Abstract
The Rac1/Cdc42 effector, p21-activated kinase (PAK), is activated by various signaling cascades, including receptor-tyrosine kinases and integrins, and regulates a number of processes such as cell proliferation and motility. PAK activity has been shown to be required for maximal activation of the canonical RAF-MEK-MAPK signaling cascade, possibly because of PAK co-activation of RAF and MEK. Here we have shown that trihydrophobin 1 (TH1), originally identified as a negative regulator of A-RAF kinase, also interacted with PAK1 in cultured cells. Confocal microscopy assay indicated that TH1 colocalized with PAK1 in both the cytoplasm and nucleus, which is consistent with our previous results. GST pulldown and coimmunoprecipitation experiments demonstrated that TH1 interacted directly with PAK1 and bound selectively to the carboxyl-terminal kinase domain of PAK1, and the ability of the binding was enhanced along with activation of PAK1. The binding pattern of PAK1 implies that this interaction was mediated in part by PAK1 kinase activity. As indicated by in vitro kinase activity assays and Western blot detections, TH1 inhibited PAK1 kinase activity and negatively regulated MAPK signal transduction. Interestingly, TH1 bound with MEK1/ERK in cells and in vitro without directly suppressing their kinase activity. Furthermore, we observed that TH1 localized to focal adhesions and filopodia in the leading edge of cells, where TH1 reduced cell migration through affecting actin and adhesion dynamics. Based on these observations, we propose a model in which TH1 interacts with PAK1 and specifically restricts the activation of MAPK modules through the upstream region of the MAPK pathway, thereby influencing cell migration.
p21-activated kinases (PAKs),2 mammalian homologues of Ste20-like Ser/Thr protein kinases, are the effector for Rac1 and Cdc42 (1-3). Members of the PAK family have been implicated in a variety of intracellular signaling events, including cell cytoskeleton rearrangement, proliferation, differentiation, transformation, apoptosis, cell cycle progression, and cell migration (2). Based on their structures, the PAK family can be grouped into two subfamilies: group A (PAK1-3), which can be activated by small GTPases such as Rac-GTP or Cdc42-GTP binding (2); and group B (PAK4-6), which can interact with Cdc42-GTP but cannot be activated by this binding (2, 4). Outside of the kinase- and GTPase-binding domains, group B PAKs are quite different from group A, and their regulation may be distinct (5). From the crystal structure of PAK1 (6), it appears that the inactive state exists as autoinhibited dimers. Upon GTPase binding, PAK1 undergoes a conformational change that separates the autoinhibitory domain from the kinase domain (7). This induces kinase activity and autophosphorylation at several sites, including the Thr-423 site in the activation loop to stabilize the active state (8, 9). In resting cells, PAK1 is localized primarily to the membrane structure within the cytoplasm (10); however, activated PAK1 translocates to focal adhesions and membrane ruffles (11). Overexpression of constitutively activated PAK1 mutants induces dissolution of actin stress fibers and focal adhesions and increases membrane protrusions, cell polarization, and cell motility (12-15). Conversely, in endothelial cells, overexpression of active PAK1 results in decreased cell migration and stabilization of actin stress fibers and focal adhesions (16). These effects are mediated through the action of PAK1 on cytoskeletal regulatory proteins such as LIM kinase (17), myosin light chain kinase (18), filamin A (19), and Op18/stathmin (20). In addition, the activation of the ERK/MAPK is considered to direct the migration of numerous cell types (21-24). Recent studies indicate that PAK1 activity plays important roles for activation of the MAPK signaling pathways (25-27). However, the exact mechanism of PAK1 regulation in these pathways is not so clear.
The human trihydrophobin 1 (TH1) gene is the homologue of Drosophila TH1, which was originally identified during the positional cloning of mei-41 (28, 29). The TH1 gene lies adjacent to mei-41 and was characterized further by Bonthron et al. (30) in 2000. According to their studies, TH1 is highly conserved from Drosophila to human, as shown by sequence comparison, and is located in chromosome 20q13, which had a transcript product of 2.4 kb. Northern blots showed that TH1 is widely expressed in multiple tissues. The human TH1 protein has been predicted to have a molecular mass of 65.8 kDa and display high levels of expression in cardiac and skeletal muscle, kidney, adrenal, and thyroid. Although highly conserved and ubiquitously expressed, human TH1 is not well understood in terms of its function. Recently, data from our laboratory indicate that TH1, as a new negative regulator of A-RAF kinase in MAPK signal transduction pathways, can specifically bind to A-RAF and inhibit its kinase activity (31). TH1 also interacts with human papilloma virus E6-associated protein (E6AP) and induces ubiquitin-dependent proteolysis (32). In addition, it is also found to be identical to NELF-C/D, an integral subunit of the human negative transcription elongation factor (NELF) complex (33). Although our understanding of the function of TH1 has advanced, little is known about the relationship between TH1 and PAK1.
We report here that TH1 interacted with PAK1 in cell and in vitro, and the ability of the binding was enhanced along with the activation of PAK1. TH1 bound selectively to the carboxyl-terminal kinase domain of PAK1 and inhibited the ability of PAK1 to phosphorylate substrate, thereby reducing MAPK signal transduction. Finally, our wound healing assay and Boyden chamber assay showed that overexpression of TH1 inhibited ERK/MAPK-driven cell migration. This suggests that TH1 specifically restricted the activation of MAPK modules through the upstream of MAPK pathway, thereby influencing cell migration.
EXPERIMENTAL PROCEDURES
Cell Culture—293T, HeLa, NIH3T3, and COS-1 cells were obtained from the Institute of Cell Biology Academic Sinica and cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in 5% CO2.
Reagents and Antibodies—Protein G-agarose and anti-GFP antibody were purchased from Roche Applied Science. [γ-32P]ATP (>3000 Ci/mm) and the ECL assay kit were from Amersham Biosciences. Leupeptin, aprotinin, and phenylmethylsulfonyl fluoride were purchased from Sigma. Antibodies against PAK1, ERK1/2, phospho-MEK1 (Ser-298), phospho-ERK (E-4), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and HA peptide were from Santa Cruz Biotechnology. Antibody against phospho-PAK1 (Thr-423) was from Cell Signaling. Antibodies against paxillin were from Abcam. Antibodies against Myc, recombinant human epidermal growth factor (EGF), the vector pcDNA3.0, myelin basic protein, and Lipofectamine™ 2000 reagents were from Invitrogen. TH1 antiserum was raised against GST-TH1 protein purified from Escherichia coli. Other reagents were commercially available in China.
Plasmids—Full-length TH1 expression plasmids were constructed as described previously (31). Myc-PAK1 WT, Myc-PAK1 T423E, and Myc-PAK1 K299R (provided by J. Chernoff) were amplified by PCR and cloned into the SalI/BamHI site of pEGFP-N3 vector or the SacI/BamHI site of pDsRed-Express-C1 vector (Clontech). AU5-cdc42 and Myc-tagged Rac1 (T17N) were gifts from G. Bokoch. All of the generated sequences and plasmids were confirmed by sequencing.
Immunoprecipitation and Immunoblot Assays—Twenty-four h after transfection, cells were washed three times with ice-cold PBS and lysed with 0.5 ml of IP lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% Nonidet P-40, 5 mm EDTA, 5 mm EGTA, 15 mm MgCl2, 60 mm β-glycerophosphate, 0.1 mm sodium orthovanadate, 0.1 mm NaF, 0.1 mm benzamide, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 10 μg/ml phenylmethylsulfonyl fluoride) for 1.5 h at 4 °C. Detergent-insoluble materials were removed by centrifugation at 12,000 rpm for 10 min at 4 °C. The whole cell lysates were incubated with control rabbit normal IgG (Santa Cruz Biotechnology) or the relevant antibody at 4 °C for 1 h. Pre-equilibrated protein G-agarose beads (Roche Applied Science) were then added, collected by centrifugation after 1 h of incubation, and then gently washed three times with the lysis buffer. The cell lysates containing recombinant protein were incubated with specific antibodies and protein G-agarose beads at 4 °C. The precipitates were washed three times with ice-cold lysis buffer. The bound proteins were eluted by boiling in SDS sample buffer and resolved on a 10% SDS-polyacrylamide gel. The proteins were transferred onto a polyvinylidene difluoride membrane and probed with a 1:1000 dilution of the relevant antibodies. Then the proteins were detected using the ECL assay kit.
GST Pulldown Assay—GST-TH1 was purified from bacterial lysates using glutathione-agarose beads (Amersham Biosciences). His-tagged PAK1 protein was purified from E. coli using a nickel-Sepharose high performance column (GE Healthcare). HA-tagged PAK1 and its deletion mutations were expressed in 293T cells for 24 h. Cells were lysed with 0.5 ml of IP lysis buffer for 1.5 h at 4 °C. Detergent-insoluble materials were removed by centrifugation at 12,000 rpm for 10 min at 4 °C. The whole cell lysates were incubated with 10 μl (about 10 μg) of GST-TH1 fusion protein and rotated for 3-5 h at 4 °C. The beads were then washed three times with lysis buffer. The beads were resuspended in SDS-PAGE sample buffer, boiled for 8 min, electrophoresed on a 15% SDS-polyacrylamide gel, and analyzed by Western blot. An in vitro binding assay was also performed as follow. Namely, purified GST-TH1 and His6-PAK1 proteins were incubated in IP buffer at 4 °C, and then GST pulldown was performed.
Kinase Activity Assay—To evaluate PAK1 activity, 293T cells were transfected with various gene plasmids or with vector. After 24 h of incubation in DMEM containing 10% FBS, the cells were serum-starved for 8-20 h and then stimulated with or without 20% FBS for 15 min. The cells were washed three times with ice-cold PBS and lysed at 4 °C in IP lysis buffer. Equal total protein was immunoprecipitated with 2 μg of anti-GFP antibody or anti-PAK1 antibody at 4 °C overnight. The beads were washed twice in lysis buffer and twice in kinase buffer (50 mm HEPES, pH 7.5, 10 mm MgCl2, and 2 mm MnCl2). Kinase assays were performed in 30-μl reactions with 25 mm ATP, 10 μCi of [γ-32P]ATP, and 20 μg of myelin basic protein. After incubation at 30 °C for 30 min, the reactions were stopped by the addition of 8 μl of 5 × sample buffer, boiled, separated by 15% SDS-PAGE, and analyzed using autoradiography. Similar assays were used to measure the activity in immunoprecipitates of MEK1 and ERK from 293T cells.
Confocal Microscopy—COS-1 or HeLa cells grown to 50% confluence on coverslips were transiently transfected with GFP or GFP-TH1 along with RFP-PAK1 or RFP-TH1. After 24 h of transfection, cells were washed with PBS, fixed in 4% formaldehyde, permeabilized in 0.2% Triton X-100/PBS, and blocked in 1% bovine serum albumin for 1 h at room temperature. The coverslips were stained with anti-paxillin antibody for 2 h at room temperature or overnight at 4 °C followed by incubation with Cy5-conjugated goat anti-mouse secondary antibody for 1 h at room temperature. Cells were washed three times with PBS, stained for F-actin and DNA with fluorescein isothiocyanate-phalloidin (5 μg/ml) and Hoechst 33258 (50 μg/ml) solution, respectively, in a dark chamber. The coverslips were washed as described above, inverted, mounted on slides, and examined in a Zeiss or Leica TCS SP5 confocal microscope.
RNA Interference—RNA interference was undertaken using the pSilencer2.0 vector (Ambion Inc.). RNA interference target sequences were selected from the human TH1L sequence (GenBank™ accession number NM_198976). Target oligonucleotides were synthesized (5′-GATCCGCAGAATTGAGCACACTTTATCGAAATAAAGTGTGCTCAATTCTGCTTTTTTGGAAA-3′ and 3′-GCGTCTTAACTCGTGTGAAATAGCTTTATTTCACACGAGTTAAGACGAAAAAACCTTTTCGA-5′), annealed, and cloned into pSilencer vector between the BamHI and HindIII sites. For shRNA expression, HeLa cells were transfected with non-targeting shRNA vector or vector encoding TH1 shRNA, and 24 h later the cells were treated.
Wound Healing Assay and Boyden Chamber Assay—An in vitro wound healing assay and Boyden chamber assay were performed as described previously (34, 35). Briefly, HeLa cells in DMEM containing 10% FBS were seeded into wells of 6-well plates. After the cells grew to confluency, wounds were made with sterile pipette tips. Wells were rinsed three times with DMEM without serum, and media containing 10% FBS and 100 ng/ml EGF were added. After overnight incubation at 37 °C, cells were fixed with 4% formaldehyde, stained with 0.1% crystal violet, and photographed. Cell migration assay was performed using Boyden chambers (tissue culture-treated, 6.5-mm diameter, 10-μm thickness, 8-μm pores; Transwell®, Costar Corp., Cambridge, MA) containing polycarbonate membranes. Serum-starved cells were trypsinized and counted. Then 100 μl of 1 ∼ 3 × 106 cells in serum-free medium was added to the upper chamber, and 600 μl of the appropriate medium with 10% FBS or 100 ng/ml EGF was added to the lower chamber. The Transwell was incubated for 12-18 h at 37 °C. Nonmigratory cells on the upper membrane surface were removed with a cotton swab, and the migratory cells on the undersurface of the membrane were fixed and stained with 0.1% crystal violet for 20 min at room temperature. Photographs of three random regions were taken, and the number of cells was counted to calculate the average number of cells that had transmigrated.
RESULTS
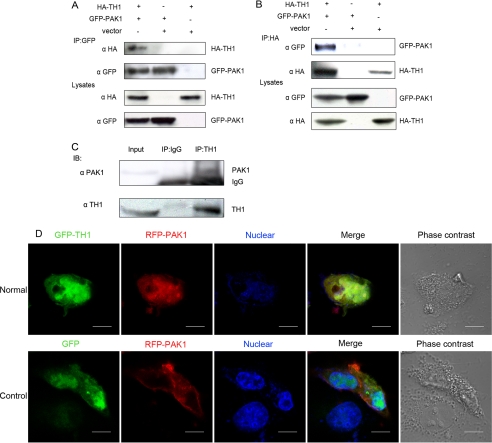
TH1 Interacts with PAK1 in Cells—Previous studies on the biological function of TH1 and PAK1 in the RAF-MEK-MAPK pathway suggested a functional linkage between TH1 and PAK1 (25, 27, 31). To determine whether the link was physical, the interaction of TH1 and PAK1 was examined. We expressed GFP-tagged PAK1 and HA-tagged TH1 in 293T cells. Although an anti-GFP antibody coimmunoprecipitated HA-TH1 in GFP-PAK1 and HA-TH1 cotransfected cells, this antibody did not coimmunoprecipitate HA-TH1 in control cells cotransfected with HA-TH1 and GFP vector or with HA vector and GFP-PAK1 (Fig. 1A). Conversely, anti-HA antibody also coimmunoprecipitated GFP-PAK1 in GFP-PAK1 and HA-TH1 cotransfected 293T cells (Fig. 1B). Supporting the importance of this interaction, TH1-PAK1 association also took place between endogenous proteins as well as during protein overexpression. Thus immunoprecipitation of endogenous TH1 from HeLa cells resulted in coimmunoprecipitation of endogenous PAK1 (Fig. 1C).
FIGURE 1.
Interaction of TH1 and PAK1 in cells. A and B, coimmunoprecipitation of TH1 and PAK1. 293T cells were cotransfected with HA-TH1, vector, and GFP-PAK1. 24 h after transfection, the cells were lysed, and the tagged proteins were immunoprecipitated with 2 μg of anti-HA or anti-GFP antibody. Anti-GFP or anti-HA antibody was used to detect the protein expression of transfected genes in whole cell extracts. C, interaction of endogenous TH1 and PAK1.HeLa cells were lysed in IP buffer, and lysates were immunoprecipitated with an agarose-conjugated rabbit anti-TH1 serum or a control agarose-conjugated IgG overnight at 4 °C. Immunoblots (IB) were probed with a polyclonal anti-PAK1 antibody. D, colocalization of TH1 and PAK1 in both the nucleus and the cytoplasm. COS-1 cells, which were cultured on coverslips, were transiently transfected with RFP-PAK1 and GFP-TH1 or GFP. 24 h later, the cells were immobilized and observed using a confocal fluorescence microscope. Normal, colocalization of GFP-TH1 and RFP-PAK1; Control, no colocalization of GFP and RFP-PAK1. GFP fluorescence is shown in green; RFP fluorescence is shown in red; an overlay image is shown in yellow; the nucleus (stained with Hoechst) is shown in blue; cell morphology is shown in phase contrast. Scale bar:10 μm. Data are representative of three to four independent experiments.
To further confirm the above results, we examined subcellular localization of TH1 and PAK1 using confocal fluorescence microscopy analysis. We selected COS-1 cells to transfect with pEGFP-TH1 or pEGFP-N3 along with pDsRed-PAK1, respectively. At exactly 24 h after transfection, the cells were fixed and analyzed under confocal microscopy. As shown in Fig. 1D, in control cells, GFP was distributed in whole cellular compartments, especially in the nucleus, whereas red fluorescent protein-fused PAK1 (RFP-PAK1) was widely distributed in the cytoplasm, and there was no colocalization between GFP and RFP-PAK1. However, in cells coexpressing GFP-TH1 and RFP-PAK1, we observed that the double-transfected cells contained yellow granules, indicating colocalization of TH1 and PAK1 in both the cytoplasm and nucleus (Fig. 1D, Normal). Taken together, these data indicate that TH1 interacts with PAK1 in cultured cells.
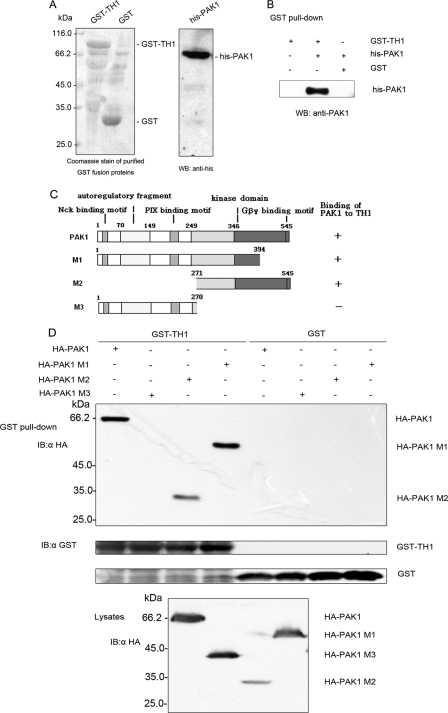
TH1 Interacts Directly with PAK1 in Vitro—To ascertain whether TH1 interacts directly with PAK1, recombinant proteins GST-TH1 and His6-tagged PAK1 were purified from E. coli (Fig. 2A), and an in vitro binding assay was conducted in a cell-free system. The purified recombinant protein His6-PAK1 was incubated with GST-TH1 or GST fusion protein. Glutathione-agarose beads were used to pull down GST or GST-TH1 from the formed complex. Western blots with anti-PAK1 antibody revealed that PAK1 interacted only with GST-TH1 but not GST (Fig. 2B). These results demonstrate that TH1 formed a complex with PAK1 in a cell-free system.
FIGURE 2.
Direct interaction of TH1 and PAK1 and identification of the TH1 binding site on PAK1. A, left panel, Coomassie staining of purified GST-TH1 and GST proteins used in in vitro interaction assays is shown. Right panel, Western blot with anti-His antibody to determine the expression of purified His6-PAK1 protein. B, direct interaction of TH1 and PAK1. Purified GST-TH1 and GST proteins were mixed with purified His6-PAK1 protein. As compared with GST, recombinant GST-TH1 fusion proteins were pulled down by GSH beads, and His6-PAK1 protein in the complex were examined with anti-PAK1 antibody. C, schematic diagram of deletion mutations of PAK1. Constructs were made by polymerase chain reaction, and PCR products were subcloned into pcDNA3.0-HA (region and residue numbers are indicated). The column on the right summarizes whether constructs did (+) or did not (-) bind to TH1. D, TH1 binds to the carboxyl-terminal kinase domain of PAK1. HA-tagged full-length PAK1 or M1 (aa 1-394), M2 (aa 271-545), or M3 (aa 1-270) were expressed in 293T cells. Cells were lysed, and the lysates were incubated with GST or GST-TH1 fusion proteins. As compared with GST, recombinant GST-TH1 fusion proteins were pulled down by GSH beads, and HA-PAK1 mutations in the precipitates were probed with anti-HA antibody. The relative amounts of GST fusion protein and HA-PAK1 mutations were also examined by immunoblotting with anti-GST antibody. Data are representative of three independent experiments.
TH1 Binding Site Is Located in the Carboxyl-terminal Kinase Domain of PAK1—The regulatory domain of PAK1 spans residues 1-248 of its amino terminus, whereas amino acids 248-545 comprise the kinase domain. The kinase domain of PAK1 is structurally composed of two lobes. The small lobe mainly involves binding to ATP, and the large lobe provides a binding site for peptide substrates (Fig. 2C). A phosphotransfer reaction was achieved by coordination between the two lobes of the kinase domain and their interaction with substrates. To determine where the TH1-binding domain resided on PAK1, we initially engineered constructs encoding aa 1-394, the amino-terminal regulatory region (aa 1-270), and the carboxyl-terminal kinase domain (aa 271-545) and expressed them in 293T cells. Then, the whole cell lysates were incubated with E. coli-expressed GST-TH1 or GST fusion protein to perform the in vitro binding assay. As shown in Fig. 2D, a GST pulldown assay showed that TH1 could bind directly with M1 (aa 1-394) and M2 (aa 271-545), whereas no binding with M3 (aa 1-270) was detected. Thus, the carboxyl-terminal 271-545 amino acids of PAK1 were necessary and sufficient for the interaction of TH1.
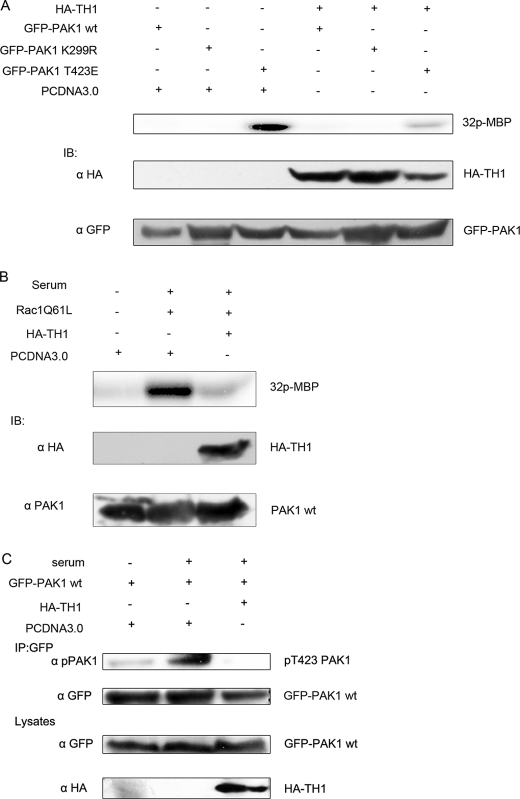
Overexpression of TH1 Inhibits PAK1 Kinase Activity—Because TH1 preferentially associates with the carboxyl-terminal kinase domain of PAK1, we hypothesized that TH1 could influence PAK1 kinase activity. To verify the hypothesis, 293T cells were transiently transfected with GFP-PAK1 WT, GFP-PAK1-K299R (a kinase-dead version of the protein (36, 37)), the active PAK1 mutant, GFP-PAK1-T423E (the threonine to glutamic acid substitution mimicked the phosphorylation of Thr-423 and partially activated the kinase (2, 8, 37)) along with HA-TH1 or vector. At 24 h after transfection, the cells were serum-starved for 16 h. The immunoprecipitated PAK1 proteins were tested for kinase activity using myelin basic protein as a substrate. As shown in Fig. 3A, the kinase activity of the active mutant PAK1-T423E was inhibited by TH1, whereas the kinase activity of PAK1 WT disappeared under serum-starved conditions. These data also suggest that TH1 directly affected PAK1 itself to lead to the inhibition of PAK1 kinase activity instead of the blockade of the signaling pathway leading to PAK1 activation.
FIGURE 3.
TH1 inhibits PAK1 kinase activity. A, TH1 inhibits T423E PAK1 activity. Immunoprecipitates were made from 293T cells transfected with the indicated constructs. Top panel, the immunoprecipitates were used in in vitro kinase assays using myelin basic protein (MBP) as a substrate. Middle panel, lysates were blotted with anti-HA antibody. Bottom panel, lysates were blotted with anti-GFP antibody. B, TH1 inhibits serum-stimulated endogenous PAK1 activity. Immunoprecipitates were made from 293T cells transfected with HA-TH1, RAC1 (Q61L), or PCDNA3.0. The cells shown in lanes 2 and 3 were starved for 8-16 h and stimulated with 20% serum for 15 min. Top panel, immunoprecipitates were used in in vitro kinase assays as described in A. Middle panel, lysates were immunoblotted (IB) with anti-HA antibody. Bottom panel, lysates were blotted with anti-PAK1 antibody. C, TH1 inhibits Thr-423 phosphorylation of PAK1. 293T cells were cotransfected with GFP-PAK1 and HA-TH1 or PCDNA3.0. At 24 h after transfected, the cells were starved for 8 h and then stimulated with or without 20% serum for 15 min. The lysates were used to immunoprecipitate with anti-GFP antibody, and the immunoprecipitates were used to detect the phospho-PAK1 (Thr-423) level. The gene expression level in the lysates was also blotted with the relevant antibody. Data are representative of three to four independent experiments.
To further study whether the overexpression of TH1 also inhibited the endogenous PAK1 kinase activity in cells, we transfected 293T cells with the active mutant Rac1 (Q61L) and HA-TH1 or vector. Immunoprecipitated endogenous PAK1 proteins were subjected to in vitro kinase assays. As seen in Fig. 3B, serum stimulation strongly increased PAK1 kinase activity, whereas overexpression of TH1 blocked the increase in PAK1 activity.
Because threonine 423 phosphorylation of the PAK1 is critical for the conformational change of the activation loop, assembly of the active configuration of catalytic domain, and activation of PAK1 (2, 6, 7, 38), we analyzed the phosphorylation state of PAK1 at site 423 in 293T cells cotransfected with GFP-PAK1 and HA-TH1 or vector. As depicted in Fig. 3C, serum stimulation strongly increased the phosphorylation level of PAK1 at site 423, whereas coexpression of HA-TH1 blocked the increase. In summary, these data strongly suggest that TH1 inhibits the activation of PAK1 in cells.
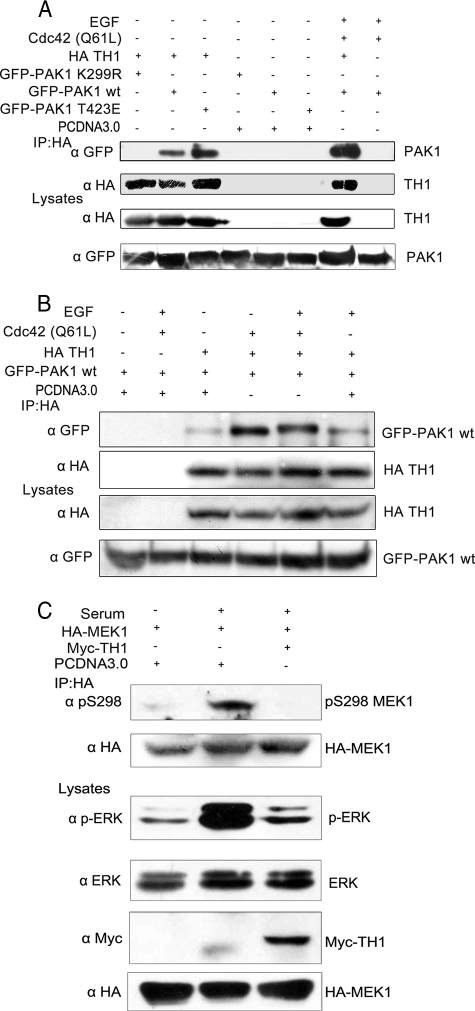
TH1 Binds with Active PAK1 Strongly and Interferes with MAPK Signaling—Because of the binding site of TH1 to the kinase domain of PAK1, we reasoned that their interaction might be related to the active state of the PAK1 kinase. To further explore this issue, HA-TH1 was coexpressed with GFP-PAK1-K299R, GFP-PAK1 WT, or GFP-PAK1-T423E. TH1 immunoprecipitated via its HA tag showed enhanced co-precipitation of PAK1-T423E as compared with the wild-type PAK1, whereas in serum-starved cells, co-precipitation of kinase-dead PAK1 was hardly found (Fig. 4A). Notably, coimmunoprecipitation of the wild-type PAK1 dramatically increased from cells cotransfected with the active mutant Cdc42 (Q61L) and stimulated with EGF as compared with that from serum-starved cells (Fig. 4A). Identical results were also obtained with different stimulation conditions (Fig. 4B). These data indicate that the activation of PAK1 facilitates its interaction with TH1.
FIGURE 4.
TH1 binds with active PAK1 strongly and impairs MAPK signal transduction. A, TH1 binds with active PAK1 strongly. GFP-PAK1 WT, PAK1 T423E, and PAK1 K299R were each transfected into 293T cells with HA-TH1. Cells were serum-starved for 16-24 h and then with or without EGF stimulation (100 ng/ml for 15 min). Coimmunoprecipitations were performed with anti-HA antibody. B, binding of TH1 and PAK1 increases with enhanced activation. 293T cells cotransfected with GFP-PAK1 WT and HA-TH1 were serum-starved and stimulated under different conditions as indicated. coimmunoprecipitations were performed with anti-HA antibody. C, overexpression of TH1 suppresses MEK1 Ser-298 phosphorylation and ERK phosphorylation. 293T cells were transfected with HA-MEK1 together with myc-TH1 or vector. Then cells were serum-starved for 8 h and stimulated with or without 20% serum for 15 min. Anti-HA immunoprecipitates were analyzed and blotted with anti-pSer-298 MEK1 or anti-HA antibody. Cell lysates were also blotted with antibodies against phospho-ERK, total ERK, Myc-TH1, and HA-MEK1, respectively. Data are representative of three to four similar experiments.
Recent studies have demonstrated that PAK1-dependent phosphorylation of MEK1 on Ser-298 is necessary for activation of MEK1 and subsequent activation of MAPK (25). To investigate whether the inhibition of PAK1 activity by TH1 affects the MAPK signal transduction pathway, 293T cells expressing HA-MEK1 together with Myc-TH1 (or vector control) were starved and then stimulated with or without 20% serum for 15 min. The cells were immunoprecipitated with anti-HA antibody, and the immunoprecipitates were used to detect the pSer-298 MEK1 level. Immunoblotting with anti-pSer-298 MEK1 revealed that serum stimulation led to an increase in MEK1 Ser-298 phosphorylation, whereas MEK1 Ser-298 phosphorylation disappeared in TH1-overexpressing cells and in serum-starved cells (Fig. 4C).
At the same time, the level of ERK phosphorylation in lysates was also examined in parallel. In serum-stimulated cells, expression of Myc-TH1 significantly decreased ERK phosphorylation as compared with the control (Fig. 4C). Our results indicate that overexpression of TH1 strongly impairs MAPK signal transduction.
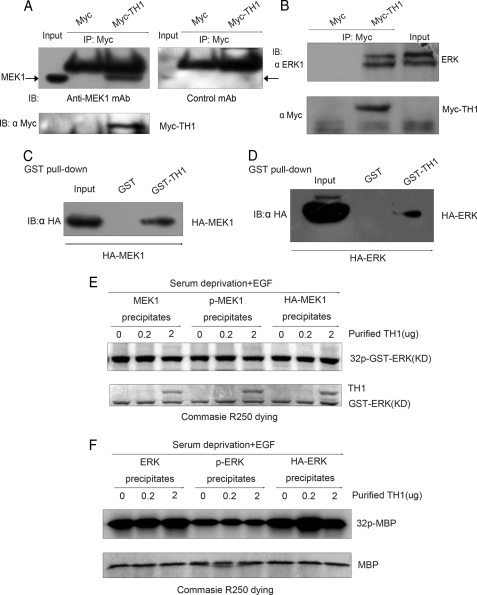
TH1 Binds with MEK1/ERK but Does Not Directly Suppress Their Kinase Activity—To further study the mechanism by which TH1 regulates the MAPK pathway, we examined whether TH1 directly suppressed several downstream targets of PAK1 in this regulatory process. We found that TH1 could pull down MEK1 and ERK in coimmunoprecipitation assays (Fig. 5, A and B). At the same time, immunofluorescence analysis also revealed that MEK1/ERK colocalized with TH1 in PC12 cells (data not shown). These data show that TH1 could bind with MEK1 and ERK in cells. To identify whether TH1 and MEK1/ERK could associate directly, we performed GST pulldown assays (Fig. 5, C and D). Recombinant protein GST-TH1 was able to associate with MEK1/ERK, indicating that the binding between TH1 and MEK1/ERK seen in coimmunoprecipitation assays (Fig. 5, A and B) was likely to be direct.
FIGURE 5.
TH1 binds with MEK1/ERK but does not directly inhibit their kinase activity. A and B, interaction of TH1 with MEK1/ERK. 293T cells were transfected with myc-TH1 or vector. At 24 h after transfected, the cells were lysed, and the tagged proteins were immunoprecipitated with 2 μg of anti-Myc antibody. Immunoblots (IB) were probed with an anti-MEK1 antibody, an irrelevant monoclonal antibody (mAb), or an anti-ERK1 antibody. C and D, in vitro interaction of TH1 with MEK1 and ERK1. GST or GST-TH1 fusion proteins were incubated with the lysates from cells transfected with HA-MEK1 or HA-ERK1. Then GST pulldown assays were performed, and the precipitates were probed with an anti-HA antibody. E and F, TH1 did not directly inhibit the kinase activity of MEK1/ERK. Cells were serum-starved and stimulated with EGF. MEK1, p-MEK1, and HA-MEK1 precipitates were prepared by anti-MEK1, anti-pMEK1, and anti-HA antibodies. These precipitates were tested for their kinase activity using purified GST-ERK1 protein as a substrate. In in vitro kinase activity assays, different weight of purified TH1 protein (0, 0.2, and 2 μg) was added. A similar method was used to measure the activity of ERK. Data are representative of three to four independent experiments.
To rule out the possibility that TH1 directly suppressed the kinase activity of MEK1/ERK, the immunoprecipitated MEK1/ERK proteins were tested for their kinase activity using purified GST-ERK protein or myelin basic protein as substrates. As shown in Fig. 5, E and F, in in vitro kinase activity assays increasing the amount of purified TH1 protein failed to affect MEK1/ERK activity. These experiments demonstrate that TH1 could bind with MEK1/ERK, but did not directly inhibit their kinase activity.
TH1 Localizes to Focal Adhesions and Affects Actin and Adhesion Dynamics—Activated PAK1, MEK1, and ERK1 are all located in peripheral membrane ruffles within the cytoplasm or focal adhesions at the leading edge of motile cells (11, 25, 27, 36, 39). To test whether TH1 could also locate to these sites, which are involved in the organization of the actin cytoskeleton and cell migration, we used a TH1 construct expressing an RFP-tagged protein to localize TH1 and examined the change of the actin cytoskeleton and focal adhesions.
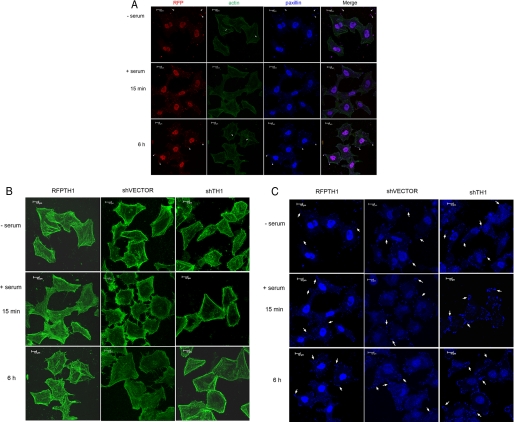
As depicted in Fig. 6A, in HeLa cells transfected with RFP-TH1, we consistently observed that RFP-TH1 localized to paxillin-containing focal adhesions and nucleus area in resting cells, suggesting that TH1 might also play an important role in nucleus. Remarkably, in serum-starved cells, TH1 localized mainly to the nucleus area, but after cells were stimulated by serum, TH1 localization was no longer restricted to the nucleus. TH1 could, in fact, along with paxillin, which formed focal adhesions, translocate from the nucleus to filopodia in the leading edge of the cell upon stimulation with serum.
FIGURE 6.
TH1 localizes to focal adhesions and affects actin and adhesion dynamics. A, localization of TH1 and its effects on focal adhesions and actin cytoskeleton. Serum-starved HeLa cells transfected with RFP-TH1 were stimulated or not by serum-containing full medium with EGF for 15 min or 6 h. Thereafter, immunofluorescence analysis was performed using the fluorescent signals of RFP (red), fluorescein isothiocyanate-conjugated phalloidin (green), and a mouse anti-paxillin monoclonal antibody (blue). An overlay of the three fluorescent signals, in which a magenta color is generated in areas of colocalization, is shown in the rightmost column. The arrows indicate stress fibers, and arrowheads indicate TH1-containing focal adhesions. In retracting regions of these cells, paxillin was left behind on the substrate (top row, arrowheads). By contrast, in normal group cells, paxillin was seldom detected on the substrate as the rear of the cell retracted. Scale bars:10 μm. B, effects of TH1 on actin cytoskeleton organization. Serum-starved HeLa cells transfected with RFP-TH1, shVECTOR, or shTH1 were stimulated or not by serum-containing full medium with EGF for the indicated time prior to immunofluorescence analysis. The fluorescent signals of actin (green) and paxillin (blue) are shown. For ease of analysis, these pictures were rearranged separately according to actin cytoskeleton (B) and focal adhesions (C). Scale bars:10 μm. C, effects of TH1 on focal adhesions. The arrows indicate the representation of the focal adhesions Data are representative of three to four independent experiments.
To test the effect of TH1 overexpression on actin polymerization and focal adhesions, the actin was also marked with fluorescein isothiocyanate-phalloidin. As compared with the normal cells or the cells receiving the non-targeting short hairpin RNA vector (Fig. 6C, shVECTOR), whether HeLa cells were stimulated by serum or not, ectopic expression of RFP-TH1 caused an increase in actin polymerization and stress fibers (Fig. 6B). In particular, actin polymerization at the cell margin drastically increased when the TH1-overexpressing cells were stimulated for 15 min by serum. Notably, the number of focal adhesions in HeLa cells overexpressing TH1, which were used to anchor stress fibers, also decreased, and the size of the focal adhesions became smaller than those in control cells (Fig. 6C). Interestingly, in serum-starved HeLa cells expressing TH1, paxillin-containing focal adhesions were readily detected on the substrate as the cells retracted (Fig. 6A, top row).
To further confirm the results, we depleted endogenous TH1 using shRNA against human TH1 (shTH1; Fig. 7A). In contrast to the TH1-overexpressing cells, deletion of TH1 in HeLa cells also resulted in the reduction of actin polymerization and stress fibers (Fig. 6C). On the other hand, the number of focal adhesions also increased. Intriguingly, deletion of TH1 induced the spike-like filopodia formation, even in the absence of serum. These data indicate that ectopic expression of TH1 affected actin and adhesion dynamics, which plays a critical role in cytoskeleton turnover and cell migration guidance (40-42).
FIGURE 7.
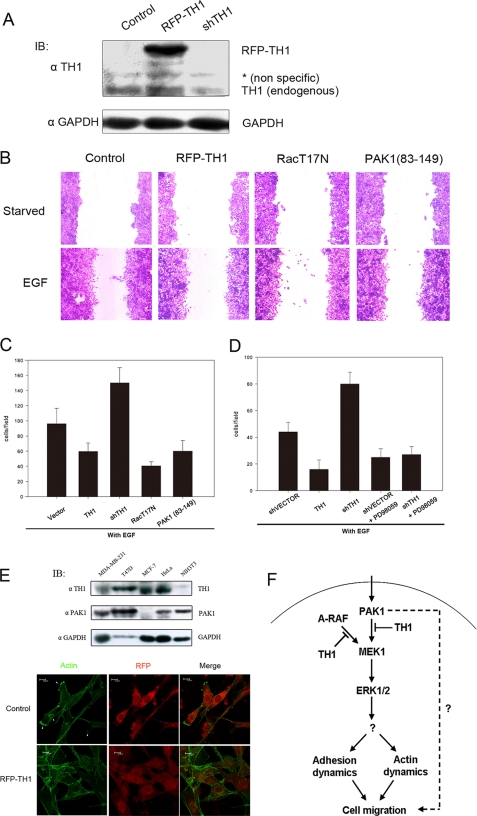
Role of TH1 in ERK/MAPK-driven cell migration. A, immunoblot (IB) with anti-TH1 serum antibody showing the protein level of TH1 in cells transfected with shTH1 or RFP-TH1 compared with cells treated with a control. The middle band (*) was a nonspecific protein recognized by rabbit TH1 antiserum. In the bottom panel, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein level is also shown. B, expression of RFP-TH1, dominant-negative Rac1 (T17N), or PAK1-(83-149) reduced the cell migration by EGF in a wound healing assay. C, expression of RFP-TH1, shTH1, dominant-negative Rac1 (T17N), or PAK1-(83-149) affected cell migration by EGF in a Boyden chamber assay. D, inhibition of MEK1 reduced cell migration as measured by a Boyden chamber assay. HeLa cells transfected with shVECTOR, RFP-TH1, or shTH1 were pretreated either with or without the MEK1 inhibitor PD98059 (75 μm) for 4 h and allowed to migrate for another 20 h. During cell migration, the inhibitor (PD98059) was also added. Data are shown as mean ± S.D. of three to four experiments. E, TH1 overexpression inhibited the formation of lamellipodia induced by EGF in NIH3T3 cells. Transfected cells were indentified with RFP-tagged TH1 or RFP vector. Examples of lamellipodia (arrowheads, lower panels) are shown. Western blot of TH1 protein and PAK1 protein levels in various cell lines are also shown (upper panels). Data are representative of three independent experiments. F, schematic diagram of TH1 inhibiting ERK/MAPK-driven cell migration.
TH1 Inhibits ERK/MAPK-driven Cell Migration—As discussed above, it was suggested that TH1 might have an effect on cell migration. To confirm this suggestion, we used different approaches to investigate whether the increase or decrease of the TH1 protein level would interfere with cell migration. First, cells were subjected to serum starvation and then allowed to migrate by EGF stimulation. As shown in Fig. 7B, in an in vitro wound healing assay, migration was inhibited in TH1-overexpressing cells. The TH1 protein level is shown in Fig. 7A (to differentiate from endogenous TH1, RFP-TH1-fused protein was expressed). These data demonstrate that TH1 is important for EGF-induced cell migration. Second, we further confirmed the effect of TH1 on EGF-induced cell migration by using a Boyden chamber assay (Fig. 7C). Similar results were observed when TH1 was overexpressed. Furthermore, EGF-induced cell migration depended on Rac1 and PAK1, as the expression of the kinase-dead mutant Rac1 (T17N) and the PAK1 autoinhibitory domain PAK1-(83-149) reduced the cell migration. In particular, the successful reduction of the TH1 by shTH1 strongly increased the cell migration. To further investigate whether MEK1/ERK activity was also involved in cell migration, we used the MEK1 inhibitor PD98059 to block MEK1 activity. As shown in Fig. 7D, cell migration induced by EGF was also blocked by the MEK1 inhibitor. Importantly, cell migration induced by shTH1 was also blocked with the MEK1 inhibitor PD98059. These studies demonstrate that MAPK signaling events were involved in cell migration progresses. Finally, we studied the potential role of TH1 in lamellipodium formation. Lamellipodia are protruding membrane structures at the leading edge of migrating cells. We first examined the endogenous TH1 protein and PAK1 protein level in different cell lines, including MDA-MB-231, T47D, MCF-7, HeLa, and NIH3T3 cells. Western blot analysis with anti-TH1 serum antibody revealed a very low TH1 protein level in NIH3T3 cells (Fig. 7E, upper panel). We thus transfected RFP-TH1 or RFP into NIH3T3 cells to investigate lamellipodium formation. As shown in Fig. 7E, migrating NIH3T3 cells (transfected with a control RFP) in the presence of EGF developed lamellipodia (∼80% of the cell). However, RFP-TH1 treatment led to the reduction of lamellipodia in the presence of EGF (∼10% of cells with lamellipodia). In summary, these data show that TH1, like Rac1 and PAK1, played a critical role in cell migration.
DISCUSSION
Cell migration is an intricate process involving several signal transduction pathways (2), including a key role for members of the PAK family of kinases. Thus it is important that PAK1 activity in different signal pathways be orchestrated in a coordinated manner. In this study, our data provided strong support to the notion that the interaction of TH1 and PAK1, by inhibiting PAK1 kinase activity, leads to a negative regulation of the MAPK signal transduction, which affects cell migration (Fig. 7F).
TH1 was originally recognized as the specific suppressor of A-RAF kinase in the MAPK-ERK cascade (31). Our evidence strongly demonstrates that TH1 could also interact with PAK1 (Fig. 1). Interestingly, immunofluorescence confocal microscopy analysis shows that colocalization of TH1 and PAK1 was found not only in the cytoplasm but also in the nucleus (Fig. 1D). The reason that TH1 and PAK1 colocalize in the nucleus is not so clear, but one explanation is that their interaction may be involved in the transcriptional regulation of some genes. Consistent with this hypothesis, recent data from our laboratory indicate that TH1 is also a potent transcriptional repressor to suppress androgen receptor and estrogen receptor activity in nucleus (data not shown). At the same time, PAK is also reported to have a relation to androgen and estrogen receptors (43-45). further study is needed to ascertain whether the correlation between TH1 and PAK1 exists in nucleus.
TH1 binds to the carboxyl-terminal kinase domain of PAK1 and strongly inhibits PAK1 kinase activity. Apart from the autoinhibitory role of the PAK1 amino terminus (7), several other mechanisms for the regulation of PAK1 kinase activity are recognized. Thus, various kinases, such as cAMP-dependent protein kinase (PKA), can inactivate PAK by phosphorylation of key residues (46), whereas certain phosphatases can inactivate PAK1 through dephosphorylation (47). The mechanism for TH1 inhibition of PAK1 kinase activity, however, has not yet been worked out in detail. Our initial results suggest that TH1 could hinder the autophosphorylation and activation of PAK1 and interfere with the ability of PAK1 to phosphorylate substrates. In contrast, TH1 did not seem to be a very good substrate for PAK1, and thus it was less likely that TH1 would block PAK1 by simply competing with other substrates for phosphorylation.
Overexpression of TH1 blocks MAPK signal transduction. PAK1 affects MAPK pathway activation through several aspects. On the one hand, PAK1 activity is essential for maximal activation of the mitogenic RAF-MEK-ERK signaling cascade (2), because active PAK1 directly associates with C-RAF to phosphorylate on Ser-338, an essential regulatory site for C-RAF activation (48, 49). On the other hand, PAK1-mediated phosphorylation of MEK1 on Ser-298, which is important for MEK1-C-RAF interaction, is central to the organization and localization of active RAF-MEK1-MAPK signaling complexes, and this phosphorylation is necessary for efficient activation of MEK1 and subsequent MAPK activation (25, 50). Our study clearly shows that the inhibition of PAK1 kinase activity by TH1 hindered MEK1 and ERK phosphorylation in cells. However, whether TH1 impairs the interaction of PAK1-RAF and phosphorylation of RAF remains to be studied. In addition, TH1 as the specific suppressor of A-RAF kinase also inhibited A-RAF to phosphorylate MEK1 in in vitro kinase activity assays using purified GST-MEK1 protein as substrate (data not shown). Therefore the suppression of A-RAF activity by TH1 may have also contributed to the negative regulation of MAPK signal.
TH1 can associate with MEK1/ERK in cells and in vitro. Our previous investigations have shown that TH1 interferes with the interactions of A-RAF/MEK1 and MEK1/ERK in vitro (data not shown). However, TH1 does not directly suppress MEK1/ERK kinase activity. The reason that TH1 associates with MEK1/ERK but does not inhibit their activity is not so clear, but there are possibilities that the MAPKs are located on a scaffold protein to which TH1 and PAK1 also dock or that TH1 works partially as a scaffold protein. Whether the phenomenon correlates with the TH1 function remains to be illustrated.
It is clear that TH1 plays a key role in the negative regulation of MAPK signaling and the control of cell migration. However, the precise mechanisms involved are not yet fully understood. One hypothesis that is consistent with current data is as follows. ERK and PAK activities have both been found to be important for cell migration (13, 16, 23, 51, 52). The inhibition of the MAPK pathway impairs phosphorylation of ERK substrates such as myosin light chain kinase (51), FAK (53-55), paxillin (56), and the protease calpain (57), which localize to focal adhesions and are direct regulators of this process, thus affecting the assembly and turnover of focal adhesions (24, 39, 57-62) and the change in the cytoskeleton. Although our results indicate a role of TH1 in cell migration through negative regulation of the MAPK pathway, other mechanisms may also regulate cell motility.
In summary, in this study we have demonstrated that TH1 could interact with PAK1 by binding to the carboxyl-terminal kinases domain of PAK1. Meanwhile, PAK1 activity was inhibited, which affected cell migration through negatively regulating the MAPK signal transduction. However, the precise mechanism by which MAPKs and their substrates affect cell migration needs to be elucidated further.
Acknowledgments
We thank Drs. Gary Bokoch and Jonathon Chernoff for providing critical plasmids for the completion of these studies. We also thank Yun Hu for secretarial work.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PAK, p21-activated kinase; TH1, trihydrophobin 1; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; GFP, green fluorescent protein; EGFP, enhanced green fluorescent protein; shRNA, short hairpin RNA; shTH1, short hairpin RNA against human TH1; GST, glutathione S-transferase; HA, hemagglutinin; EGF, epidermal growth factor; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; IP, immunoprecipitation; PBS, phosphate-buffered saline; WT, wild type; RFP, red fluorescent protein; aa, amino acid(s).
References
- 1.Knaus, U. G., and Bokoch, G. M. (1998) Int. J. Biochem. Cell Biol. 30 857-862 [DOI] [PubMed] [Google Scholar]
- 2.Bokoch, G. M. (2003) Annu. Rev. Biochem. 72 743-781 [DOI] [PubMed] [Google Scholar]
- 3.Kumar, R., and Vadlamudi, R. K. (2002) J. Cell. Physiol. 193 133-144 [DOI] [PubMed] [Google Scholar]
- 4.Jaffer, Z. M., and Chernoff, J. (2002) Int. J. Biochem. Cell Biol. 34 713-717 [DOI] [PubMed] [Google Scholar]
- 5.Dan, C., Nath, N., Liberto, M., and Minden, A. (2002) Mol. Cell. Biol. 22 567-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lei, M., Lu, W., Meng, W., Parrini, M. C., Eck, M. J., Mayer, B. J., and Harrison, S. C. (2000) Cell 102 387-397 [DOI] [PubMed] [Google Scholar]
- 7.Parrini, M. C., Lei, M., Harrison, S. C., and Mayer, B. J. (2002) Mol. Cell 9 73-83 [DOI] [PubMed] [Google Scholar]
- 8.Chong, C., Tan, L., Lim, L., and Manser, E. (2001) J. Biol. Chem. 276 17347-17353 [DOI] [PubMed] [Google Scholar]
- 9.Buchwald, G., Hostinova, E., Rudolph, M. G., Kraemer, A., Sickmann, A., Meyer, H. E., Scheffzek, K., and Wittinghofer, A. (2001) Mol. Cell. Biol. 21 5179-5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dharmawardhane, S., Sanders, L. C., Martin, S. S., Daniels, R. H., and Bokoch, G. M. (1997) J. Cell Biol. 138 1265-1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sells, M. A., Pfaff, A., and Chernoff, J. (2000) J. Cell Biol. 151 1449-1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sells, M. A., Knaus, U. G., Bagrodia, S., Ambrose, D. M., Bokoch, G. M., and Chernoff, J. (1997) Curr. Biol. 7 202-210 [DOI] [PubMed] [Google Scholar]
- 13.Sells, M. A., Boyd, J. T., and Chernoff, J. (1999) J. Cell Biol. 145 837-849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manser, E., Huang, H. Y., Loo, T. H., Chen, X. Q., Dong, J. M., Leung, T., and Lim, L. (1997) Mol. Cell. Biol. 17 1129-1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frost, J. A., Khokhlatchev, A., Stippec, S., White, M. A., and Cobb, M. H. (1998) J. Biol. Chem. 273 28191-28198 [DOI] [PubMed] [Google Scholar]
- 16.Kiosses, W. B., Daniels, R. H., Otey, C., Bokoch, G. M., and Schwartz, M. A. (1999) J. Cell Biol. 147 831-844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards, D. C., Sanders, L. C., Bokoch, G. M., and Gill, G. N. (1999) Nat. Cell Biol. 1 253-259 [DOI] [PubMed] [Google Scholar]
- 18.Sanders, L. C., Matsumura, F., Bokoch, G. M., and de Lanerolle, P. (1999) Science 283 2083-2085 [DOI] [PubMed] [Google Scholar]
- 19.Vadlamudi, R. K., Li, F., Adam, L., Nguyen, D., Ohta, Y., Stossel, T. P., and Kumar, R. (2002) Nat. Cell Biol. 4 681-690 [DOI] [PubMed] [Google Scholar]
- 20.Wittmann, T., Bokoch, G. M., and Waterman-Storer, C. M. (2003) J. Cell Biol. 161 845-851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krueger, J. S., Keshamouni, V. G., Atanaskova, N., and Reddy, K. B. (2001) Oncogene 20 4209-4218 [DOI] [PubMed] [Google Scholar]
- 22.Jo, M., Thomas, K. S., Somlyo, A. V., Somlyo, A. P., and Gonias, S. L. (2002) J. Biol. Chem. 277 12479-12485 [DOI] [PubMed] [Google Scholar]
- 23.Huang, C., Jacobson, K., and Schaller, M. D. (2004) J. Cell Sci. 117 4619-4628 [DOI] [PubMed] [Google Scholar]
- 24.Webb, D. J., Donais, K., Whitmore, L. A., Thomas, S. M., Turner, C. E., Parsons, J. T., and Horwitz, A. F. (2004) Nat. Cell Biol. 6 154-161 [DOI] [PubMed] [Google Scholar]
- 25.Slack-Davis, J. K., Eblen, S. T., Zecevic, M., Boerner, S. A., Tarcsafalvi, A., Diaz, H. B., Marshall, M. S., Weber, M. J., Parsons, J. T., and Catling, A. D. (2003) J. Cell Biol. 162 281-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin, S., Zhuo, Y., Guo, W., and Field, J. (2005) J. Biol. Chem. 280 24698-24705 [DOI] [PubMed] [Google Scholar]
- 27.Sundberg-Smith, L. J., Doherty, J. T., Mack, C. P., and Taylor, J. M. (2005) J. Biol. Chem. 280 2055-2064 [DOI] [PubMed] [Google Scholar]
- 28.Banga, S. S., Yamamoto, A. H., Mason, J. M., and Boyd, J. B. (1995) Mol. Gen. Genet. 246 148-155 [DOI] [PubMed] [Google Scholar]
- 29.Hari, K. L., Santerre, A., Sekelsky, J. J., McKim, K. S., Boyd, J. B., and Hawley, R. S. (1995) Cell 82 815-821 [DOI] [PubMed] [Google Scholar]
- 30.Bonthron, D. T., Hayward, B. E., Moran, V., and Strain, L. (2000) Hum. Genet. 107 165-175 [DOI] [PubMed] [Google Scholar]
- 31.Liu, W., Shen, X., Yang, Y., Yin, X., Xie, J., Yan, J., Jiang, J., Liu, W., Wang, H., Sun, M., Zheng, Y., and Gu, J. (2004) J. Biol. Chem. 279 10167-10175 [DOI] [PubMed] [Google Scholar]
- 32.Yang, Y., Liu, W., Zou, W., Wang, H., Zong, H., Jiang, J., Wang, Y., and Gu, J. (2007) J. Cell. Biochem. 101 167-180 [DOI] [PubMed] [Google Scholar]
- 33.Narita, T., Yamaguchi, Y., Yano, K., Sugimoto, S., Chanarat, S., Wada, T., Kim, D. K., Hasegawa, J., Omori, M., Inukai, N., Endoh, M., Yamada, T., and Handa, H. (2003) Mol. Cell. Biol. 23 1863-1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shan, D., Chen, L., Njardarson, J. T., Gaul, C., Ma, X., Danishefsky, S. J., and Huang, X. Y. (2005) Proc. Natl. Acad. Sci. U. S. A 102 3772-3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, S., and Huang, X. Y. (2005) J. Biol. Chem. 280 27130-27137 [DOI] [PubMed] [Google Scholar]
- 36.Alahari, S. K., Reddig, P. J., and Juliano, R. L. (2004) EMBO J. 23 2777-2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei, M., Robinson, M. A., and Harrison, S. C. (2005) Structure (London) 13 769-778 [DOI] [PubMed] [Google Scholar]
- 38.Guo, D., Tan, Y. C., Wang, D., Madhusoodanan, K. S., Zheng, Y., Maack, T., Zhang, J. J., and Huang, X. Y. (2007) Cell 128 341-355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fincham, V. J., James, M., Frame, M. C., and Winder, S. J. (2000) EMBO J. 19 2911-2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koestler, S. A., Auinger, S., Vinzenz, M., Rottner, K., and Small, J. V. (2008) Nat. Cell Biol. 10 306-313 [DOI] [PubMed] [Google Scholar]
- 41.Nemethova, M., Auinger, S., and Small, J. V. (2008) J. Cell Biol. 180 1233-1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serrels, B., Serrels, A., Brunton, V. G., Holt, M., McLean, G. W., Gray, C. H., Jones, G. E., and Frame, M. C. (2007) Nat. Cell Biol. 9 1046-1056 [DOI] [PubMed] [Google Scholar]
- 43.Rayala, S. K., Talukder, A. H., Balasenthil, S., Tharakan, R., Barnes, C. J., Wang, R. A., Aldaz, M., Khan, S., and Kumar, R. (2006) Cancer Res. 66 1694-1701 [DOI] [PubMed] [Google Scholar]
- 44.Schrantz, N., da Silva Correia, J., Fowler, B., Ge, Q., Sun, Z., and Bokoch, G. M. (2004) J. Biol. Chem. 279 1922-1931 [DOI] [PubMed] [Google Scholar]
- 45.Lee, S. R., Ramos, S. M., Ko, A., Masiello, D., Swanson, K. D., Lu, M. L., and Balk, S. P. (2002) Mol. Endocrinol. 16 85-99 [DOI] [PubMed] [Google Scholar]
- 46.Howe, A. K., and Juliano, R. L. (2000) Nat. Cell Biol. 2 593-600 [DOI] [PubMed] [Google Scholar]
- 47.Koh, C. G., Tan, E. J., Manser, E., and Lim, L. (2002) Curr. Biol. 12 317-321 [DOI] [PubMed] [Google Scholar]
- 48.Zang, M., Hayne, C., and Luo, Z. (2002) J. Biol. Chem. 277 4395-4405 [DOI] [PubMed] [Google Scholar]
- 49.King, A. J., Sun, H., Diaz, B., Barnard, D., Miao, W., Bagrodia, S., and Marshall, M. S. (1998) Nature 396 180-183 [DOI] [PubMed] [Google Scholar]
- 50.Frost, J. A., Steen, H., Shapiro, P., Lewis, T., Ahn, N., Shaw, P. E., and Cobb, M. H. (1997) EMBO J. 16 6426-6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klemke, R. L., Cai, S., Giannini, A. L., Gallagher, P. J., de Lanerolle, P., and Cheresh, D. A. (1997) J. Cell Biol. 137 481-492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, W. S., Wu, J. R., and Hu, C. T. (2008) Cancer Metastasis Rev. 27 303-314 [DOI] [PubMed] [Google Scholar]
- 53.Hunger-Glaser, I., Salazar, E. P., Sinnett-Smith, J., and Rozengurt, E. (2003) J. Biol. Chem. 278 22631-22643 [DOI] [PubMed] [Google Scholar]
- 54.Frodin, M., and Gammeltoft, S. (1999) Mol. Cell. Endocrinol. 151 65-77 [DOI] [PubMed] [Google Scholar]
- 55.Deak, M., Clifton, A. D., Lucocq, L. M., and Alessi, D. R. (1998) EMBO J. 17 4426-4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu, Z. X., Yu, C. F., Nickel, C., Thomas, S., and Cantley, L. G. (2002) J. Biol. Chem. 277 10452-10458 [DOI] [PubMed] [Google Scholar]
- 57.Glading, A., Bodnar, R. J., Reynolds, I. J., Shiraha, H., Satish, L., Potter, D. A., Blair, H. C., and Wells, A. (2004) Mol. Cell. Biol. 24 2499-2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carragher, N. O., Westhoff, M. A., Fincham, V. J., Schaller, M. D., and Frame, M. C. (2003) Curr. Biol. 13 1442-1450 [DOI] [PubMed] [Google Scholar]
- 59.Pullikuth, A., McKinnon, E., Schaeffer, H. J., and Catling, A. D. (2005) Mol. Cell. Biol. 25 5119-5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vial, E., Sahai, E., and Marshall, C. J. (2003) Cancer Cell 4 67-79 [DOI] [PubMed] [Google Scholar]
- 61.Stahle, M., Veit, C., Bachfischer, U., Schierling, K., Skripczynski, B., Hall, A., Gierschik, P., and Giehl, K. (2003) J. Cell Sci. 116 3835-3846 [DOI] [PubMed] [Google Scholar]
- 62.Sahai, E., Olson, M. F., and Marshall, C. J. (2001) EMBO J. 20 755-766 [DOI] [PMC free article] [PubMed] [Google Scholar]