Abstract
Xanthine oxidoreductase is a ubiquitous cytoplasmic protein that catalyzes the final two steps in purine catabolism. We have previously investigated the catalytic mechanism of the enzyme by rapid reaction kinetics and x-ray crystallography using the poor substrate 2-hydroxy-6-methylpurine, focusing our attention on the orientation of substrate in the active site and the role of Arg-880 in catalysis. Here we report additional crystal structures of as-isolated, functional xanthine oxidase in the course of reaction with the pterin substrate lumazine at 2.2 Å resolution and of the nonfunctional desulfo form of the enzyme in complex with xanthine at 2.6 Å resolution. In both cases the orientation of substrate is such that the pyrimidine subnucleus is oriented opposite to that seen with the slow substrate 2-hydroxy-6-methylpurine. The mechanistic implications as to how the ensemble of active site functional groups in the active site work to accelerate reaction rate are discussed.
Xanthine oxidoreductase (XOR)3 is a molybdenum-containing enzyme that is the prototypical member of the molybdenum hydroxylase family of proteins (1, 2). It is one of four molybdenum-containing enzymes encoded by the human genome and catalyzes the sequential hydroxylation of hypoxanthine to xanthine and xanthine to uric acid. Under normal physiological circumstances the enzyme is thought to be present as a dehydrogenase (XDH) but can be readily converted to an oxidase (XO) by the oxidation of sulfhydryl residues or by limited proteolysis (3). XDH shows a preference for NAD+ as the oxidizing substrate (although it is also able to react with O2), whereas XO cannot react with NAD+ and can only use O2; both forms generate significant amounts of both hydrogen peroxide and superoxide when reacting with O2 (3). Conversion of XDH to XO is thought to play a role in ischemia-reperfusion injury associated with heart attack and stroke (4). The enzyme is also the target of antihyperuricemia drugs and is often targeted in tandem chemotherapeutic regimens (5). Excellent reviews describing XOR in pharmacology and human pathology are available (6, 7).
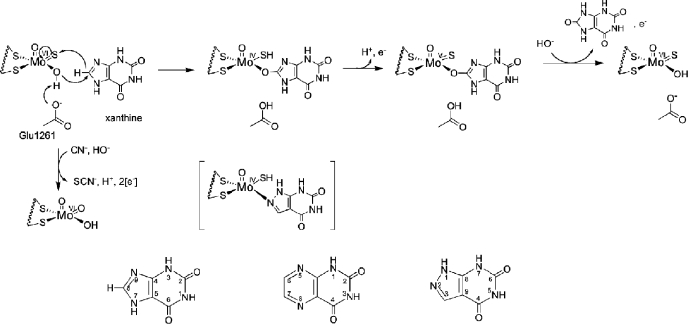
Bovine xanthine oxidase is a 290-kDa homodimer, with each independently acting monomer possessing a molybdenum center at which the oxidative hydroxylation of substrates takes place. Hydroxylation of substrate results in the two-electron reduction of the molybdenum center from Mo(VI) to Mo(IV). Following the initial reduction of the Mo, electrons are passed via two [2Fe-2S] clusters to an FAD cofactor, at which reducing equivalents pass out of the enzyme. The crystal structure of the bovine enzyme has been determined previously (8), showing that the four redox-active centers of each monomer are found in separate domains of the polypeptide. The molybdenum center possesses a square-pyramidal coordination geometry with an apical Mo=O group and as shown in Fig. 1 can be formulated as LMoVIOS(OH), with L being a bidentate enedithiolate ligand contributed by a unique pyranopterin cofactor that is common to all molybdenum- and tungsten-containing enzymes (with the exception of nitrogenase) (1).
FIGURE 1.
The catalytic mechanism at the molybdenum site of xanthine oxidase. Shown is the hypothesized orientation of xanthine during catalysis. Also shown is the MoV state that gives rise to the well studied “very rapid” EPR signal. The structure in brackets is that of the complex of reduced enzyme with the inhibitor alloxanthine. Inactivation of the enzyme by KCN results from replacement of the planar Mo=S by oxygen from solvent water, forming an unreactive LMoVIO2(OH) form of the active site. Also shown is the numbering scheme for purines, pteridines, and ayrazolopyrimidines, as exemplified by xanthine, lumazine, and alloxanthine, respectively.
The now generally accepted mechanism of XOR begins with proton abstraction from the Mo-OH group by an active site glutamate that is universally conserved in the molybdenum hydroxylase family of enzymes (9). This is followed by nucleophilic attack of the resulting Mo-O- unit on the carbon center to be hydroxylated with concomitant hydride transfer to the Mo=S of the molybdenum center (Fig. 1). The reaction yields an intermediate that can be represented as LMoIV(SH)(OR), with OR being the now hydroxylated product coordinated to the molybdenum via the newly introduced hydroxyl group. Catalysis is completed by displacement of the bound product from the molybdenum coordination sphere by hydroxide from solvent water, electron transfer out of the molybdenum center to the flavin site, and deprotonation of the MoIV-SH to give the original oxidized LMoVIOS(OH) form of the molybdenum center. The relative rates of product displacement on the one hand and electron transfer from the molybdenum center to the other redox-active centers on the other is dependent on the particular substrate being hydroxylated and also the pH. When electron transfer takes place prior to product dissociation, a transient LMoVS(OR) species is generated that gives rise to the well characterized “very rapid” EPR signal (10, 11).
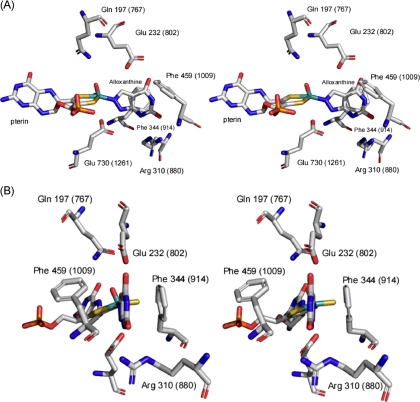
In addition to the glutamate residue that is thought to function as a general base, the active sites of all xanthine-utilizing enzymes (but not those otherwise related enzymes that act on aldehyde substrates) have a conserved arginine residue, Arg-310 in the numbering for XDH from Rhodobacter capsulatus, Arg-880 in the bovine enzyme (12). This distal residue is positioned some 10 Å from the molybdenum center, on the opposite side of bound substrate to the molybdenum center, and presumably too far to participate directly in catalysis. In the structure of the R. capsulatus xanthine dehydrogenase with the mechanism-based inhibitor alloxanthine, Arg-310 is hydrogen-bonded to the pyrimidine subnucleus of the heterocycle via the C-6 carbonyl group of the inhibitor (equivalent to the C-2 of xanthine and other purines, given the different numbering system in the two classes of heterocycles), as shown in Fig. 2.
FIGURE 2.
The active site of alloxanthine-inhibited xanthine dehydrogenase from R. capsulatus (Ref. 12; Protein Data Bank code 1JRP). Shown is the numbering convention for the bacterial enzyme, and bovine numbering is shown in parentheses. B is a 90° clockwise rotation about the vertical axis from A. The planar sulfur and apical oxygen were switched given recent evidence that it is the oxygen rather than the sulfur that occupies the apical position in the square pyramidal coordination sphere of the molybdenum (30).
In previous work we have shown that the distal Arg-310 of the R. capsulatus enzyme contributes at least 4.5 kcal/mol toward transition state stabilization; its mutation to methionine results in a 2 × 104-fold decrease in reaction rate with xanthine as substrate (13). A comparable effect is seen with the human enzyme (14). We have suggested that this distal arginine lowers the activation energy for the reaction by stabilizing the negative charge that accumulates on the heterocycle in the course of nucleophilic attack via an electrostatic interaction with the C-6 carbonyl oxygen of substrate. We have also examined the reaction of wild-type and R310M mutants of the R. capsulatus xanthine dehydrogenase with a homologous series of purine substrates (13). Those that are effective substrates of the wild-type enzyme were strongly affected by mutation of Arg-310 to methionine, whereas those that react slowly with wild-type enzyme were much less affected by the mutation. Noting that all the good substrates possessed a carbonyl or thio group at C-6, whereas poor substrates did not, we concluded that poor substrates bound in an inverted orientation relative to that of xanthine and other good substrates and were unable to utilize Arg-310 for transition state stabilization. We suggested that this inverted orientation accounted for both the low intrinsic reactivity of the poor substrates with wild-type enzyme and also their relative insensitivity to loss of the arginine (13).
We have recently reported the crystal structure of bovine xanthine oxidase in the course of the reaction with the poor substrate 2-hydroxy-6-methylpurine (HMP) (15) (Protein Data Bank code 3B9J), in which the bound HMP molecule is clearly oriented with the C-2 rather than the C-6 position oriented toward Arg-880 (in an orientation analogous to that seen with the inhibitor alloxanthine). This work also showed strong electron density (in one of the two active sites in the asymmetric unit) between the planar Mo-O and the C-8 position of the HMP; thus this structure showed the orientation of a true catalytic intermediate. To further test our hypothesis regarding substrate orientation and to gain further insight into the function of the catalytic machinery of xanthine oxidase, we have here determined the x-ray crystal structures of functional bovine xanthine oxidase with the pteridine substrate lumazine, as well as the inactive desulfo form of the enzyme in complex with xanthine. In both cases, the orientation of substrate is such that the pyrimidine subnucleus is oriented opposite to that seen with the poor substrate HMP and the inhibitor alloxanthine (bound to reduced enzyme), consistent the proposed role of Arg-880 in catalysis. The implications regarding the function of the catalytic machinery of the enzyme are discussed.
EXPERIMENTAL PROCEDURES
Materials—Magnetic bases for use with the automated express crystallography system at SGX-CAT were obtained from Rayonix (Evanston, IL). Mounted cryoloops, magnetic cryovials, and crystal growth materials were obtained from Hampton Research (Aliso Viejo, CA). All of the chemicals and reagents were obtained at the highest quality/purity available from Sigma-Aldrich or Fisher and were used without further purification.
Isolation of Xanthine Oxidase—Xanthine oxidase was isolated from fresh, unpasteurized bovine milk (Waterman Farm, The Ohio State University or Scott Brothers Dairy, Chino, CA) according to published methods (16). The milk was obtained from a single animal, so as to minimize any heterogeneity in the qualities and characteristics of the isolated protein. Isolated enzyme was stored in N2(l) and passed down a Sephadex G-25 column to remove salicylate prior to use.
Crystallization, Data Acquisition, and Structure Determination—For the structure of xanthine oxidase with lumazine, crystals were grown via the batch method according to previously published methods using microbridges to hold the batch solutions in the sealed wells of a 24-well tray (15). The batch solutions contained 20 μl of a 10 mg/ml (34.5 μm) enzyme solution mixed with 10 μl of the precipitant solution (see further below).
To obtain the structure of xanthine oxidase with xanthine, the enzyme had to first be inactivated by replacing the planar Mo=S with a second Mo=O by reaction with cyanide, as shown in Fig. 1 (17). Here, enzyme stocks were incubated on ice for 1 h with 0.1 m KCN. The concentrated enzyme was then passed down a Sephadex G-25 column to remove excess cyanide. After initial attempts to obtain the structure using synchrotron radiation indicated dissociation of the dimer into separate monomers, we decided to treat the cyanolyzed enzyme with 10 mm oxidized glutathione. After removal of excess reagent by G-25 chromatography, the enzyme was used for crystal growth according to previous methods (15). The batch solutions contained 10 μl of the enzyme solution and 5 μl of the precipitant solution.
The final conditions for optimal crystal growth with both lumazine and xanthine were a precipitant solution of 12% polyethylene glycol 8000, 0.1 m potassium phosphate at initial pH 6.5, and 0.2 mm EDTA. The enzyme solution contained 10 mg/ml XO in 40 mm Tris-HCl, initial pH 7.8, 20 mm pyrophosphate, initial pH 8.5, 0.2 mm EDTA, and 5 mm dithiothreitol. The enzyme solution was allowed to sit on ice for 1 h following the addition of dithiothreitol, prior to beginning the construction of the well solutions. The final pH of the well solutions was 7.2 ± 0.1. The crystals grew after 2-3 days at 25 °C in darkness. XO crystals grew in the form of rectangular plates, which ranged from 0.1 to 1.0 mm in their longest dimension.
A 42% polyethylene glycol 200 solution at pH 8.0 for lumazine or 8.3-8.4 for xanthine containing all proportions of the enzyme and precipitant solutions was used as the cryoprotectant solution. This was introduced by buffer exchanges of 2 μl using a micropipette. Following an exchange of volume greater than the original batch volume, the substrate (xanthine at 33.3 mm and lumazine at 10 mm in the same cryoprotectant solution) was introduced from stock solutions to give either a 10 mm concentration of xanthine or a 1 mm concentration of lumazine in the new well solution. The crystals were mounted and frozen after 1-5 min. Final diffraction data were collected at Argonne National Laboratory on the SGX Pharmaceuticals, Inc. beamline using a wavelength of 0.9793 Å and a MARCCD 165 detector. Data sets were collected at 2.2 Å resolution for the XO-lumazine complex and 2.6 Å for the desulfo-XO-xanthine complex.
The data were processed using the MOSFLM package of the CCP4 program suite (18, 19). The structure of the enzyme was determined by molecular replacement using the MOLREP package of CCP4, with Protein Data Bank code 1FIQ as the search model (8). Following rigid body refinement in REFMAC of the CCP4 suite, the structure was refined using the restrained refinement protocol in REFMAC (20-24). The weighting term for geometric restraints was adjusted in REFMAC to minimize Rcryst while at the same time minimizing the difference between Rcryst and Rfree. Noncrystallographic symmetry restraints (set on “tight” in REFMAC) were used between the monomers of each respective homodimer in refining the structure of the desulfo enzyme, because this also minimized the divergence between Rcryst and Rfree.
The lumazine and xanthine molecules were constructed using the PRODRG2 server (25), and the respective Protein Data Bank codes were built into the corresponding 2Fo-Fc and Fo-Fc omit electron density maps observed in the XO active sites using COOT (26). Following the merging of the substrates with the respectively refined XO structures, the resulting structures were refined again using restrained refinement in REFMAC. Water molecules were subsequently added to the XO-lumazine electron density maps prior to the addition of the lumazine molecules. All of the images were rendered using PyMol (27).
RESULTS
Overall Structure of Xanthine Oxidase with Lumazine—The overall structure seen in the present XO-lumazine complex was very similar to that seen previously for the oxidized enzyme (8) and the complex with HMP (15). As shown in Table 1, the space group was again P21, and the unit cell dimensions and structure factors were comparable with our previous structure (15). As before, the asymmetric unit contained one dimer, and the unit cell was monoclinic. The overall dimensions of the unit cell were a = 133.2 Å, b = 73.5 Å, and c = 146.5 Å with angles of 90, 98.7, and 90 degrees. As in the case of our previous crystal form with bound HMP (15), in one subunit of the asymmetric unit, electron density for residues 1316-1328 (disordered in the search model used for molecular replacement) became apparent in the electron density map of our final structure. As also seen previously (15), this electron density was rather poorer in quality than throughout the remainder of the map. In addition, after refinement three of the 13 residues in this region were found to assume Ramachandran “disallowed” configurations. As discussed previously (15), given that this additional electron density was manifestly evident in the map, we felt obligated to model it to the best of our ability, and such residues were included in the statistical analysis (below). Still, we emphasize that our confidence in this (very limited) region is somewhat lower than throughout the model at large.
TABLE 1.
Statistics for data collection and refinement
Ramachandran statistics indicate the percentage of residues in the most favored, additionally allowed, generously allowed, and disallowed regions of the Ramachandran diagram as defined by the program PROCHECK (31).
| Statistics | XO with lumazine | Desulfo-XO with xanthine | XO with bound HMP (15) |
|---|---|---|---|
| Protein Data Bank code | 3ETR | 3EUB | 3B9J |
| Space group | P21 | P1 | P21 |
| Resolution (Å) | 26.4-2.2 | 33.1-2.6 | 33.6-2.3 |
| Wavelength (Å) | 0.9793 | 0.9793 | 0.9793 |
| Unique reflections (test set) | 133,695 (7052) | 119,503 (5403) | 114,520 (5785) |
| Completeness % (highest resolution shell, Å) | 98.7 (96.1) | 72.7 (46.8) | 96.3 (90.5) |
| I/σ (highest resolution shell) | 5.0 (1.6) | 6.2 (5.0) | 13.5 (1.8) |
| Rcryst (highest resolution shell) | 19.7 (23.6) | 21.4 (26.5) | 19.4 (25.1) |
| Rfree (highest resolution shell) | 26.7 (34.2) | 26.8 (39.8) | 26.3 (32.5) |
| Ramachandran statistics (%) | 88.2, 10.1, 1.0, 0.7 | 86.5, 12.0 0.9, 0.6 | 87.2, 11.0, 1.0, 0.7 |
| Mean coordinate error based on free R value (Å) | 0.234 | 0.457 | 0.260 |
| Mean coordinate error based on maximum likelihood (Å) | 0.176 | 0.263 | 0.196 |
| Root mean square deviation bond length (Å) | 0.020 | 0.015 | 0.017 |
| Root mean square deviation bond angles (°) | 2.0 | 1.7 | 1.8 |
| Average B factor (A2) | 29.5 | 13.9 | 22.7 |
| Number of nonhydrogen atoms in refinement | 20,307 | 38,070 | 19,852 |
| Number of waters | 1232 | 0 | 861 |
As in previously reported structures of XOR, each monomer consisted of four domains; two N-terminal domains each with a [2Fe-2S] cluster, an FAD-binding domain, and a molybdenum-binding portion of the protein with the molybdenum center sandwiched between two large domains at the C-terminal end of the polypeptide. All of the cofactors in the two subunits of the asymmetric unit were accounted for by both the refined electron density maps and those following omission of the cofactors. The root mean square displacements for the α-carbons of each monomer in the final refined structure of the present complex with lumazine relative to that of oxidized enzyme (Protein Data Bank accession number 1FIQ) were 0.366 and 0.338 Å, respectively (as compared with 0.345 and 0.340 Å for our structure with bound HMP). The relative orientations and distances between the redox-active centers was essentially identical to those reported previously (8, 15).
The final R factors for the XO-lumazine structure were an Rcryst of 19.7% and an Rfree of 26.7% (Table 1), comparable with our previous results with the complex of xanthine oxidase with HMP (Rcryst, 19.4%; Rfree, 26.3%; Table 1). The mean B factor for the structure was 29.5 Å2, as compared with 22.7 Å2 seen with the HMP complex (15). The Ramachandran statistics and structure factors of the structure with lumazine were also very similar to our previous structure of the enzyme complexed with HMP (15). Again, disallowed residues were largely scattered at random throughout the >2500-residue dimer, although in all of the cases the residues were located on the surface of the protein in regions of weaker electron density. Also, as with our structure with HMP (15) and the limazine structure described above, the model for the additional residues 1316-1328 in the one monomer resulted in several disallowed orientations. Again, these residues were included in the statistical analysis, given the clear positive density that was present in the electron density map. Still, including the residues in this small, poorly defined region, only 0.7% of all amino acid residues fell in the disallowed region of the Ramachandran plot.
A MolProbity report on the structure yielded a score of 2.69 with a Clashscore of 17.59 following the addition of hydrogens (28). These metrics compare favorably with those for our previous structure with HMP (15), which had a MolProbity report of 2.66 and Clashscore of 17.58. These values, particularly the MolProbity scores, must be understood in the context of the particularly large size of the dimer in the asymmetric unit (290 kDa) as compared with the mean size of the cohort of structures at comparable resolution and are taken to reflect an acceptable quality of the model here for the structure with lumazine.
Overall Structure of Desulfo Xanthine Oxidase with Xanthine—The structure of the desulfo enzyme with xanthine proved considerably more difficult to obtain. The initial data sets were of low resolution and completeness, and the molecular replacement solution showed that the dimer had dissociated into monomers having little other than crystallographic contacts. However, treatment of the cyanolyzed enzyme with oxidized glutathione resulted in triclinic crystals of space group P1 that had two dimers in the asymmetric unit. An asymmetric unit containing two enzyme molecules has precedent in previous structures of XOR (12), and thus we proceeded to process the data set from 33.7 to 2.6 Å despite a rather low completeness of 73% over that resolution range. The unit cell dimensions for the desulfo structure were a = 73.30 Å, b = 133.18 Å, and c = 142.63 Å with angles of 96.9, 93.1, and 90.0 degrees (Table 1). As was the case for our structures of enzyme in complex with HMP and lumazine, one monomer of each dimer displayed positive density for previously disordered residues. These were residues 1316-1324 and 1316-1326, of the two dimers. Again, all cofactors in the two dimers of the asymmetric unit were accounted for by both the refined electron density maps and those following omission of the cofactors, and again the overall protein fold was essentially identical to that seen previously. The α-carbon root mean square xyz displacements of the four monomers in our asymmetric unit as compared with the 1FIQ search model were 0.314, 0.299, 0.291, and 0.300 Å, somewhat better than our structures with bound lumazine or HMP.
The final R factors for the desulfo-XO-xanthine structure were Rcryst of 21.4% and an Rfree of 26.5%, comparable with the values seen with the HMP- and lumazine-bound structures (Table 1). Ramachandran statistics and structure factors were comparable with our previous structures, with 0.6% of all residues lying in Ramachandran-disallowed geometries, three of these again in the rather poorly defined region 1316-1326 (15). The MolProbity report for our structure of the desulfo enzyme/xanthine complex gave a score of 2.57 and a Clashscore of 15.81. These metrics placed our structure of the xanthine complex in the 78th and 88th percentiles, respectively, of all structures at comparable resolution. The mean B factor for this structure was very low at 13.9 Å2, an apparent manifestation of the incompleteness of the data set; there was no manipulation of TLS parameters in the refinement process, for example, that might lowered the B factor. We note that the average B factor as determined from a Wilson plot, was a more reasonable 27.3 Å2.
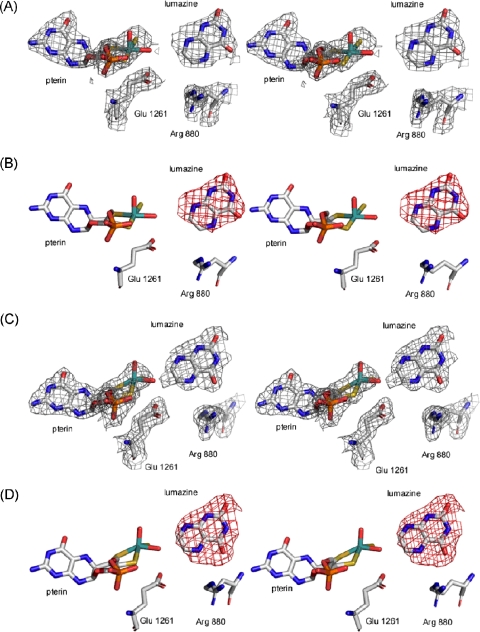
Lumazine and Xanthine Bound in the Active Site of Xanthine Oxidase—Fig. 3 shows the 2Fo-Fc and Fo-Fc omit density maps for the structure of the lumazine complex, contoured at 1.0 and 3.0 σ, respectively, in both active sites of the asymmetric unit. It is clearly evident that the C-4 carbonyl (i.e. the proximal carbonyl, equivalent to C-6 in xanthine given the changed numbering pattern for pteridines relative to purines) is oriented toward Arg-880, with the distal C-2 carbonyl (equivalent to C-2 in purines) directed away from this residue. This orientation of the pyrimidine subnucleus is opposite that seen previously for both HMP (15) and alloxanthine (12), where it is the distal rather than proximal carbonyl of the pyrimidine subnucleus that interacts with Arg-880. The orientation of xanthine in the active site as seen here fully supports our previous hypothesis that effective purine substrates bind in an orientation with the C-6 rather than C-2 oriented toward Arg-880. Attempts to fit the lumazine into the omit density in an inverted orientation places several atoms on the pyrazine ring outside the electron density associated with the heterocycle. In the structure shown in Fig. 3, the C-7 of lumazine (the site ultimately hydroxylated) is positioned 4.0 ± 0.02 and 2.3 ± 0.02 Å from the Mo-OH, and the C-4 carbonyl is 5.9 ± 0.02 and 3.5 ± 0.02 Å from Arg-880 at its nearest point in the two active sites in the final refined structure. The same orientation of lumazine is seen in the two active sites of the asymmetric unit, although the first site (in the subunit showing the resolved electron density for residues 1316-1328) appears relatively less occupied than the second. In the second site, there is some evidence of electron density forming between the C-7 and the Mo-OH, suggesting some formation of a reduced enzyme-product complex with a Mo-OR structure (Fig. 1). Although the lumazine crystal was frozen at approximately the same time (2-3 min) after the addition of lumazine to the mother liquor than was the case with HMP previously, the kinetic behavior of the two substrates resulted in the observation of different species. With HMP, the reaction conditions are known to lead to nearly quantitative accumulation of the reduced enzyme product complex that is evident in the crystal structure (15). With lumazine, the rate constants for formation and decay of this intermediate are such that it does not accumulate to nearly the same degree during turnover.
FIGURE 3.
Stereo images of the active sites of xanthine oxidase with lumazine. Shown are the 2Fo-Fc (panels A and C) and Fo-Fc (panels B and D) maps following omission of the xanthine molecules. 2Fo-Fc maps were contoured at 1.0 σ, and Fo-Fc maps at 3.0 σ. Omit maps were overlaid with the final model (Protein Data Bank code 3ETR). The active site depicted in A and B was the less occupied of the two active sites in the asymmetric unit, and in C and D the more highly occupied (see text). The molybdenum atom is in teal, sulfurs are in yellow, oxygens are in red, and nitrogens are in blue.
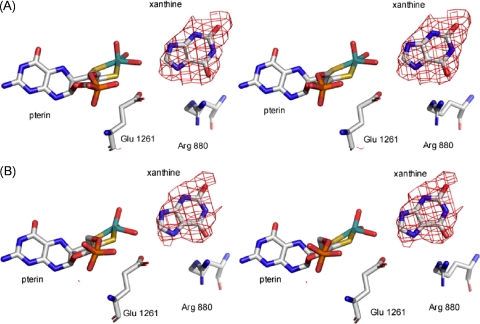
The structure of the desulfo enzyme was much more challenging to obtain than was that of the wild-type enzyme with lumazine. Incubation of XO with KCN has long been known to inactivate the enzyme, although any additional effects on the enzyme structure have not been documented (cyanide can, for example, add across disulfide bonds to give a thiocyanate derivative and a free thiol). Our initial attempts at obtaining a useable data set resulted in several structures that showed the dimer dissociated into two monomers that were ∼20 Å apart. We found empirically that treatment of cyanolyzed enzyme with oxidized glutathione resulted in protein that yielded satisfactory crystals. As shown in Fig. 4, the Fo-Fc omit maps, contoured at 3.0 σ consistently showed the same orientation for xanthine in each active site. The clear electron density omit maps for the xanthine molecules and molybdenum center and the acceptable R factors and overall structural features of the two 290 kDa dimers in the asymmetric unit lead us to conclude that, despite the fact that the diffraction pattern was only 73% complete, the structure here is valid and a true representation of the orientation of xanthine bound in the active site of the enzyme.
FIGURE 4.
Stereo images of the active sites of desulfo-XO with xanthine. Shown in A and B are the more highly occupied active site in each of the two dimers found in the asymmetric unit. Fo-Fc maps were constructed with omission of xanthine and overlaid with the final model (Protein Data Bank code 3EUB). The Fo-Fc maps were contoured at 3.0 σ. The molybdenum atom is in teal, sulfurs are in yellow, oxygens are in red, and nitrogens are in blue.
As in the structure with lumazine, one active site of each dimer showed lower occupancy, again in the subunit showing the additional electron density for residues 1316-1328. As in the case of lumazine above, attempts to fit the xanthine in an orientation opposite that shown resulted in several atoms on the imidazole subnucleus lying outside the electron density for the heterocycle. This was particularly true of the two most highly occupied active sites of the asymmetric unit, where the positive density contoured at 3.0 σ could only be fit by xanthine oriented such that the C-6 carbonyl was directed at Arg-880 as shown. In these two sites, the C-8 of xanthine is positioned 4.6 ± 0.03 and 4.3 ± 0.03 Å from the Mo and 2.7 ± 0.03 and 2.6 ± 0.03 Å from the Mo-OH. The C-6 = O is 2.8 ± 0.03 and 2.7 ± 0.03 Å from the guanidinium group of Arg-880. No evidence for catalysis was seen in any of the four active sites, as expected for the desulfo form of the enzyme (17).
DISCUSSION
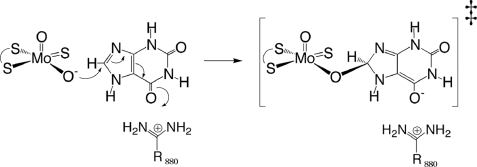
We find here that the orientation of both lumazine and xanthine in the active site of xanthine oxidoreductase is opposite that seen with HMP (15) and the inhibitor alloxanthine (12), with the proximal carbonyl group (C-4=O in the case of the pteridine lumazine and C-6=O in the case of the purine xanthine) rather than the distal carbonyl of the pyrimidine subnucleus oriented toward the active site arginine. We have previously examined the reactivities of a homologous series of purine substrates and their sensitivity to mutation at Arg-310 in the R. capsulatus XDH and predicted that poor substrates with no proximal carbonyl group will bind oppositely to good substrates that do have a proximal carbonyl (or C=S). It is because of their inverted orientation that the poor substrates are less reactive toward enzyme but also much less sensitive to mutation of Arg-310 (Arg-880 in the bovine enzyme). The structures with lumazine and xanthine seen here demonstrate that xanthine, a good substrate, does indeed bind in the opposite orientation to that seen with HMP, as we have predicted. This observation is consistent with a catalytic role for the arginine in which it stabilizes negative charge accumulation on the heterocycle in the course of nucleophilic attack by charge complementation, as shown in Fig. 5. We note that this conclusion implies that the orientation of the inhibitor alloxanthine complexed with the reduced enzyme (12), which is analogous to the orientation seen with HMP rather than xanthine, does not reflect the more catalytically effective orientation of substrate in the active site. It appears that with the alloxanthine lying some 1.6 Å closer to the molybdenum than substrate by virtue of its direct coordination to the metal (displacing the Mo-OH of the molybdenum coordination sphere), its distal rather than proximal carbonyl is better positioned to interact optimally with Arg-880.
FIGURE 5.
Transition state stabilization by Arg-880. The figure was adapted from Ref. 13.
The above crystallographic results provide important information concerning the working of the ensemble of active site residues of XOR in accelerating reaction rate. Glu-1261 of the bovine XOR (Glu-730 in the R. capsulatus XDH) lies at the bottom of the substrate-binding site and is universally conserved in the purine- and aldehyde-hydroxylating enzymes. It is thought to act as a general base, deprotonating the Mo-OH of the molybdenum center to facilitate nucleophilic attack on substrate. Indeed, mutation of Glu-730 to alanine in the R. capsulatus enzyme reduces the limiting rate of enzyme reduction by at least a factor of 107, amounting to ∼10 kcal/mol of transition state stabilization contributed by this residue (32). Furthermore, XAS analysis of the bovine enzyme at pH 10 indicates a shortening of the Mo-O bond that is consistent with spontaneous deprotonation to Mo-O- (33), and indeed the R. capsulatus E730A mutant does gain some activity at high pH.4
Glu-802 of the bovine enzyme (Glu-232 in the R. capsulatus enzyme) lies opposite Glu-1261/730 from substrate, atop the active site. It is strictly conserved only among enzymes that hydroxylate xanthine and is usually absent in aldehyde-oxidizing enzymes. Mutation of this residue to an alanine results in a 12-fold decrease in the limiting rate of enzyme, kred, reduction at high xanthine concentration, and a similar 12-fold increase in Kd for xanthine (13). This residue thus contributes ∼1.5 kcal/mol to transition state stabilization and a similar amount to substrate affinity (32). A comparable effect is seen in the human enzyme when this residue is mutated to a valine (14). A computational study comparing the relative energies of tautomeric forms of xanthine in free solution and in intermediates encountered in the course of catalysis (34) showed that the tautomer with protons on nitrogens 1, 7, and 9 was significantly stabilized relative to the predominant tautomer in solution (with protons on nitrogens 1, 3, and 7) once nucleophilic attack had occurred. Glu-802/232 was proposed to facilitate this tautomerization and thus contribute to rate acceleration. The orientation of substrate seen here in the crystal structure of the desulfo bovine enzyme in complex with xanthine is consistent with such a role, with the unprotonated N-9 of free substrate oriented “up” toward Glu-802/232. It had alternatively been proposed that xanthine bound inverted to the orientation seen here and that this tautomerization was facilitated by Glu-1261/730 (33). It would indeed be elegant were Glu-1261/730 to simultaneously deprotonate the Mo-OH and transfer the proton thus obtained to N-9 of substrate once nucleophilic attack has been initiated. Such a role, however, is predicated on a substrate orientation in the active site opposite to that seen crystallographically here, with the unprotonated N-9 of the imidazole subnucleus oriented “down” rather than up, and now seems unlikely. In the case of HMP, inversion of substrate orientation in the active site also precludes the involvement of Glu-802/232 altogether in facilitating its hydroxylation.
In summary, the present work demonstrates that the orientation of substrate in the active site of xanthine oxidoreductase plays a critical role in determining the catalytic effectiveness of Glu-802/232 and Arg-880/310 in hydroxylation at C-8. Properly oriented, with N-7 toward the glutamate and C-6=O toward the arginine, the reaction proceeds much more rapidly than when substrate binds (as in the case of HMP) in the inverted orientation.
Acknowledgments
We thank Dr. Charles Bell (Department of Molecular and Cellular Biochemistry, The Ohio State University) for consultation and advice regarding the refinement procedures, and for a critical reading of the manuscript.
The atomic coordinates and structure factors (codes 3ETR and 3EUB) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant GM 075036 (to R. H.). This work was also supported in part by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-06CH11357 and by SGX Pharmaceuticals, Inc. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: XOR, xanthine oxidoreductase; XDH, xanthine dehydrogenase; XO, xanthine oxidase; HMP, 2-hydroxy-6-methylpurine.
A. Ibdah and R. Hille, unpublished observations.
References
- 1.Hille, R. (1996) Chem. Rev. 96 2757-2816 [DOI] [PubMed] [Google Scholar]
- 2.Hille, R. (2002) Trends Biochem. Sci. 27 360-367 [DOI] [PubMed] [Google Scholar]
- 3.Hille, R., and Nishino, T. (1995) FASEB J. 9 995-1003 [PubMed] [Google Scholar]
- 4.Nishino, T., Okamoto, K., Kawaguchi, Y., Hori, H., Matsumura, T., Eger, B. T., Pai, E. F., and Nishino, T. (2005) J. Biol. Chem. 280 24883-24894 [DOI] [PubMed] [Google Scholar]
- 5.Elion, G. B. (1989) Science 244 41-47 [DOI] [PubMed] [Google Scholar]
- 6.Pacher, P., Nivorozhkin, A., and Szabo, C. (2006) Pharmacol. Rev. 58 87-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry, C. E., and Hare, J. M. (2004) J. Physiol. 3 589-606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enroth, C., Eger, B. T., Okamoto, K., Nishino, T., Nishino, T., and Pai, E. F. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 10723-10728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huber, R., Hof, P., Duarte, R. O., Moura, J. J. G., Moura, I., LeGall, J., Hille, R., Archer, M., and Romao, M. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 8846-8851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bray, R. C., and Vänngard, T. (1969) Biochem. J. 114 725-734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bray, R. C. (1998) Q. Rev. Biophys. 21 299-329 [DOI] [PubMed] [Google Scholar]
- 12.Truglio, J. J., Theis, K., Leimkühler, S., Rappa, R., Rajagopalan, K. V., and Kisker, C. (2002) Structure (Camb.) 10 115-125 [DOI] [PubMed] [Google Scholar]
- 13.Pauff, J. M., Hemann, C. F., Jünemann, N., Leimkühler, S., and Hille, R. (2007) J. Biol. Chem. 282 12785-12790 [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi, Y., Matsumura, T., Ichida, K., Okamoto, K., and Nishino, T. (2007) J. Biochem. (Tokyo) 141 513-524 [DOI] [PubMed] [Google Scholar]
- 15.Pauff, J. M., Zhang, J., Bell, C. E., and Hille, R. (2008) J. Biol. Chem. 283 4818-4824 [DOI] [PubMed] [Google Scholar]
- 16.Massey, V., Brumby, P. E., Komai, H., and Palmer, G. (1969) J. Biol. Chem. 244 1682-1691 [PubMed] [Google Scholar]
- 17.Massey, V., and Edmondson, D. (1970) J. Biol. Chem. 245 6595-6598 [PubMed] [Google Scholar]
- 18.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 19.Potterton, E., Briggs, P., Turkenburg, M., and Dodson, E. J. (2003) Acta Crystallogr. Sect. D Biol. Crystallogr. 59 1131-1137 [DOI] [PubMed] [Google Scholar]
- 20.Murshudov, G., Vagin, A., and Dodson, E. (1996) The Refinement of Protein Structures, Proceedings of Daresbury Study Weekend, Daresbury, UK
- 21.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 22.Pannu, N. J., Murshudov, G. N., Dodson, E. J., and Read, R. J. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 1285-1294 [DOI] [PubMed] [Google Scholar]
- 23.Murshudov, G. N., Lebedev, A., Vagin, A. A., Wilson, K. S., and Dodson, E. J. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 55 247-255 [DOI] [PubMed] [Google Scholar]
- 24.Winn, M., Isupov, M., and Murshudov, G. N. (2001) Acta Crystallogr. Sect. D Biol. Crystallogr. 57 122-133 [DOI] [PubMed] [Google Scholar]
- 25.Schuettelkopf, A. W., and van Aalten, D. M. F. (2004). Acta Crystallogr. Sect. D Biol. Crystallogr. 60 1355-1363 [DOI] [PubMed] [Google Scholar]
- 26.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 27.DeLano, W. L. (2004) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA
- 28.Lovell, S. C., Davis, I. W., Arendall, W. B., de Bakker, P. I. W., Word, J. M., Prisant, M. G., Richardson, J. S., and Richardson, D. C. (2003) Proteins Struct. Funct. Genet. 50 437-450 [DOI] [PubMed] [Google Scholar]
- 29.Deleted in proof
- 30.Hille, R. (2006) Eur. J. Inorg. Chem. 2006 1913-1926 [Google Scholar]
- 31.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 32.Leimkühler, S., Stockert, A. L., Igarashi, K., Nishino, T., and Hille, R. (2004) J. Biol. Chem. 279 40437-40444 [DOI] [PubMed] [Google Scholar]
- 33.Doonan, C. J., Stockert, A. L., Hille, R., and George, G. N. (2005) J. Am. Chem. Soc. 127 4518-4522 [DOI] [PubMed] [Google Scholar]
- 34.Ilich, P., and Hille, R. (1997) Inorg. Chim. Acta 263 87-94 [Google Scholar]