Abstract
Rhodobacter capsulatus xanthine dehydrogenase (XDH) is an (αβ)2 heterotetrameric cytoplasmic enzyme that resembles eukaryotic xanthine oxidoreductases in respect to both amino acid sequence and structural fold. To obtain a detailed understanding of the mechanism of substrate and inhibitor binding at the active site, we solved crystal structures of R. capsulatus XDH in the presence of its substrates hypoxanthine, xanthine, and the inhibitor pterin-6-aldehyde using either the inactive desulfo form of the enzyme or an active site mutant (EB232Q) to prevent substrate turnover. The hypoxanthine- and xanthine-bound structures reveal the orientation of both substrates at the active site and show the importance of residue GluB-232 for substrate positioning. The oxygen atom at the C-6 position of both substrates is oriented toward ArgB-310 in the active site. Thus the substrates bind in an orientation opposite to the one seen in the structure of the reduced enzyme with the inhibitor oxypurinol. The tightness of the substrates in the active site suggests that the intermediate products must exit the binding pocket to allow first the attack of the C-2, followed by oxidation of the C-8 atom to form the final product uric acid. Structural studies of pterin-6-aldehyde, a potent inhibitor of R. capsulatus XDH, contribute further to the understanding of the relative positioning of inhibitors and substrates in the binding pocket. Steady state kinetics reveal a competitive inhibition pattern with a Ki of 103.57 ± 18.96 nm for pterin-6-aldehyde.
Rhodobacter capsulatus xanthine dehydrogenase (XDH4; EC 1.17.1.4) is a cytoplasmic enzyme that is highly identical to eukaryotic xanthine oxidoreductases. Despite differences in subunit composition, the folds of bovine XDH and R. capsulatus XDH are very similar (1). The bacterial enzyme can be described as a butterfly-shaped (αβ)2 heterotetramer. Each (αβ) dimer represents one half of the active molecule and is encoded by two separate gene products, termed XdhA and XdhB, unlike the (α)2 dimeric eukaryotic protein, which is derived from a single polypeptide chain (2). Each subunit of the (αβ) heterodimer carries a specific set of cofactors, which are crucial for catalysis and electron transfer. The 50-kDa XdhA subunit harbors two [2Fe2S] clusters as well as a FAD cofactor; the 85-kDa XdhB subunit contains the molybdenum cofactor harboring a catalytically essential terminal sulfido ligand (1, 2). This cofactor is part of the active site binding pocket and catalyzes the oxidative hydroxylation of hypoxanthine to xanthine and further to uric acid. Most XDHs, with the exception of R. capsulatus and avian XDH, can be converted to the oxidase form (XO) while losing their ability to use NAD+ as the electron acceptor (3, 4).
The catalytic sequence of R. capsulatus XDH is initiated by abstraction of a proton from the Mo-OH group by the highly conserved active site residue GluB-730 (where B indicates the XdhB subunit), followed by nucleophilic attack of the resulting Mo-O- on the carbon center of the substrate (C-2 in hypoxanthine and C-8 in xanthine) and concomitant hydride transfer to the Mo=S of the molybdenum center (3). Residue GluB-232, on the other hand, is involved in both substrate binding and transition state stabilization (3, 5). Mutation of GluB-232 to alanine leads to a 12-fold increase in the KD for xanthine (3). It has been suggested that interaction of ArgB-310 with the C-6 carbonyl group of the substrate xanthine stabilizes negative charge accumulation on the heterocycle that accompanies nucleophilic attack at C-8, thus stabilizing the transition state and accelerating the reaction of substrate oxidation (6). However, oxypurinol and 2-hydroxy-6-methylpurine were shown to bind in the opposite orientation in the active site, with the C-4 of oxypurinol facing GluB-232 in the R. capsulatus enzyme and C-2 of 2-hydroxy-6-methylpurine facing Arg-880 in bovine XO (7, 8).
Allopurinol (1-H-pyrazolo [3,4-d] pyrimidine-4-one), developed in 1963, is the current clinical treatment option for patients exhibiting symptoms of hyperuricemia, indicative of gout. The main drawback to allopurinol administration in humans is the possible onset of a toxicity syndrome manifested as eosinophilia, vasculitus, rash hepatitis, and progressive renal failure (9). This is most likely due to the inhibitory effect of allopurinol and its metabolites on other enzymes, such as purine nucleoside phosphorylase and orotidine-5′-monophosphate decarboxylase (10). Allopurinol is oxidized by XDH to oxypurinol (1,2-dihydropyrazolo [4,3-e] pyrimidine-4, 6-dione) (Fig. 1), which commits suicide inhibition of XDH by replacing the hydroxyl ligand of the molybdenum ion and thereby inhibiting further catalysis (8).
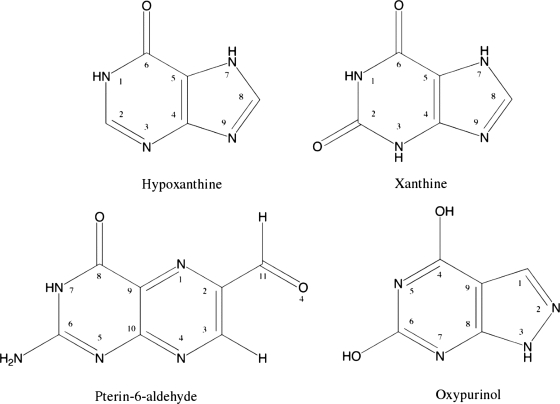
FIGURE 1.
Schematic representation of the substrates hypoxanthine and xanthine and the two inhibitors pterin-6-aldehyde and oxypurinol analyzed in this study.
Pterin-6-aldehyde (Fig. 1) is a potent inhibitor of XO and is only found in the urine of cancer patients, a discovery that could play a key role in early cancer detection (11, 12). Inhibition of XO by pterin-6-aldehyde is on the same order as that for oxypurinol, with a Ki = 10-9m (13-17). Because of its very slow rate of conversion to pterin-6-carboxylic acid, pterin-6-aldehyde is considered to be a very good inhibitor for XO (11, 14, 18).
Here we describe the first structures of the desulfo form and an active site variant EB232Q of R. capsulatus XDH in the presence of its substrates xanthine and hypoxanthine as well as the pterin-6-aldehyde inhibited structure of the wild-type protein. The structures reveal an intricate hydrogen-bonding network that can accommodate the substrates in the orientation required for catalysis and allow a tight approach of the inhibitor pterin-6-aldehyde toward the molybdenum cofactor. Furthermore, the comparison between the xanthine- and oxypurinol-bound structures provides an explanation of why the inhibitor is bound in an opposite orientation relative to the xanthine molecule.
EXPERIMENTAL PROCEDURES
Expression and Purification of R. capsulatus XDH in Escherichia coli Cells—R. capsulatus wild-type XDH was expressed and purified as described previously (3). The mutation EB232Q was introduced through PCR mutagenesis into R. capsulatus XDH. The generated variant in addition to the EB232A variant was purified by nickel-nitrilotriacetic acid chromatography, ion exchange chromatography (Q-Sepharose; GE Healthcare), and size exclusion chromatography (Superose 12; GE Healthcare). To obtain the desulfo form of XDH, XdhAB were expressed from plasmid pMS3 (19) in the absence of XdhC in E. coli TP1000 cells (20). The cells were grown for 24 h at 210 rpm in LB medium supplemented with 1 mm Na2MoO4, 150 μg/ml ampicillin, and 20 μm isopropyl β-d-thiogalactopyranoside. Purification was achieved by nickel-nitrilotriacetic acid chromatography, and ion exchange chromatography using Q-Sepharose. To separate Moco-containing XDH from the enzyme lacking the cofactor, affinity chromatography on a Sepharose 4B/folate gel was used (3). Finally, the protein was purified by size exclusion chromatography. The different XDH variants in 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 200 mm NaCl, and 2.5 mm dithiothreitol were concentrated to 15 mg/ml. Crystallization trials followed immediately, and the remaining protein was stored for up to 5 days at -80 °C.
Enzyme Assays—Enzyme assays were carried out at 25 °C in 50 mm Tris and 1 mm EDTA, pH 7.5, in a final volume of 1 ml. Routine assay mixtures contained 1 mm NAD+ and various concentrations of xanthine or pterin-6-aldehyde. XDH activities were determined by monitoring the absorbance changes at 340 nm for NAD+ reduction. The kinetic parameters were obtained by nonlinear fitting using the Microcal Origin 6.0 program. The data for enzyme inhibition were additionally obtained through a double reciprocal plot (1/v ≈ 1/[S]).
Crystallization—Wild-type XDH, the EB232A and EB232Q variants as well as the desulfo XDH were crystallized via the hanging drop vapor diffusion method equilibrating equal volumes of the wild-type protein with the reservoir solution containing 6-8 mm BaCl2, 6-8% polyethylene glycol 8000, 100 mm Tris-HCl, pH 8.3, 5-25 mm dithiothreitol, and 3-4% isopropanol, whereas the EB232Q variant was mixed in a 1:2 ratio with the same reservoir solution. Prior to crystallization set-ups desulfo XDH was rebuffered into 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, and 2.5 mm dithiothreitol. The structures of the inhibitor-bound wild type, the hypoxanthine-bound EB232Q crystals, as well as the hypoxanthine-bound desulfo XDH were obtained through soaking experiments. The xanthine-bound desulfo XDH was obtained by cocrystallization with a saturated xanthine solution. The inhibitor experiment was performed in the dark because of its light sensitivity. Because of its low solubility, the inhibitor was first suspended in 60 mm NaOH and then mixed in a 1:50 ratio with the reservoir solution. The final concentration is unknown because pterin-6-aldehyde failed to dissolve completely. The EB232Q and desulfo XDH crystals were soaked in a solution containing 60 mm NaOH and 10 mm hypoxanthine. After 12 h of soaking, the crystals were transferred into a cryoprotectant containing 30% glycerol and the mother liquor components and subsequently cryo-cooled in liquid nitrogen.
Data Collection and Structure Solution—Data of the EB232Q, the EB232A mutant, and the wild-type protein were collected at beamline X26C at the National Synchrotron Light Source (Brookhaven National Laboratory) using an ADSC Quantum 4 charged coupled device. The diffraction data were indexed, integrated, and scaled using the HKL suite (21). Data for the desulfo XDH crystals were collected at the microfocus beamline ID23-2 at the European Synchrotron Radiation Facility using a MarCCD 225 detector. The diffraction data were indexed and scaled using Mosflm and Scala from the CCP4 suite (22). All of the crystals belonged to space group P1 and contained two (αβ)2 heterotetramers in the asymmetric unit. Representative unit cell dimensions, taken from the EB232Q-hypoxanthine structure, are a = 92.7 Å, b = 140.6 Å, c = 157.6 Å, α = 109.5°, β = 106.1°, and γ = 101.1°. Difference Fourier methods were sufficient to solve the new structures because all R. capsulatus crystals belong to the same space group as the previously solved apo structure (Protein Data Bank code 1JRO) (8). Model building was initiated through rigid body refinement of the four monomers, with the XdhA and XdhB subunits of each monomer considered as separate domains. Non-crystallographic symmetry restraints between the XdhA domains and XdhB domains of each monomer were subsequently applied. The EB232A and EB232Q variant structures were further refined using XPLOR (23) followed by REFMAC (TLS refinement) (22), separating the monomer further into three domains: the [2Fe2S] domain, the FAD domain, and the molybdenum cofactor domain, to aid in modeling anisotropic motion of the individual components. The structures of the desulfo form were refined with REFMAC without the utilization of TLS. The models were analyzed in O (24) and Coot (25) to check for difference density at the location of the cofactors and the proposed location of the substrates or inhibitor. Cofactors were added at this point, and restrained refinement continued, employing non-crystallographic symmetry averaging or/and TLS refinement, to obtain the best nonmodel biased difference density and facilitate substrate or inhibitor insertion. After model completion, the different complexes were refined using REFMAC (see Table 1) using medium weighted positional and thermal non-crystallographic symmetry restraints for the protein moiety excluding ligands. To support the refinement of the alternate xanthine conformations, xanthine was included in the non-crystallographic symmetry restraints using the medium weighting scheme. The B factors for all structures were refined isotropically. Outliers of the Ramchandran plot being in proximity to the active site are the residues SerB-458, ArgB-342, and SerB-233 (and all corresponding copies) and are clearly defined in the electron density. Because they were already observed in 1JRO, it is unlikely that these are of functional significance related to substrate binding. Calculation of omit maps was performed omitting all cofactors, substrates, and the active site residues GlnB-232, ArgB-310, ThrB-460, GluB-730, PheB-344, and PheB-459.
TABLE 1.
Data collection and refinement statistics
The values in parentheses refer to the highest resolution shell. Rsym = ∑hkl{∑|I - <I>|/∑<I>}. Rcryst = ∑∥Fo| - |Fc∥/|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively. I/σI indicates the average of the intensity divided by its average standard deviation. Rfree is the same as Rvalue for the 5% of the data randomly omitted from refinement. Ramachandran statistics indicate the fraction of residues in the most favored, additionally allowed, generously allowed, and disallowed regions of the Ramachandran diagram, as defined by PROCHECK.
| E232Q-hypoxanthine | Wild-type pterin-6-aldehyde | Desulfo XDH hypoxanthine | Desulfo XDH xanthine | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P1 | P1 | P1 | P1 |
| Cell dimensions | ||||
| a, b, c (Å) | 92.7, 140.6, 157.6 | 92.6, 140.7, 157.9 | 92.8, 139.9, 158.1 | 93.8, 140.6, 158.2 |
| α, β, γ (°) | 109.5, 106.1, 101.1 | 109.6, 105.8, 101.2 | 109.3, 106.2, 101.1 | 109.6, 105.9, 101.2 |
| Resolution (Å) | 50-3.40 (3.52-3.40) | 50-3.30 (3.36-3.30) | 51-2.90 (3.06-2.90) | 50.6-2.60 (2.74-2.60) |
| Completeness (%) | 93.9 (69.0) | 99.1 (98.6) | 98.2 (97.9) | 97.8 (97.3) |
| Rsym | 0.152 (0.443) | 0.185 (0.651) | 0.122 (0.567) | 0.128 (0.672) |
| I/σI | 7.8 (1.9) | 8.9 (2.6) | 7.4 (1.4) | 7.7 (1.2) |
| Redundancy | 3.5 (2.7) | 4.0 (4.0) | 2.0 (2.0) | 2.0 (1.9) |
| Refinement | ||||
| Resolution (Å) | 50-3.40 | 50-3.30 | 51.2-2.90 | 50.1-2.60 |
| No. reflections | 83,695 | 96,884 | 141476 | 196212 |
| Rcryst/Rfree | 0.22/0.27 | 0.18/0.22 | 0.24/0.28 | 0.23/0.28 |
| No. atoms | ||||
| Protein/cofactors/ions | 36,699 | 36,708 | 36853 | 36858 |
| Ligand | 40 | 56 | 40 | 44 |
| Water | 43 | 0 | 81 | 67 |
| Root mean square deviations | ||||
| Bond lengths (Å) | 0.007 | 0.006 | 0.006 | 0.011 |
| Bond angles (°) | 1.08 | 0.99 | 0.98 | 1.43 |
| Ramachandran statistics (%) | 84.4/14.2/0.9/0.1 | 87.6/11.3/0.7/0.2 | 87.5/11.5/0.8/0.2 | 86.2/12.5/1.1/0.2 |
RESULTS
Steady State Kinetics of the XDH Variant EB232Q—The crystal structure of R. capsulatus XDH with oxypurinol bound at the active site revealed that the six-membered ring of the inhibitor interacts with GluB-232, ArgB-310, and GluB-730, suggesting that these amino acids are either involved in substrate binding or in substrate oxidation (8). Kinetic analyses clearly established GluB-232 to be important for both substrate binding and transition state stabilization and GluB-730 to be essential for catalysis (3). These results favored a reaction mechanism, in which GluB-730 acts as an active site base that abstracts the proton from the Mo-OH group, which in turn undertakes a nucleophilic attack on the C-8 position of the substrate xanthine. We generated the EB232Q XDH variant to reduce turnover of the substrate while maintaining the geometric environment of the active site. Analysis of the cyanolyzable sulfur of the purified protein showed that the EB232Q variant was purified with a Mo=S content of about 56%. Steady state kinetics of the recombinant protein with varying concentrations of xanthine and NAD+ were obtained by monitoring absorbance increases at 340 nm for NADH production. The kcat (xanthine) and Km (xanthine) for the EB232Q variant were found to be 1.16 ± 0.08 s-1 and 55.21 ± 3.10 μm, respectively. This indicates considerably slower rates for substrate turnover of the EB232Q mutant, because kcat was decreased by a factor of 90 from wild type (108 ± 1.5) (3). However, the Km (xanthine) remained the same (64.4 ± 0.9 for the wild type). In comparison with the EB232A variant (3), the activity was further decreased almost 3-fold. Thus the EB232Q variant seems to be better suitable to obtain structural information of a substrate-bound complex in contrast to the EB232A variant.
Structural Features of the EB232Q-Hypoxanthine Complex—Crystals of the EB232Q variant were incubated with its substrate hypoxanthine. The mutant crystallized in the same space group as the wild-type enzyme and displayed two (αβ)2 heterotetramers per asymmetric unit. The 3.4 Å structure was refined to an R factor of 0.22 and an Rfree of 0.27 (Table 1). A superposition of the EB232Q variant with the native enzyme yielded a root mean square deviation of 0.5 Å for all Cα atoms. The electron density for the substrate was best defined in the first monomer (chains A and B; Fig. 2A), and therefore the subsequent analysis is based on this molecule. The hydrogen bonds discussed for this structure have, because of the low resolution, a qualitative character and should be seen as possible hydrogen bonds. However, the clear electron density for the involved side chains and ligands allowed us to unambiguously identify the position and interacting residues of the ligand.
FIGURE 2.
A, the active site of the EB232Q variant with bound hypoxanthine. The Fo-Fc electron density omit map is shown in blue contoured at 2.5 σ for the GlnB-232, ArgB-310, ThrB-460, and GluB-730 residues, the Moco, and the hypoxanthine. The panel is shown in stereo. The substrate is located within bonding distance to the Mo-O ligand of the cofactor and thus poised for catalysis. B, possible hydrogen bonding contacts between the substrate, the protein backbone, and the Moco are shown as blue dashed lines. The locus for hypoxanthine to xanthine catalysis is depicted as a red dashed line between the Mo-O ligand and the C-2 of the substrate.
The six-membered ring of the substrate is sandwiched between two phenyalanine residues: about 3.4 Å from and parallel to PheB-344 and perpendicular to PheB-459 (Fig. 2B), forming van der Waals' interactions with both. The N-1 atom of the substrate is in hydrogen bonding distance to the Mo-O moiety of the molybdenum cofactor. The O-6 atom could form a hydrogen bond to the guanidinium group of ArgB-310. The five-membered ring forms hydrogen bonding contacts via its N-7 to the side chain hydroxyl group of ThrB-460. Residue GlnB-232 serves to anchor the substrate by forming hydrogen-bonding contacts to N-3. The most important contact made is that between the C-2 of hypoxanthine and the Mo-O with a distance ranging from 2.4 to 3.3 Å in the four molecules in the asymmetric unit. The C-2 to oxygen interaction distance is closer than the 3.4 Å extent of the van der Waals' radii for the two atoms, indicating that the substrate is poised to abstract the O from the Mo-O. However, the reaction has not proceeded far enough to enable formation of a C-2=O bond, which would be indicated by a distance on the order of 1.2 Å. The substrate is thus strongly coordinated to residues within the active site and thereby ideally oriented for catalysis to form the product xanthine.
We also solved the structure of the EB232A variant in the presence of xanthine (Rsym = 0.13, Resolution = 2.7 Å Completeness = 98.7%, Rcryst/Rfree = [0.242/0.286]). The structure revealed that the size of the amino acid is also a critical factor for positioning the substrate correctly. Multiple orientations of the bound xanthine molecule were observed in the active site, which could not be modeled correctly (data not shown). This, however, underlines the importance of this side chain for substrate binding and positioning in the active site.
Structure of Desulfo XDH in the Presence of Hypoxanthine—For further analysis of substrate binding and to exclude changes in substrate binding caused by the EB232Q amino acid exchange, we pursued the structural characterization of the R. capsulatus desulfo XDH in the presence of hypoxanthine (Fig. 3). The 2.9 Å structure was refined to an R factor of 0.24 and an Rfree of 0.28 (Table 1). In all of the active sites in the asymmetric unit, the hypoxanthine is positioned with its C-2 toward the molybdenum and forms van der Waals' interactions with PheB-344 and PheB-459 and hydrogen bond interactions with ArgB-310, ThrB-460, and GluB-232, with similar distances as observed in the EB232Q variant. The observed hydrogen bonding pattern supports our observation from the low resolution EB232Q structure in complex with hypoxanthine and underlines the significance of the structural data. A superposition of the EB232Q and the desulfo hypoxanthine containing XDH structures reveals that the substrate orientation is similar in the two structures (Fig. 3C). In both structures the C-2 of the hypoxanthine points toward the molybdenum; however, the C-2 to Mo-O distance is increased approximately 0.6 Å in the desulfo form. Nevertheless both structures clearly reveal the orientation of the hypoxanthine in the active site with O-6 pointing toward ArgB-310 and C-2 being oriented toward the molybdenum cofactor.
FIGURE 3.
A, Fo-Fc electron density omit map contoured at 2.5 σ is shown for the active site residues, the substrate, and the molybdenum cofactor. B, hypoxanthine bound to the active site of desulfo XDH. Interactions are shown as described in the legend to Fig. 2. C, superposition of the hypoxanthine-bound complexes to the active site of the EB232Q (yellow) and the desulfo XDH (blue).
Structure of Desulfo XDH in the Presence of Xanthine—The 2.6 Å structure of desulfo XDH in the presence of xanthine was refined to an R factor of 0.23 and an Rfree of 0.28 (Table 1). During the refinement process the electron density maps indicated two possible orientations of the xanthine molecule. The first orientation represents the substrate complex in which xanthine is positioned with its C-8 toward the molybdenum cofactor. The second orientation provides a view of the product complex after hypoxanthine has been oxidized to xanthine, i.e. the O-2 of xanthine is positioned toward the molybdenum cofactor. The resolution of 2.6 Å does not support the full refinement of alternate conformations. However, because only these two conformations are supported by the electron density maps, we opted to refine them with an occupancy of 0.5 each. Both orientations reveal again an intricate hydrogen-bonding network with active site residues. In the substrate orientation the xanthine molecule forms interactions with ArgB-310, ThrB-460, and GluB-232 (Fig. 4A), albeit with different atoms of the substrate because of the altered orientation compared with the hypoxanthine molecule. O-6 of xanthine forms hydrogen bonds to the main chain N of ThrB-460 as well as to the guanidinium group of ArgB-310. The opposite side of the substrate is held in place through a hydrogen bond formed between GluB-232 to N-9. Further, PheB-344 and PheB-459 form van der Waals' interactions to the substrate as observed in the hypoxanthine structure. In all four active sites the C-8 atom of xanthine is positioned toward the molybdenum cofactor at a distance of 2.5-3.2 Å, indicating that the substrate is poised to abstract the O from the Mo-O, but the reaction cannot proceed because of the utilization of the desulfo form of XDH.
FIGURE 4.
Xanthine bound in the substrate-bound form (A) and in the product-bound form (B) to the active site of desulfo XDH. Possible hydrogen bonds are shown as blue dashed lines, and the proximity of the C-8 atom of xanthine to the Mo-O ligand is depicted by a red dashed line. C, Fo-Fc electron density omit map contoured at 2.5 σ is shown in stereo for the active site residues, the substrate in both conformations, and the molybdenum cofactor. D, superposition of the xanthine-bound (green) and the oxypurinol-bound (cyan) XDH structures.
In the product-bound form, in which the xanthine molecule points with its O-2 atom toward the molybdenum cofactor, the molecule is held in place by the same interactions as described for the hypoxanthine structure, although the two molecules are slightly shifted with respect to each other so that the O-2 of xanthine occupies approximately the same position as the C-2 in hypoxanthine, thus providing a glance how the substrate hypoxanthine is repositioned once it is oxidized to xanthine and is subsequently released from the substrate-binding pocket (Fig. 4B). Because the more relevant structure is revealed by the substrate complex, all comparisons with other complexes and structures were carried out using the substrate structure with the C-8 positioned toward the Mo-O.
Despite the differences of the two substrate complexes as well as the product complex, the same amino acids within the active site are utilized to form hydrogen bonds. The hypoxanthine and the xanthine-bound structures thus clearly identify GluB-232, ArgB-310, and ThrB-460 as important residues to position the substrates optimally for catalysis.
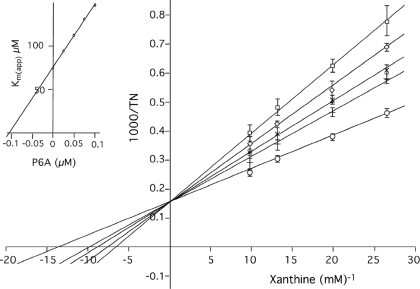
Steady State Kinetics and Structural Features of Wild-type XDH with Pterin-6-aldehyde—To analyze the inhibitory effects of pterin-6-aldehyde on R. capsulatus XDH, steady state kinetics were performed with varying concentrations of xanthine and pterin-6-aldehyde using NAD+ as the electron acceptor. A representative Lineweaver-Burk plot is shown in Fig. 5. As reported previously for XO, pterin-6-aldehyde exhibits a competitive inhibition pattern. Analysis of the kinetic results showed a Ki value of 103.57 ± 18.96 nm, which is 100-fold higher than that reported for XO (1.78 nm) (26).
FIGURE 5.
Lineweaver-Burk plots for the inhibition of xanthine-NAD+ activity in the presence of pterin-6-aldehyde. ○, without pterin-6-aldehyde; +, 25 nm; ×, 50 nm; ⋄, 70 nm, □, 100 nm pterin-6-aldehyde. Inset, the Ki value was obtained from a secondary plot of the apparent Km values versus the inhibitor concentration.
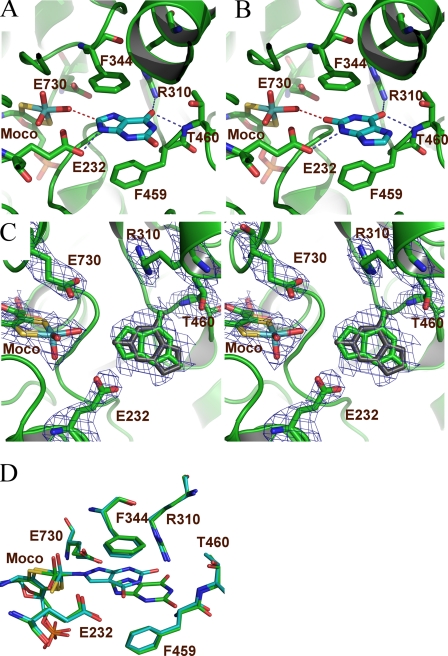
The 3.3 Å crystal structure of wild-type XDH in complex with pterin-6-aldehyde reveals that the two rings of the inhibitor are positioned in a slightly different orientation compared with hypoxanthine and xanthine; pterin-6-aldehyde has undergone a small lateral translation as well as an almost 30° rotation away from the cofactor, effectively blocking access to the binding pocket (Fig. 6). The active site maintains its architecture, and the overall superposition reveals a 0.4 Å root mean square deviation compared with the EB232Q-hypoxanthine structure for all Cα atoms and a root mean square deviation of 0.3 Å for the xanthine-bound structure. The inhibitor can be well placed within the electron density, which is best defined in the third monomer, and all distance calculations derive from this complex. Like the substrate-bound structures, the inhibitor is sandwiched between the two phenyl rings PheB-344 and PheB-459. The hydrogen bond network is increased compared with the xanthine and hypoxanthine-bound structures, leading to an involvement of GluB-730, being in hydrogen bonding distance via its carboxylate group to the N-6 of pterin-6-aldehyde. GluB-730 does not form any substrate interactions in the other complexes within our analysis. In addition the N-6 of pterin-6-aldehyde is likely to form a hydrogen bond to the Mo-O of the Moco. Unlike hypoxanthine and xanthine, which have no amino group jutting out from the ring structures, pterin-6-aldehyde can also form a hydrogen bond through this moiety to the Mo-O ligand, thereby locking the inhibitor tightly into place. N-4 can contact the other catalytically important glutamic acid, GluB-232, via its carboxylate group. O-8, like O-6 of hypoxanthine and xanthine, is in hydrogen bonding distance to the guanidinium group of ArgB-310 and to the main chain N of ThrB-460. The aldehyde group only forms hydrophobic interactions with the neighboring ProB-306 and LeuB-461. The high number of contacts indicates that the inhibitor is firmly situated within the active site, thereby inhibiting access of native substrates.
FIGURE 6.
A, pterin-6-aldehyde (cyan) binds in a similar fashion as hypoxanthine within the active site of wild-type XDH. The Fo-Fc electron density omit map is shown in dark blue contoured at 2.5 σ for the inhibitor, the Moco, and the key residues GluB-232, ArgB-310, ThrB-460, PheB-459, AlaB-529, and GluB-730. B, possible hydrogen bonding interactions observed for pterin-6-aldehyde are shown as blue dashed lines.
DISCUSSION
We cocrystallized the R. capsulatus XDH variant EB232Q with hypoxanthine in addition to the desulfo enzyme with hypoxanthine and xanthine. These structures provide a thorough insight into the binding of both substrates in the active site. The structure of the desulfo enzyme in the presence of xanthine revealed two possible xanthine orientations. One of them being the substrate orientation the other one being xanthine in a product-bound state from the conversion of hypoxanthine to xanthine. Because the desulfo enzyme is catalytically incompetent, and the xanthine concentration in the experiment is a thousand times above the KD for xanthine, both orientations are likely to occur. This is further supported by the well known feature of substrate inhibition of eukaryotic XORs (27, 28). The wild-type enzyme shows substrate inhibition at xanthine concentrations >200 μm (data not shown), which is well in line with previously measured xanthine concentrations for substrate inhibition of XORs from various organisms. Thus our structure describes a feasible mechanism for the observed xanthine substrate inhibition of XORs. Furthermore, the structure of the wild-type enzyme was solved with the inhibitor pterin-6-aldehyde. Crystallization involving the R. capsulatus XDH EB232A variant in the presence of xanthine revealed that the position of the substrate could not be clearly defined because of the openness of the active site resulting from the substitution of the glutamate by an alanine side chain (data not shown). This result indicates the importance of GluB-232 for substrate binding and positioning at the active site. In contrast, the EB232Q variant preserved the active site architecture and has lost the biochemical properties of a glutamate, and thus the variant maintained binding of hypoxanthine in a similar manner as the inactive desulfo enzyme.
The R. capsulatus XDH EB232Q variant and the desulfo form allowed the first structural characterization of substrate-bound complexes and demonstrate that both molecules are ideally positioned for catalysis within the active site through a combination of stacking interactions and hydrogen bonds. Intriguingly both substrates utilize the same amino acids for the formation of hydrogen bonds despite their different orientation. Compared with pterin-6-aldehyde, however, both are more loosely held in place in terms of overall interactions within the active site, thereby facilitating product release and subsequent oxidation to uric acid. This suggests that the inhibitor is situated more tightly within the active site and thereby prevents the binding of native substrates.
The EB232Q and the desulfo hypoxanthine complex both reveal that the C-2 atom approaches the Mo-O moiety at distances of 2.9-3.9 Å, respectively (Figs. 2 and 3). The C-8 atom in the xanthine-bound structure approaches the Mo-O at a distance of ∼3.0 Å (Fig. 4). In contrast, the previously determined 3.0 Å structure of R. capsulatus XDH with oxypurinol (Fig. 7) showed that its N-2 atom coordinates directly to the molybdenum and obscures the hydroxyl moiety of the cofactor (oxypurinol numbering scheme taken from Massey (8, 15)). All three molecules form contacts to residues GluB-232 and ArgB-310 at similar distances, whereas oxypurinol also interacts with the catalytic GluB-730. Most striking, however, is the direct comparison between the orientation of the bound substrate xanthine and the inhibitor oxypurinol. Although the O-6 of xanthine forms a hydrogen bond to ArgB-310, it is the O-6 of oxypurinol that interacts with the same residue, whereas O-4 of the inhibitor forms hydrogen bonds to GluB-232. This orientation is stabilized through additional interactions formed between GluB-730 with the N-3 and through a water-mediated hydrogen bond to N-7 of the inhibitor. These interactions are only possible through the tight approach of the inhibitor toward the molybdenum and thereby stabilize an orientation, which is opposite to the one observed in the xanthine-bound structure (Fig. 4C). Xanthine cannot form these interactions with a carbon at position 8 and therefore does not enter the active site as deeply to generate direct interactions to GluB-730.
FIGURE 7.
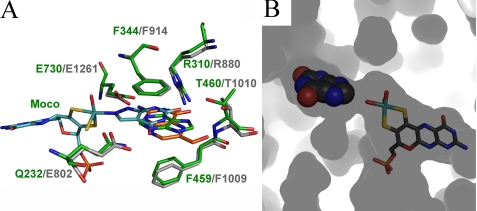
A, superpositions of hypoxanthine, oxypurinol and pterin-6-aldehyde, with respect to the hypoxanthine E232Q structure. Hypoxanthine is shown in green, whereas oxypurinol is shown in cyan. The new inhibitor structure of pterin-6-aldehyde is shown in orange, surrounded by the key residues GlnB-232, ArgB-310, ThrB-460, and GluB-730 as well as the Moco. Residues of the bovine enzyme are indicated in gray. B, inside out view of the binding pocket surface of the active site with xanthine shown in spheres and the Moco shown in all-bonds representation.
Prior to our structural studies it has been proposed that the O-6 of xanthine interacts with ArgB-310 in the active site (6). The orientation of xanthine was supported by kinetic studies utilizing a homologous series of purines as substrates in addition to mutant studies of ArgB-310 (6). Our analysis of the hypoxanthine and xanthine-bound structures and the previous study clearly support the importance of ArgB-310 for correct positioning of the substrates in the active site. ArgB-310 does not interact directly with the two carbon atoms that undergo the hydroxylation reaction but has been proposed to lower the activation energy for the reaction by stabilizing negative charge accumulation on the heterocycle through an electrostatic interaction with the C-6 carbonyl oxygen of the substrate. All of the structures clearly reveal hydrogen bonds between the O-6 of the substrate and the guanidinium group of ArgB-310, thus providing further support for the importance of this residue. Our results are, however, in contrast to a model proposed for the human xanthine oxidoreductase by Yamaguchi et al. (29), who suggested an orientation of the substrate similar to that observed for oxypurinol. Based on the structural identity of the active sites, however, and because the orientation of their substrate is hypothetical without supporting structural data, we believe that eukaryotic XORs bind the substrate in the same orientation as observed in our bacterial XDH complexes.
In contrast, the crystal structure of bovine XO in the course of the reaction with the poor substrate 2-hydroxy-6-methylpurine has been solved (7), in which the bound 2-hydroxy-6-methylpurine molecule was clearly oriented with the C-2 rather than the C-6 position oriented toward Arg-880 (corresponding to ArgB-310 in R. capsulatus XDH). However, because 2-hydroxy-6-methylpurine contains a methyl group at the C-6 position, it was expected to bind in the opposite direction at the active site because of steric hindrance, as suggested previously by kinetic studies (6).
The small 5 × 3 × 5 Å rectangular prismic nature of the active site-binding channel dictates that substrates, key inhibitors, and drugs are small planar molecules. All of the molecules evaluated here superimpose well in a small area sandwiched between two phenylalanine rings (PheB-344 and PheB-459 in R. capsulatus and Phe-913 and Phe-1008 in the bovine enzyme), which dictate the necessity for a planar molecule (Fig. 7). GluB-730, GluB-232, ArgB-310, and ThrB-460 in R. capsulatus and the corresponding residues Glu-1261, Glu-802, Arg-880, and Thr-1010 in the bovine enzyme define the other two walls of the binding pocket and provide the necessary atoms to form hydrogen bonds, which position the substrates for catalysis and as proposed for ArgB-310 (Arg-880 in the bovine enzyme) lower the activation energy of the reaction. One of the ring moieties of each of the six substrates and inhibitors therefore assume similar positions and stack on top of each other. The other ring is either in close proximity to the molybdenum cofactor, in the case of hypoxanthine, xanthine, and oxypurinol, or pushed back toward the entrance of the active site-binding channel, as observed for pterin-6-aldehyde.
Because all of the structures analyzed here contain four molecules in the asymmetric unit, a comparison of the substrates bound in the active site is possible. In both the hypoxanthine and xanthine structures, variability in the hydrogen bond distances was observed, thus indicating that the orientation of the substrate is well defined, but the position is not exactly maintained. This observation supports the hypothesis that a balance has to be achieved, which allows the substrates to be ideally positioned for catalysis but also held loosely enough in place to facilitate subsequent product release. The C-2 of the hypoxanthine is located in close proximity to an oxo ligand within the coordination sphere of the Mo. This would enable oxygen transfer from the Mo-O to the C-2 of the hypoxanthine, converting it to xanthine. ArgB-310 and GluB-232 serve to orient the substrate in the proper manner for oxidation. pKa value calculations for GluB-232 (calculated with PROPKA) (30) indicate a pKa of 8.1 for this residue, underlining its ability to form hydrogen bonds to the corresponding substrates. The surrounding residues anchor the substrate during catalysis while maintaining an unobstructed substrate channel, which facilitates substrate reemergence into the solvent. The space within the substrate-binding pocket is ideally designed to coordinate and position the substrates for catalysis. This tight binding pocket, however, obstructs reorientation of the substrate within the pocket to allow oxidation at the C-2 position, followed by oxidation at C-8. Our structures therefore suggest that after the first round of oxidation, xanthine has to retreat at least 16 Å from the Moco to acquire adequate space to rotate the C-8 into position close to the Mo atom for further oxidation. The xanthine-bound structure clearly shows that the C-8 is again located in close proximity to the planar oxo ligand of the molybdenum cofactor, allowing further oxidation to uric acid. Our crystal structures corroborate the canonical mechanism of substrate turnover within the bacterial XDH system. Hypoxanthine and xanthine are optimally positioned to coordinate their C-2 or C-8, respectively, to one of the oxo ligands within the molybdenum cofactor coordination sphere.
Pterin-6-aldehyde displays multiple binding contacts to key residues within the active site cavity. This double ringed moiety is firmly situated for enzyme inhibition, unlike the substrates hypoxanthine and xanthine, which are more loosely held to allow product release after oxidation.
Acknowledgments
We thank the staff of beamline ID23-2 at the European Synchrotron Radiation Facility for technical support.
The atomic coordinates and structure factors (codes 2w55, 2w3r, 2w3s, and 2w54) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Grant GM070873. This work was also supported by Deutsche For schungsgemeinschaft Grant LE1171/3 (to S. L.), Forschungszentrum Grant FZ-82 (to C. K.), and in part by the State University of New York and its Research Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: XDH, xanthine dehydrogenase; XO, xanthine oxidase; Moco, molybdenum cofactor.
References
- 1.Leimkuhler, S., Kern, M., Solomon, P. S., McEwan, A. G., Schwarz, G., Mendel, R. R., and Klipp, W. (1998) Mol. Microbiol. 27 853-869 [DOI] [PubMed] [Google Scholar]
- 2.Leimkuhler, S., and Klipp, W. (1999) J. Bacteriol. 181 2745-2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leimkuhler, S., Stockert, A. L., Igarashi, K., Nishino, T., and Hille, R. (2004) J. Biol. Chem. 279 40437-40444 [DOI] [PubMed] [Google Scholar]
- 4.Nishino, T., Okamoto, K., Kawaguchi, Y., Hori, H., Matsumura, T., Eger, B. T., Pai, E. F., and Nishino, T. (2005) J. Biol. Chem. 280 24888-24894 [DOI] [PubMed] [Google Scholar]
- 5.Okamoto, K., Matsumoto, K., Hille, R., Eger, B. T., Pai, E. F., and Nishino, T. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 7931-7936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pauff, J. M., Hemann, C. F., Junemann, N., Leimkuhler, S., and Hille, R. (2007) J. Biol. Chem. 282 12785-12790 [DOI] [PubMed] [Google Scholar]
- 7.Pauff, J. M., Zhang, J., Bell, C. E., and Hille, R. (2008) J. Biol. Chem. 283 4818-4824 [DOI] [PubMed] [Google Scholar]
- 8.Truglio, J. J., Theis, K., Leimkuhler, S., Rappa, R., Rajagopalan, K. V., and Kisker, C. (2002) Structure 10 115-125 [DOI] [PubMed] [Google Scholar]
- 9.Reginato, A. J. (2005) in Harrison's Principles of Internal Medicine (Kasper, D. L., Braunwald, E., Hauser, S., Longo, D., Jameson, J. L., and Fauci, A. S., eds) 16th Ed., pp. 2046-2050, McGraw-Hill, Columbus, OH
- 10.Takano, Y., Hase-Aoki, K., Horiuchi, H., Zhao, L., Kasahara, Y., Kondo, S., and Becker, M. A. (2005) Life Sci. 76 1835-1847 [DOI] [PubMed] [Google Scholar]
- 11.Boss, G. R., Ragsdale, R. A., Zettner, A., and Seegmiller, J. E. (1980) J. Lab. Clin. Med. 96 783-789 [PubMed] [Google Scholar]
- 12.Halpern, R., Halpern, B. C., Stea, B., Dunlap, A., Conklin, K., Clark, B., Ashe, H., Sperling, L., Halpern, J. A., Hardy, D., and Smith, R. A. (1977) Proc. Natl. Acad. Sci. U. S. A. 74 587-591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowry, O. H., Bessey, O. A., and Crawford, E. J. (1949) J. Biol. Chem. 180 389-398 [PubMed] [Google Scholar]
- 14.Lowry, O. H., Bessey, O. A., and Crawford, E. J. (1949) J. Biol. Chem. 180 399-410 [PubMed] [Google Scholar]
- 15.Massey, V., Komai, H., Palmer, G., and Elion, G. B. (1970) J. Biol. Chem. 245 2837-2844 [PubMed] [Google Scholar]
- 16.Spector, T., and Johns, D. G. (1970) J. Biol. Chem. 245 5079-5085 [PubMed] [Google Scholar]
- 17.Williams, J. W., and Bray, R. C. (1981) Biochem. J. 195 753-760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalckar, H. M., Kjeldgaard, N. O., and Klenow, H. (1950) Biochim. Biophys. Acta 5 586-594 [DOI] [PubMed] [Google Scholar]
- 19.Neumann, M., Schulte, M., Junemann, N., Stocklein, W., and Leimkuhler, S. (2006) J. Biol. Chem. 281 15701-15708 [DOI] [PubMed] [Google Scholar]
- 20.Palmer, T., Santini, C. L., Iobbi-Nivol, C., Eaves, D. J., Boxer, D. H., and Giordano, G. (1996) Mol. Microbiol. 20 875-884 [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski, Z., and Minor, W. (1997) in Methods in Enzymology (Carter, C. W., and Sweet, R. M., eds) Vol. 276, pp. 307-326, Academic Press, New York [DOI] [PubMed] [Google Scholar]
- 22.Bailey, S. (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-763 [DOI] [PubMed] [Google Scholar]
- 23.Brunger, A. T. (1992) XPLOR Version 3.1: A System for X-ray Crystallography and NMR, Yale University Press, New Haven, CT
- 24.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Crystallogr. Sect. A 47 110-119 [DOI] [PubMed] [Google Scholar]
- 25.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 26.Watanabe, K., Arai, T., Mori, H., Nakao, S., Suzuki, T., Tajima, K., Makino, K., and Mori, K. (1997) Biochem. Biophys. Res. Commun. 233 447-450 [DOI] [PubMed] [Google Scholar]
- 27.Dixon, M., and Thurlow, S. (1924) Biochem. J. 18 976-988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofstee, B. H. (1955) J. Biol. Chem. 216 235-244 [PubMed] [Google Scholar]
- 29.Yamaguchi, Y., Matsumura, T., Ichida, K., Okamoto, K., and Nishino, T. (2007) J. Biochem. (Tokyo) 141 513-524 [DOI] [PubMed] [Google Scholar]
- 30.Bas, D. C., Rogers, D. M., and Jensen, J. H. (2008) Proteins 73 765-783 [DOI] [PubMed] [Google Scholar]