)NH, HNCACB, and 15N-edited NOESY-HSQC. Analysis of the Cα, Hα, Cβ, and C′ secondary chemical shifts (supplemental Fig. S2) supported the presence of two short α-helices and two random coil regions flanking the central domain (29). Side chain assignments were completed using two-dimensional 1H-13C HMQC, three-dimensional HBHA-(CO)NH, and three-dimensional 13C-edited HMQC-NOESY. There were four conserved histidines (at positions 552, 553, 564, and 590) that could potentially coordinate Zn2+ ion; however, long distance NOEs (between Cys548-His564 and Cys573-His590) indicated that His564 and His590 were involved in Zn2+ coordination. A two-dimensional 15N HMQC (33) spectrum showed Zn2+ coordination through the His564 Nδ1 and His590 Nε2 atoms (supplemental Fig. S3). Manual analysis of three-dimensional 15N- and 13C-edited NOESY spectra identified 859 unambiguous interproton distance restraints. The NOE restraints together with 24 and 15 Ψ dihedral angle restraints and zinc coordination restrains (Table 1) were used in structure calculation and refinement with CYANA and AMBER. The 20 lowest energy NMR structures converged into a family of structures (Fig. 4A) with low restraint violations and good Ramachandran plot statistics (Table 1).
FIGURE 3.
The Andes virus G1 tail zinc finger domain (residues 543–599) shows a well dispersed two-dimensional 1H-15N HSQC spectrum, which facilitated the acquisition of additional NMR data sets and allowed resonance assignments and NMR structure determination of this domain. Shown are the complete assignments for backbone and side chain amides.
TABLE 1.
Restraints and structural statistics for 20 NMR structures
| Total NOE-derived distance restraints | 859 |
| Intraresidue (i,i) | 323 |
| Sequential (i, i+1) | 207 |
| Medium Range (2 ≥ |i – j| ≤ 4) | 161 |
| Long Range (|i – j| > 4) | 168 |
| Total dihedral angle restraints | 39 |
| Phi | 24 |
| Psi | 15 |
| Root mean square difference from mean structure | |
| Backbone atoms (N, Cα, C′) (Å) | 0.20 |
| All heavy atoms (C, N, O) (Å) | 0.64 |
| Violation analysis | |
| Max distance violation (Å) | 0.5 |
| Max dihedral angle violation (°) | 40 |
| Energies | |
| Mean generalized Born potential-AMBER energy (kcal mol–1) | –2113 |
| Mean restraint energy (kcal mol–1) | 13 |
| Ramachandran plot (%) | |
| Most favorable region | 81.5 |
| Additionally allowed regions | 18.0 |
| Generously allowed regions | 0.3 |
| Disallowed regions | 0.1 |
FIGURE 4.
The NMR structure of the Andes virus G1 tail zinc-binding domain reveals two classical ββα fold zinc fingers that are joined together. A, stereoview of the superposition of 20 lowest energy NMR structures. B and C, ribbon structures of the lowest energy NMR structure showing the residues involved in the first (ZF1) (B) and second (ZF2) (C) zinc fingers. Shown are the cysteine and histidine residues (yellow) that coordinate Zn2+ ions (gray) as well as the secondary structures (α1-α2, β1-β2). The dual hantaviral G1 zinc fingers interact with each other and form a single domain with a novel fold as revealed by DALI (40) and TM-align (41) structural homology searches.
Structure of Individual Zinc Finger—Each G1 zinc finger folded similarly to the ββα fold of classical zinc fingers. In the first CCHC array (ZF1), residues Met546–Cys555 formed a β-hairpin that encompassed the first two coordinating cysteines (Cys548 and Cys551; Fig. 4B). Asp549 and Val550 formed the loop apex with the coordinating cysteines on either side of the β-hairpin. The structured region terminated at Lys559 where helix α1 began and folded back toward the β-hairpin and allowed the completion of ZF1 with His564 and Cys568 on the interior face of helix α1.
In ZF2, the β-hairpin (Gly571–Thr580) contained the first two coordinating cysteines (Cys573 and Cys576; Fig. 4, B and C). The coordination site on the loop was partly formed by the positioning of Pro574 between the two. Strong Cys573 Hα to Pro574 Hδ NOEs indicated that Pro574 was in the trans configuration. A structured loop followed the β-hairpin and terminated at Glu584, where helix α2 began, and folded back toward the β-hairpin to complete ZF2 by coordinating His590 and Cys594 to the Zn2+ ion.
A Novel Dual CCHC Zinc Finger—Unlike classical ββα zinc fingers, which fold independently of each other forming a “beads-on-a-string” configuration, the two G1 zinc fingers interacted with each other, forming a compact structure in which the two zinc atoms were located a mere 10 Å apart (Fig. 4). Two short, parallel helices of 8 and 9 residues in length were linked by a 15-residue β-hairpin extending between Cys568 of helix α1 and Thr583 of helix α2. Another loop preceded helix 1 and an unstructured tail of 7 residues followed helix 2. Both zinc coordination sites were formed at the junction of a loop and the face of a proximal α-helix. Structural searches using DALI (40) and TM-align (41) returned no homologous structures; thus, the G1 zinc finger domain has a novel fold.
Mutations of Zn2+-coordinating Residues—To confirm the Zn2+ coordination topology indicated by the NMR structure, we created point mutants in each of the cysteine and histidine residues within the Andes virus zinc finger domain. Of the 8 residues expected to coordinate zinc, only C594S and H590F expressed as soluble proteins, the rest (C548S, C551S, H564F, C568S, C573S, and C576S) could be expressed only as inclusion bodies despite the presence of the GB1 solubility tag. This result suggested that zinc coordination was necessary for stabilizing the overall fold of the zinc finger domain. For further analysis, all of the inclusion bodies were solubilized overnight in 8 m urea, purified by nickel affinity chromatography, and refolded by stepwise dialysis to remove urea. Refolding of the zinc finger domain was determined by the proper refolding of the attached GB1 tag using two-dimensional 15N HSQC, which served as a control to show that the refolding conditions would have properly refolded a native protein. The spectra of the mutated G1 zinc finger domain in the GB1 fusion proteins consisting of C548S, C551S, H564F, C573S, C576S, H590F, and C594S all showed narrowly dispersed spectra consistent with an unfolded domain (supplemental Fig. S4). These results suggested that the two zinc fingers did not fold independently of each other (supplemental Fig. S4). Of these eight positions, only C568S showed any peak dispersion at all (supplemental Fig. S4). In each instance, the peaks corresponding to the attached GB1 tag were well dispersed, thus indicating that the fusion protein was refolded properly (supplemental Fig. S4). These results suggested that, in the dual zinc finger domain, mutation of a Zn2+-coordinating residue in either ZF1 or ZF2 lead to the unfolding of the entire domain.
Mutations of Non-Zn2+-coordinating Residues—The domain contains three histidines (His552, His553, and His542, the cloning artifact) and a cysteine (Cys555) (Fig. 1) that are not involved in Zn2+ coordination. To eliminate the possibility that Zn2+ could coordinate these other cysteine and histidine residues, we generated four additional point mutants corresponding to H542F, H552F, H553F, and C555S. Three of the four mutants (H552F, H553F, and H542F) gave a dispersed spectrum consistent with a folded domain (supplemental Fig. S5). Only the C555S mutant gave an unfolded spectrum (supplemental Fig. S5). Analysis of the structure reveals that the side chain of Cys555 was oriented toward the interior of the structure and therefore played a role in stabilizing the hydrophobic core of the overall domain. These data hence confirmed that Zn2+ was coordinated to the predicted zinc finger residues (Cys548, Cys541, Cys568, Cys574, Cys576, and Cys594 and His564 and His590).
Hantaviral G1 Zinc Fingers Does Not Bind RNA—Classical ββα-fold zinc fingers are well known nucleic acid-binding motifs (15–18). However, our attempts to verify the ability of the Andes virus G1 zinc finger domain to bind RNA by electrophoretic mobility shift assay revealed that under the conditions used, the G1 zinc finger domain did not bind RNA obtained from Andes virus-infected Vero E6 cells (supplemental Fig. S7). Although a known RNA-binding protein PACT (22) showed smearing of the RNA bands, which suggested nonspecific PACT-RNA interaction, increasing amounts of the Andes virus G1 zinc finger failed to demonstrate even nonspecific binding of RNA (supplemental Fig. S7).
DISCUSSION
The G1 tail of Bunyaviriade viruses is important in viral assembly (7, 8) and host-pathogen interaction (9–13). Our results showed that a conserved cysteine/histidine-rich region in the hantavirus G1 tail (Fig. 1) required Zn2+ binding to fold properly (Fig. 2). This region formed an independently folded domain that gave excellent NMR data (Fig. 3) and NMR structure determination revealed dual CCHC-type zinc fingers where Zn2+ ligands were sequential and nonoverlapping (Fig. 4). The folding of each G1 zinc finger is related to the classical ββα zinc finger fold (42), which are among the most abundant protein motifs in eukaryotic genomes (reviewed in Ref. 43).
Implication of the Zinc Finger Structure in the Biology of Hantavirus—It has been suggested that the conserved cysteine-histidine region in the G1 tail of hantaviruses forms a RING finger motif (14). This assumption is based on the following observations: (i) the G1 tail is ubiquitinated and proteasomally degraded as part of the host-pathogen interaction of hantaviruses (13), (ii) RING fingers are structural domains of ubiquitin ligases, which are part of the ubiquitin degradation pathway, and (iii) some viruses contain RING finger motifs that are involved in the ubiquitin degradation pathway as part of their host evasion mechanism (14).
Instead of forming a RING finger motif (14), however, our results showed that the conserved cysteine/histidine region in the G1 tail of hantaviruses formed a dual classical type ββα-fold zinc fingers. Classical zinc fingers are well known DNA- and RNA-binding domains (17, 18, 44). Recent reports indicate that proteins containing classical zinc fingers are also involved in protein-protein interaction (43, 45–47). Thus, instead of functioning as a domain of a ubiquitin ligase (as a RING finger), the classical ββα-fold of the hantaviral zinc fingers suggests nucleic acid binding and/or protein-protein interaction. This is consistent with the observations that the G1 tail is important in binding the ribonucleoprotein during viral assembly of Bunyaviridae (7, 8).
Thus, the ββα-fold implies that the hantaviral zinc fingers may interact with the RNA genome or the protein component of the ribonucleoprotein during viral assembly. Our electrophoretic mobility shift assay showed that the hantaviral zinc fingers did not interact with RNA (supplemental Fig. S7). Additionally, the hantaviral zinc domain has a theoretical pI of 5.8, which is too acidic to be a nucleic acid-binding motif. Further, many of the dual zinc fingers that have been characterized to date (RING, MYND, and LIM) are involved in protein-protein interaction (reviewed in Ref. 47). Therefore, a similar protein binding function for the hantaviral zinc finger is likely rather than RNA binding. It has also been suggested by others that during the assembly of hantaviruses, the G1 tail binds the nucleocapsid protein (48), which is a key component of the viral ribonucleoprotein. Efforts are now underway to identify the protein binding partners of the hantaviral zinc fingers.
Unique Properties of Hantaviral Zinc Fingers—Although hantaviral zinc fingers have classical ββα zinc finger fold (Fig. 4), they differ from classical zinc fingers in two aspects. First, the two hantaviral zinc fingers fold together as a single domain, which is likely due to a short 4-residue linker between the two zinc fingers. Commonly, classical zinc fingers fold independently of each other, forming a beads-on-a-string configuration. However, multiple classical zinc fingers can interact with each other when bound to DNA (44) or RNA (17, 18). Another example of a dual classical ββα zinc finger that folds together as one unit is the yeast Zap1 transcription factor (49). Second, the folding of one hantaviral zinc finger affects the folding of the other zinc finger. For example, mutations in cysteine and histidine residues that disrupted the first or second zinc finger disrupted the folding of the entire dual zinc finger domain (supplemental Fig. S4). Because classical zinc fingers fold independently of each other, disrupting the folding of one zinc finger domain does not affect the folding of the other zinc fingers.
Other Viral Zinc Fingers—Among viruses, the CCHC zinc fingers of the nucleocapsid proteins of retroviruses (50) have been studied extensively because of their critical role in binding and packaging the RNA genomes. Examples of zinc fingers in viral glycoproteins, however, are scarce. Our structure presented here is the first atomic resolution structure of a zinc finger domain from a viral glycoprotein. The sequence homology of the hantaviral zinc finger region with other Bunyaviridae (Fig. 1) also suggests that that the G1 tail in nairoviruses and orthobunyaviruses will also form zinc finger motifs. Therefore, our results form the structural framework for future studies aimed at elucidating the precise role of the G1 tail in the viral assembly and immune evasion of Bunyaviridae.
Supplementary Material
Acknowledgments
We are grateful to Yu Wang (University of Kansas), Peter Gegenheimer (University of Kansas), Brian Lee (Southern Illinois University), and Gaya Amarasinghe (Iowa State University) for helpful discussion.
The atomic coordinates and structure factors (code 2K9H) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant AI065359 (to S.C.S.J.) and AI057160 (to R. N. D.). This work was also supported by American Heart Association Grant 0755724Z (to R. N. D.), National Science Foundation Grant 0326999 (to S. C. S.), the Madison and Lila Self Graduate Fellowship (to D. F. E.), and the Reno Cancer Foundation (to D.M.B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
Footnotes
The abbreviations used are: DTT, dithiothreitol; HSQC, heteronuclear single quantum coherence; HMQC, heteronuclear multiple quantum coherence; NOE, nuclear Overhauser effect; NOESY, NOE spectroscopy.
References
- 1.Elliott, R. M., Bouloy, M., Calisher, C. H., Goldbach, R., Moyer, J. T., Nichol, S. T., Pettersson, R., Plyusnin, A., and Schmaljohn, C. (2000) in Virus Taxonomy: The classification and Nomenclature of Viruses: The Seventh Report of the International Committee on Taxonomy of Viruses. (Van Regenmortel, M. H. V., Fauquet, C. M., Bishop, D. H. L., Carsten, E. B., Estes, M. K., Lemon, S. M., Maniloff, J., Mayo, M. A., McGeoch, D. J., Pringle, C. R., and Wickner, R. B., eds) pp. 599-621, Academic Press, San Diego
- 2.Khaiboullina, S. F., Morzunov, S. P., and St. Jeor, S. C. (2005) Curr. Mol. Med. 5 773-790 [DOI] [PubMed] [Google Scholar]
- 3.Mertz, G. J., Hjelle, B., Crowley, M., Iwamoto, G., Tomicic, V., and Vial, P. A. (2006) Curr. Opin. Infect. Dis. 19 437-442 [DOI] [PubMed] [Google Scholar]
- 4.Elliott, R. M. (1990) J. Gen. Virol. 71 501-522 [DOI] [PubMed] [Google Scholar]
- 5.Flint, S. J., Enquist, L. W., Krug, R. M., Racaniello, V. R., and Skalka, A. M. (2000) Principles of Virology: Molecular Biology, Pathogenesis, and Control, p. 92, ASP Press, Washington, D.C.
- 6.von Bonsdorff, C. H., and Pettersson, R. (1975) J. Virol. 16 1296-1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Overby, A. K., Pettersson, R. F., and Neve, E. P. (2007) J. Virol. 81 3198-3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi, X., Kohl, A., Li, P., and Elliott, R. M. (2007) J. Virol. 81 10151-10160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alff, P. J., Gavrilovskaya, I. N., Gorbunova, E., Endriss, K., Chong, Y., Geimonen, E., Sen, N., Reich, N. C., and Mackow, E. R. (2006) J. Virol. 80 9676-9686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alff, P. J., Sen, N., Gorbunova, E., Gavrilovskaya, I. N., and Mackow, E. R. (2008) J. Virol. 82 9115-9122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spiropoulou, C. F., Albarino, C. G., Ksiazek, T. G., and Rollin, P. E. (2007) J. Virol. 81 2769-2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geimonen, E., LaMonica, R., Springer, K., Farooqui, Y., Gavrilovskaya, I. N., and Mackow, E. R. (2003) J. Virol. 77 1638-1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geimonen, E., Fernandez, I., Gavrilovskaya, I. N., and Mackow, E. R. (2003) J. Virol. 77 10760-10868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sen, N., Sen, A., and Mackow, E. R. (2007) J. Virol. 81 4323-4330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wuttke, D. S., Foster, M. P., Case, D. A., Gottesfeld, J. M., and Wright, P. E. (1997) J. Mol. Biol. 273 183-206 [DOI] [PubMed] [Google Scholar]
- 16.Nolte, R. T., Conlin, R. M., Harrison, S. C., and Brown, R. S. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2938-2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, D., Searles, M. A., and Klug, A. (2003) Nature 426 96-100 [DOI] [PubMed] [Google Scholar]
- 18.Lee, B. M., Xu, J., Clarkson, B. K., Martinez-Yamout, M. A., Dyson, H. J., Case, D. A., Gottesfeld, J. M., and Wright, P. E. (2006) J. Mol. Biol. 357 275-291 [DOI] [PubMed] [Google Scholar]
- 19.Huth, J. R., Bewley, C. A., Jackson, B. M., Hinnebusch, A. G., Clore, G. M., and Gronenborn, A. M. (1997) Protein Sci. 6 2359-2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geisbrecht, B. V., Bouyain, S., and Pop, M. (2006) Protein Expression Purif. 46 23-32 [DOI] [PubMed] [Google Scholar]
- 21.Wang, Y., Boudreaux, D. M., Estrada, D. F., Egan, C. W., St. Jeor, S. C., and De Guzman, R. N. (2008) J. Biol. Chem. 283 28297-28304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel, R. C., and Sen, G. C. (1998) EMBO J. 17 4379-4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 24.Johnson, B. A. (2004) Methods Mol. Biol. 278 313-352 [DOI] [PubMed] [Google Scholar]
- 25.Grzesiek, S., and Bax, A. (1993) J. Am. Chem. Soc. 115 12593-12594 [Google Scholar]
- 26.Grzesiek, S., Dobeli, H., Gentz, R., Garotta, G., Labhardt, A. M., and Bax, A. (1992) Biochemistry 31 8180-8190 [DOI] [PubMed] [Google Scholar]
- 27.Wittekind, M., and Mueller, L. (1993) J. Magn. Reson. 101B 201-205 [Google Scholar]
- 28.Muhandiram, D. R., and Kay, L. E. (1994) J. Magn. Reson. 103B 203-216 [Google Scholar]
- 29.Wishart, D. S., and Nip, A. M. (1998) Biochem. Cell Biol. 76 153-163 [DOI] [PubMed] [Google Scholar]
- 30.Tolman, J. R., Chung, J., and Prestegard, J. H. (1992) J. Magn. Reson. 98 462-467 [Google Scholar]
- 31.Grzesiek, S., and Bax, A. (1993) J. Biomol. NMR 3 185-204 [DOI] [PubMed] [Google Scholar]
- 32.Fesik, S. W., and Zuiderweg, E. R. P. (1998) J. Magn. Reson. 78 588-593 [Google Scholar]
- 33.Pelton, J. G., Torchia, D. A., Meadow, N. D., and Roseman, S. (1993) Protein Sci. 2 543-558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marion, D., Driscoll, P. C., Kay, L. E., Wingfield, P. T., Bax, A., Gronenborn, A. M., and Clore, G. M. (1989) Biochemistry 28 6150-6156 [DOI] [PubMed] [Google Scholar]
- 35.Guntert, P. (2004) Methods Mol. Biol. 278 353-378 [DOI] [PubMed] [Google Scholar]
- 36.Hoffman, R. C., Xu, R. X., Klevit, R. E., and Herriott, J. R. (1993) J. Magn. Reson. 102B 61-72 [Google Scholar]
- 37.Dames, S. A., Martinez-Yamout, M., De Guzman, R. N., Dyson, H. J., and Wright, P. E. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 5271-5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R., and Thornton, J. M. (1996) J. Biomol. NMR 8 477-486 [DOI] [PubMed] [Google Scholar]
- 39.DeLano, W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA
- 40.Holm, L., and Sander, C. (1996) Science 273 595-603 [DOI] [PubMed] [Google Scholar]
- 41.Zhang, Y., and Skolnick, J. (2005) Nucleic Acids Res. 33 2302-2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller, J., McLachlan, A. D., and Klug, A. (1985) EMBO J. 4 1609-1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laity, J. H., Lee, B. M., and Wright, P. E. (2001) Curr. Opin. Struct. Biol. 11 39-46 [DOI] [PubMed] [Google Scholar]
- 44.Foster, M. P., Wuttke, D. S., Radhakrishnan, I., Case, D. A., Gottesfeld, J. M., and Wright, P. E. (1997) Nat. Struct. Biol. 4 605-608 [DOI] [PubMed] [Google Scholar]
- 45.Fox, A. H., Liew, C., Holmes, M., Kowalski, K., Mackay, J., and Crossley, M. (1999) EMBO J. 18 2812-2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westman, B. J., Perdomo, J., Matthews, J. M., Crossley, M., and Mackay, J. P. (2004) Biochemistry 43 13318-13327 [DOI] [PubMed] [Google Scholar]
- 47.Gamsjaeger, R., Liew, C. K., Loughlin, F. E., Crossley, M., and Mackay, J. P. (2007) Trends Biochem. Sci. 32 63-70 [DOI] [PubMed] [Google Scholar]
- 48.Kaukinen, P., Vaheri, A., and Plyusnin, A. (2005) Arch. Virol. 150 1693-1713 [DOI] [PubMed] [Google Scholar]
- 49.Wang, Z., Feng, L. S., Matskevich, V., Venkataraman, K., Parasuram, P., and Laity, J. H. (2006) J. Mol. Biol. 357 1167-1183 [DOI] [PubMed] [Google Scholar]
- 50.De Guzman, R. N., Wu, Z. R., Stalling, C. C., Pappalardo, L., Borer, P. N., and Summers, M. F. (1998) Science 279 384-388 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.