Abstract
p53 tumor suppressor activity is negatively regulated through binding to the oncogenic proteins Hdm2 and HdmX. The p53 residues Leu26, Trp23, and Phe19 are crucial to mediate these interactions. Inhibiting p53 binding to both Hdm2 and HdmX should be a promising clinical approach to reactivate p53 in the cancer setting, but previous studies have suggested that the discovery of dual Hdm2/HdmX inhibitors will be difficult. We have determined the crystal structures at 1.3 Å of the N-terminal domain of HdmX bound to two p53 peptidomimetics without and with a 6-chlorine substituent on the indole (which binds in the same subpocket as Trp23 of p53). The latter compound is the most potent peptide-based antagonist of the p53-Hdm2 interaction yet to be described. The x-ray structures revealed surprising conformational changes of the binding cleft of HdmX, including an “open conformation” of Tyr99 and unexpected “cross-talk” between the Trp and Leu pockets. Notably, the 6-chloro p53 peptidomimetic bound with high affinity to both HdmX and Hdm2 (Kd values of 36 and 7 nm, respectively). Our results suggest that the development of potent dual inhibitors for HdmX and Hdm2 should be feasible. They also reveal possible conformational states of HdmX, which should lead to a better prediction of its interactions with potential biological partners.
The tumor suppressor protein p53 plays a central role in cellular responses to stress signals (1). As a transcription factor, p53 can induce a variety of genes involved in cell cycle arrest and apoptosis. Consequently, cancers often accumulate genetic alterations to suppress p53 activity, including p53 missense mutations and deletions or amplifications of its negative regulators human Mdm2 (Hdm2)2 or human MdmX (HdmX) (1). Hdm2 inhibits p53 by acting as an E3 ubiquitin ligase, namely by binding and promoting ubiquitin-dependent degradation of p53. HdmX can also bind p53 and appears to suppress p53 transcriptional activity through an as yet undefined mechanism (for review, see Refs. 1 and 2). Notably, deletion of either Mdm2 or MdmX in the mouse results in embryonic lethality which is fully reverted by additional loss of p53, providing genetic validation for the importance of these two proteins in regulating p53 (3-7).
Reactivation of the tumor suppressor activity of p53 is an attractive targeted therapy approach currently under exploration. One example of this is p53-Hdm2 protein-protein interaction inhibitors, namely low molecular mass molecules, which are able to compete with p53 for binding to Hdm2 (for review, see Ref. 8). Nutlins are a class of molecules that have been shown to stabilize p53 through blocking its interaction with Hdm2 and inducing p53 transcriptional activity in vivo (9). However, both nutlins and the recently reported spiro-oxindole class (10) appear to lack sufficient potency to inhibit the p53-HdmX interaction (11, 12). In support of this, cellular activity of the compounds could be enhanced by additionally knocking down HdmX (12-15). The body of experimental evidence to date, therefore, suggests that targeting both Hdm2 and HdmX would provide the best anti-tumorigenic effect and that currently developed p53/Hdm2 antagonists lack the ability to inhibit HdmX.
Both Hdm2 and HdmX share significant homology in the primary structure of their N-terminal p53 binding domains (54%) (2). Nevertheless, previous biochemical studies using p53 peptides have revealed that Hdm2 binds p53 with 10-fold higher affinity than HdmX (16). This was further confirmed using a phage display approach, where the peptide identified to bind both proteins showed a 10-fold difference in affinities (17). Finally, recent work in modeling the conformational transitions of apo-Hdm2 and apo-HdmX to their p53-bound states has also reaffirmed the notion that p53 recognition is favored by Hdm2 (18). p53 binding to Hdm2 was originally described as an amphipathic α-helix binding in the deep hydrophobic cleft of Mdm2 (19). The p53 residues Leu26, Trp23, and Phe19 are crucial to mediate this interaction and bind in three Mdm2 subpockets of the same name. To date, several structures have been published, confirming the original observations of Kussie et al. (19) for both Hdm2 and HdmX. These include p53 peptidomimetic-bound Hdm2 (20), p53 peptide-bound humanized zebrafish MdmX (21), and p53 peptide-bound HdmX (22). The latter structure and its comparison with Hdm2 have led to several hypotheses (22). First, the side chain of Tyr99 of HdmX (shaping the p53 binding pocket) was proposed to be permanently in the so called “closed conformation” (χ1 ≅ -70°), in contrast to the open conformation of the corresponding Tyr100 (χ1 ≅ 180°) observed to be possible for Hdm2. This was thought to be an intrinsic property of HdmX arising from differences between HdmX and Hdm2 in helix α2′. As a consequence, it was also concluded that the binding site of HdmX is intrinsically shallower and more restricted than the one of Hdm2 (21, 22). Second, no conformational changes in the Trp and Leu pockets of HdmX were observed or predicted. Third, it was concluded from the structures that HdmX must be targeted independently from Hdm2 and that a dual inhibitor would not be feasible (22).
Here we report the first crystal structures of human HdmX complexed with 8-mer peptide analogues of p53 at 1.35 Å and 1.33 Å resolution. The synthetic peptidomimetic ligands are Ac-Phe-Met-Aib-Pmp-Trp-Glu-Ac3c-Leu-NH2 (compound 1, Fig. 1B) and Ac-Phe-Met-Aib-Pmp-6-Cl-Trp-Glu-Ac3c-Leu-NH2 (compound 2, Fig. 1B) (23). Compound 2 is a derivative of compound 1 which has an additional 6-chlorine substituent on the indole ring of the Trp side chain. Our results elucidate the molecular basis for the interaction of ligands (low molecular mass compounds or protein partners) with HdmX. Surprisingly, we have found Tyr99 in an open conformation (χ1 = 178°) in the HdmX-compound 2 structure. We have also found an unexpected cross-talk to be possible between the Trp and Leu pockets of HdmX, considerably enlarging the Leu pocket of HdmX and, thus, allowing deeper interactions of groups binding into this pocket. Finally, we determined by isothermal titration calorimetry (ITC) and differential scanning fluorimetry (DSF) that compound 2 is a potent dual inhibitor of HdmX and Hdm2 with Kd values of 36 and 7 nm, respectively (blocking binding of a p53 peptide). Unexpectedly, this suggests that high affinity binding to both Hdm2 and HdmX is compatible.
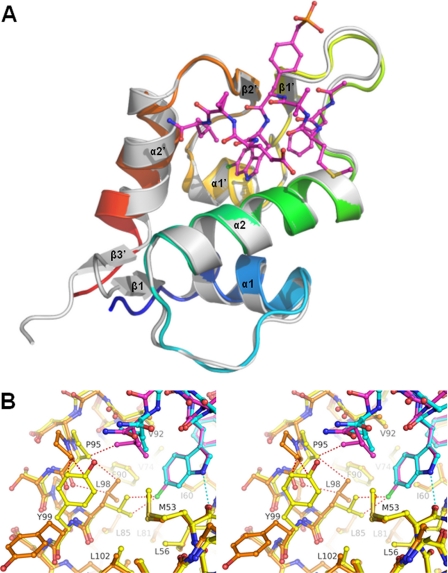
FIGURE 1.
Overall crystal structure of human HdmX in complex with the peptide-analogue Ac-Phe-Met-Aib-Pmp-Trp-Glu-Ac3c-Leu-NH2 (compound 1). A, van der Waals surface representation showing the complex between HdmX (carbons in yellow, nitrogens in blue, oxygens in red, and sulfurs in brown) and the p53 peptide analogue compound 1 (ball-and-stick-model, carbons in cyan). Selected water molecules and hydrogen bonds are shown in white. The pockets into which Phe19, Trp23, and Leu26 of p53 bind are indicated. Compound 1 makes several water-mediated interactions with HdmX and two direct hydrogen bonds (with CO-Met53 and OE1-Gln71). The phosphonate of the Pmp residue does not make direct interactions with HdmX. The most important differences with Hdm2 are seen for the Leu pocket and the bottom of the Trp pocket (Fig. 3), whereas the Phe pockets are similar. B, chemical structures of compound 1 (Ac-Phe-Met-Aib-Pmp-Trp-Glu-Ac3c-Leu-NH2) and the derivative compound 2, which has a 6-chlorine substituent on the indole ring. The orientation of the chemical structure drawing is adapted to resemble A and C, i.e. with the C terminus on the left and the N terminus on the right. C, compound 1 fitted into the 2Fo-Fc electron density map. Figs. 1, A and C, 2, and 3 were generated with PyMOL (39).
EXPERIMENTAL PROCEDURES
Protein Cloning, Expression, and Purification—The construct His-PreSc-HdmX-(14-111; C17S) was made (numbering according to Swiss-Prot O15151, i.e. equivalent residues of HdmX have residue numbers reduced by 1 compared with Hdm2). We have introduced the mutation C17S in HdmX to reduce biochemical instability problems during purification and crystallization associated with cysteine oxidation. As the x-ray structures of the HdmX complexes with both compounds 1 and 2 have shown (cf. below), the N-terminal region (Gly-Pro-Asp14-Asn25) containing this mutation C17S is flexible (no electron density visible), i.e. does not interact with the ligands. Because Cys17 could not make a disulfide bond with the only other Cys in the construct (Cys76, which is buried and too far away), this flexibility is probably not caused by the mutation. It is, thus, unlikely that the mutation C17S has a significant influence on the binding behavior of compounds 1 and 2. The mutation was introduced by site-specific mutagenesis using the QuikChange mutagenesis kit of Stratagene, and the correctness of the construction was verified by DNA sequence analysis (the correct clone was called pXI607e). The plasmid was transformed into Escherichia coli BL21 Rosetta (DE3) cells (Invitrogen) for expression. The expressed protein was found to be soluble in the cytosol of the cells. The inoculation culture in LB Medium was shaken at 37 °C overnight and was added the following day to Fernbach flask (2.8 liters) with 1 liter of LB medium with 50 mg/liter kanamycin and polypropyleneglycol. The culture was incubated at 37 °C until the A600 reached a value of ∼0.9. The temperature was lowered to 18 °C, and the culture was induced with 0.25 mm isopropyl 1-thio-β-d-galactopyranoside overnight. Frozen E. coli cells expressing His-PreSc-HdmX-(14-111; C17S) were lysed in buffer A (50 mm sodium phosphate buffer, containing 250 mm NaCl, 10% v/v glycerol, 2 mm TCEP, and Complete™ EDTA-free protease inhibitor mixture) by passing twice through an Avestin high pressure homogenizer. The resultant lysate was clarified by centrifugation (45 min at 43,000 × g) and filtration (0.45 μm) then loaded at a flow rate of 5 ml/min onto a 5-ml chelating column (Nickel Hi-Trap Crude) which had been pre-equilibrated with buffer A. The column was subsequently washed with 5 volumes of buffer A, then eluted with a 0-100% gradient of buffer B (buffer A, containing 500 mm imidazole-HCl) over 120 ml. Fractions containing the desired protein were pooled and dialyzed against 100 volumes of buffer A in the presence of PreScission™ protease (1 μl of protease solution/mg protein) to remove the histidine tag. The cleaved protein was applied as two separate aliquots to an HR10/10 Mono Q™ column pre-equilibrated with buffer C (buffer A, but containing 25 mm sodium phosphate). The column was washed with 10 volumes of buffer C and eluted with a 0-1 m gradient of NaCl in buffer C over 30 min with a flow of 3 ml/min. The peak fractions from both runs were pooled and concentrated to ∼10 ml using a 5-kDa cut-off ultrafiltration device before loading onto an XK26/60 column of Superdex™75, which was equilibrated and run in buffer A. The HdmX-containing peak was pooled and concentrated to 8.2 mg/ml in 200 mm NaCl, 1 mm TCEP, 1 mm EDTA, 10% glycerol, 50 mm Tris-HCl, pH 7.5, before crystallization. The resulting protein was estimated to be >95% pure and homogeneous by SDS-PAGE and reverse phase HPLC-coupled to mass spectrometry. The measured molecular mass of 11,174 Da of the protein corresponded to the PreScission-cleaved, des-Met form of Gly-Pro-HdmX-(Asp 14-Thr111 C17S). For the ITC measurements of compound 2 with Hdm2, the construct His-TEV-Hdm2-(17-111) was made, resulting in the protein Gly Hdm2-(Ser17-Asn111) after cleavage with TEV protease (i.e. the same protein length as described previously in Ref. 34).
Crystallization, Data Collection, and Structure Determination—Compounds 1 and 2 were synthesized in-house following protocols previously described (23). For co-crystallization, a 5:1 stoichiometric excess of compound from a 100 mm stock solution in DMSO was added to the protein solution. Crystals for compound 1 were obtained at 4 °C by the vapor diffusion method in 1-μl hanging drops containing equal volumes of protein (8.1 mg/ml Gly-Pro-HdmX-(Asp14-Thr111; C17S), 200 mm NaCl, 1 mm TCEP, 1 mm EDTA, 10% glycerol, 50 mm Tris-HCl, pH 7.5), and crystallization buffer (3.1 m ammonium sulfate, 1% 2-methyl-2,4-pentanediol, 4% polypropylene glycol, 0.1 m MES, pH 6.0). The initial crystallization hit was obtained with the AmSO4 screen from Qiagen. The additive polypropylene glycol was found with the additive screen from Hampton Research. Crystals for compound 2 were obtained as for compound 1, except that the crystallization buffer was modified slightly to 3.1 m ammonium sulfate, 1% 2-methyl-2,4-pentanediol, 0.1 m MES, pH 6.5. Crystals were directly mounted in cryoloops and flash-frozen in the nitrogen stream. Diffraction data at 100 K were collected at the Swiss Light Source (beamline X10SA) using a Marresearch CCD detector and an incident monochromatic x-ray beam with 0.99975 and 0.98371 Å wavelength for compounds 1 and 2, respectively. Raw diffraction data were processed and scaled with the HKL program suite Version 1.96.1 (24). The estimated B-factors by Wilson plot analysis are 13.4 and 16.7 Å2 for compounds 1 and 2, respectively. The structure for compound 1 was determined by molecular replacement with MOLREP (25, 26) using as starting model the coordinates of Hdm2 in complex with compound 2 (PDB access code 2GV2) refined to 1.8 Å resolution (20). The structure for compound 2 was then solved using the compound 1 complex, starting with rigid body refinement. The program REFMAC Version 5.0 (25, 27) was used for refinement. Bulk solvent correction, an initial anisotropic B-factor correction, and restrained isotropic atomic B-factor refinement were applied. The refinement target was the maximum-likelihood target using amplitudes. No σ cut-off was applied on the structure factor amplitudes. Cross-validation was used throughout refinement using a test set comprising 5.1% (1056) of the unique reflections. Water molecules were identified with the program ARP/wARP (27, 28) and selected based on difference peak height (greater than 3.0 σ) and distance criteria. Water molecules with temperature factors greater than 60 Å2 were rejected. The programs O Version 7.0 (29) and COOT (30) were used for model rebuilding, and the quality of the final refined model was assessed with the programs PROCHECK Version 3.3 (31) and REFMAC Version 5.0 (25, 27). Crystal data, data collection, and refinement statistics are shown in Table 1.
TABLE 1.
Crystallographic summary
| HdmX-cpd1 | HdmX-cpd2 | |
|---|---|---|
| Diffraction data | ||
| Space group | P41 | P41 |
| Complexes/asymmetric units | 1 | 1 |
| Unit cell dimensions | a = b = 43.3 Å, c = 65.6 Å | a = b = 42.2 Å, c = 70.4 Å |
| Source | SLS X10SA | SLS X10SA |
| Wavelength | 0.99975 Å | 0.98371 Å |
| Resolution range | 20.0-1.35 Å (1.40-1.35 Å) | 20.0-1.33 Å (1.38-1.33 Å) |
| No. of unique reflections | 25,341 | 27,490 |
| No. of observations | 175,502 | 223,722 |
| 〈I/ |
32.9 (1.60) | 39.2 (1.10) |
| Rsym on intensitiesa | 0.057 (0.27) | 0.062 (0.27) |
| Completeness | 94.90% (61.30%) | 97.10% (74.20%) |
| Refinement | ||
| Resolution range | 20.0-1.35 Å | 20.0-1.33 Å |
| Rcrystb | 0.192 | 0.193 |
| Rfree | 0.209 | 0.206 |
| Protein atoms | 665 | 665 |
| Ligand atoms (chain L) | 82 | 83 |
| Water molecules | 85 | 82 |
| Average B-factor for protein | 15.8 Å2 | 18.2 Å2 |
| Average B-factor for ligand (chain L) | 13.6 Å2 | 17.0 Å2 |
| Average B-factor for ligand (chain M) | NAc | 34.5 Å2 |
| Rmsd from target values | ||
| Bond lengths | 0.009 Å | 0.011 Å |
| Bond angles | 1.21° | 1.60° |
| Ramachandran plot | ||
| Most favorable regions | 94.30% | 94.3% |
| Allowed regions | 5.70% | 5.7% |
| Generously allowed regions | 0% | 0% |
Rsym = ∑|Iavg - II|/∑II.
Rcryst = ∑|FP - FPcalc|/∑FP, where FP and FPcalc are the observed and calculated structure factors, Rfree is calculated for a randomly chosen 5% of reflections, and Rcryst is calculated for the remaining 95% of reflections.
NA = not applicable.
Isothermal Titration Calorimetry and Differential Scanning Fluorimetry—The ITC experiments were performed using a Microcal VP-ITC instrument (Microcal, Inc., Northampton, MA). For the titrations of compounds 1 or 2 into apoHdmX, the sample cell of the calorimeter was loaded with HdmX-(14-111; C17S) at 30 μm in 50 mm Tris, pH 7.5, 200 mm NaCl, 1.0 mm TCEP, 1 mm EDTA, and 10% glycerol. The syringe was loaded with compounds 1 or 2 (300 μm) in the same buffer. For the titration of compound 2 into apoHdm2, the sample cell of the calorimeter was loaded with Hdm2-(17-111) at 30 μm in 50 mm Tris, pH 8.0, 200 mm NaCl, 1.0 mm TCEP, 1 mm EDTA, and 10% glycerol. The syringe was loaded with compound 2 (300 μm) in the same buffer. For the titrations of the 8-mer p53 peptide (Ac-18TFSDLWKLL26-NH2, synthesized in-house) into apoHdmX and apoHdm2, the protein was at 27 and 25 μm, respectively, and the peptide was at 10× higher concentration in the syringe. For the titrations of the p53 peptide into the complexes HdmX-compound 2 and Hdm2-compound 2, complexes obtained previously by ITC were used in the sample cell. All solutions were degassed for 10 min. Titrations were performed at 25 °C with injection volumes of 5-7 μl and a spacing of 240 s. For the Kd determinations, the base line was set to zero assuming that the final injections of each titration represent only the heat of dilution. The data were fit using a one-site binding model available in the Origin ITC data analysis software (Version 7.0). DSF analysis (32, 33) was performed on HdmX-compound complexes (1:13 molar ratio) containing 30 μm HdmX in 50 mm Tris-HCl, pH 7.5, 200 mm NaCl, 1 mm TCEP, 1 mm EDTA, and 10% glycerol. A stock solution (1:700 dilution in buffer) of Sypro Orange protein gel stain (Sigma) was added in a 1:7 ratio to make up 50 μl of complex solution. DSF scans were obtained using the iQ5 reverse transcription-PCR Detection System from Bio-Rad (Switzerland) at a scan rate of 1 °C/min in the temperature range from 20 to 95 °C. All measurements were performed in duplicate.
RESULTS AND DISCUSSION
The Overall Structures of the Complexes between Human HdmX and the Peptide Analogue Compounds 1 and 2 Reveal Significant Differences around Pro95 and for α2′—We have solved the crystal structures of the complexes between human HdmX (residues 14-111, containing the mutation C17S) and two 8-mer p53 peptide analogues at 1.35 Å and 1.33 Å resolution. The synthetic peptidomimetic ligands are Ac-Phe-Met-Aib-Pmp-Trp-Glu-Ac3c-Leu-NH2 (compound 1, Fig. 1B) and Ac-Phe-Met-Aib-Pmp-6-Cl-Trp-Glu-Ac3c Leu-NH2 (compound 2, Fig. 1B) (23). Compound 2 (the most potent peptide-based antagonist of the p53-Hdm2 interaction described to date) is a derivative of compound 1 which has an additional 6-chlorine substituent on the indole ring of the Trp side chain. The asymmetric unit contains one HdmX-ligand complex, but in the case of compound 2 a second ligand molecule is bound between two protein molecules, mediating crystal contacts. The results of the crystallographic refinement are summarized in Table 1. In general the electron density for the HdmX-compound 1 complex is well defined (Fig. 1C), except for the N-terminal residues Gly-Pro-Asp14-Asn25 and the C-terminal residues Leu109-Thr111 (all of which were not modeled). The protein part of the refined model consists of the amino acids Gln26-Thr108 of HdmX. The refined model also contains 85 water molecules and one compound 1 molecule. One alternate side chain conformation had to be built, namely for Met61. The electron density for the HdmX-compound 2 complex is also well defined, except for the same N-terminal and C-terminal residues as for the compound 1 complex. There is a 2-methyl-2,4-pentanediol molecule and a second compound 2 ligand bound for which the electron density is only weak at the N-terminal Phe. This second ligand molecule mediates crystal contacts between two HdmX molecules (e.g. the 6-chlorine -indole binds in a cleft between Val49 and Ala39 of two different molecules). The refined model also contains 82 water molecules. One case of alternate side chain conformations had to be built, namely for the Met side chain of compound 2 in the p53 binding site. For both complexes the backbone region with lowest B-factors is helix α2 (8 and 10 Å2 for the compound 1 and 2 complexes, respectively). The highest backbone B-factors are observed (apart from N and C termini) in the loops β1′-α1′ and α1′-β2′ (22 and 26 Å2 for the compound 1 and 2 complexes, respectively) (for nomenclature of secondary structure elements cf. Fig. 2A). Overall, the two HdmX complexes have similar backbone structures, with a root mean square deviation (r.m.s.d.) of 0.36 Å for 83 Cα atoms of residues 26-108, but importantly, a large backbone change occurs at the N terminus of the last helix α2′. In particular, Cα of Pro95 moves by 1.7 Å. This latter change considerably enlarges the Leu pocket in the HdmX-compound 2 complex (Fig. 2B). In addition Leu98 and Tyr99 have very different side chain orientations in the two complexes which leads to shape changes of the Leu and Trp pockets (cf. below and Fig. 2B). The C termini of α2′, on the other hand, do not move significantly with respect to each other (e.g. Cα of Lys104 moves only by 0.3 Å).
FIGURE 2.
The complexes HdmX-compound 2 and Hdm2-compound 2 have different tilt angles and modified positions for their helices α2′. A chlorine substituent from a ligand at the bottom of the Trp pocket of HdmX can drastically modify the shape of the Leu pocket of HdmX (i.e. there is a cross-talk between these two pockets for HdmX, mediated by Leu98). A, ribbon drawing showing the complex between HdmX (color ramped from the N terminus is in blue to the C terminus is in red) and compound 2 (ball-and-stick-model with carbons in magenta) superposed with Hdm2-compound 2 (white, ligand not shown; PDB entry code 2GV2). B, superposition of the complexes HdmX-compound 1 (color coding as in Fig. 1A, i.e. HdmX with carbons in yellow, compound 1 with carbons in cyan) and HdmX-compound 2 (HdmX with carbons in brown, compound 2 with carbons in magenta, and chlorine in green) zoomed in on the Leu and Trp pockets. The hydrogen bond between the indole nitrogen and CO-Met53 is indicated as a cyan dotted line. Steric clashes that would occur without movement of selected atoms are shown as red dotted lines. To avoid a steric clash with the 6-chlorine substituent of compound 2, Leu98 has to adopt a different side chain orientation (the distance between 6-chlorine and CD1-Leu98 would be 2.9 Å without reorientation). This leads to a movement of Pro95 and a side chain flip of Tyr99, which drastically changes the shape of the Leu pocket. As a consequence, e.g. the Leu side chain of compound 2 can penetrate more deeply (CD2 moves by 1.5 Å into the Leu pocket). The diagram is programmed for stereo viewing.
The overall fold of HdmX is quite similar to that of Hdm2 (Figs. 1A and 2A), preserving the principal architecture (19) of two elements, β1α1β2α2 and β1′α1′β2′α2′, related by an approximate 2-fold axis. A superposition (using the SSM option in Coot (30)) of HdmX-compound 1 with Hdm2-optimized p53 peptide (PDB entry code 1T4F (34)) yields an r.m.s.d. value of 1.15 Å (sequence identity is 56.6%). The largest backbone differences occur at the N terminus of the last helix α2′. The Cα atoms of Pro95-Ser96-Pro97 from HdmX move by 2.8-2.9 Å (toward β2′) with respect to the corresponding Cα atoms of His96-Arg97-Lys98 from Hdm2, which leads to modified shapes of the Leu-pockets (cf. below and Fig. 3A). At the C terminus of α2′, Cα of Lys104 in HdmX moves by 1.5 Å (toward the C terminus of α2′) with respect to Cα of Arg105 in Hdm2. Another significant backbone change occurs for α1′, for which the Cα atoms of Glu83-Leu84-Leu85 from HdmX move by 1.0-1.2 Å with respect to the corresponding Cα atoms of Asp84-Leu85-Phe86 from Hdm2, which influences the bottom of the Trp-pocket A superposition of HdmX-compound 2 with Hdm2-compound 2 (PDB entry code 2GV2), yields an r.m.s.d. value of 0.98 Å. The largest backbone differences again occur at the N terminus of the last helix α2′ (Fig. 2A). The Cα atoms of Pro95-Ser96-Pro97 from HdmX move by 1.4-2.5 Å (toward β2′) with respect to the corresponding Cα atoms of His96-Arg97-Lys98 from Hdm2. The second largest backbone difference occurs around the end of α2′, where the Cα atoms of Met101-Leu102-Arg103 from HdmX move by 1.8-2.2 Å (toward the C terminus of α2′) with respect to the corresponding Cα atoms of Met102-Ile103-Tyr104 from Hdm2. Importantly, the side chains of Met101 and Met102 (from HdmX and Hdm2, respectively) have very different orientations, pointing to two different sides of Leu85 and Phe86, respectively.
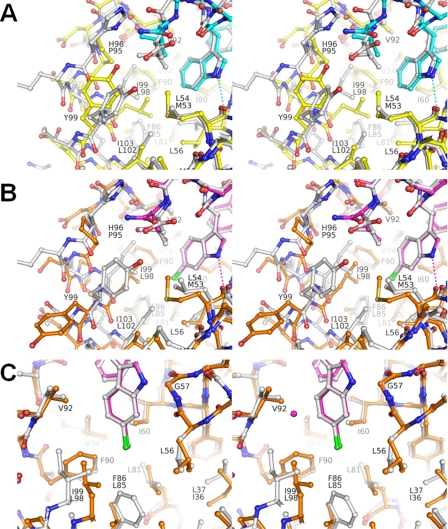
FIGURE 3.
HdmX and Hdm2 have important differences in the Leu and Trp pockets. A, superposition of the complexes HdmX-compound 1 (color coding is as in Figs. 1A and 2, i.e. HdmX with carbons are in yellow, compound 1 with carbons are in cyan) and Hdm2-optimized p53 peptide (PDB entry code 1T4F; Hdm2 and ligand with carbons in white) zoomed in on the Leu and Trp pockets. Amino acid residues that differ in identity between HdmX and Hdm2 have two labels (upper label from Hdm2), otherwise only one label (numbering for HdmX). The differences Pro95 versus His96, Met53 versus Leu54, and Leu98 versus Ile99 for HdmX versus Hdm2 modify the shape of the Leu pocket. In particular, the presence and position of CB-Pro95 for HdmX leads to a different position of the Leu side chain from the ligand. B, superposition of the complexes HdmX-compound 2 (color coding as in Fig. 2, i.e. HdmX with carbons in brown, compound 2 with carbons in magenta) and Hdm2-compound 2 (PDB entry code 2GV2; Hdm2 and ligand with carbons in white), zoomed in on the Leu and Trp pockets. The presence of a 6-chlorine substituent at the bottom of the Trp pocket leads to dramatic side chain movements of Leu98, Tyr99, and Pro95 for HdmX, where as Hdm2 shows practically no changes (cf. Figs. 2 and 3A). The Leu side chain of compound 2 can now adopt a very similar position for the HdmX and Hdm2 complexes (because the Leu pocket has been widened for HdmX), in contrast to the situation with a 6-H substituent in the Trp pocket (Fig. 3A). C, same superposition as in Fig. 3B but zoomed in on the Trp pocket. The differences Leu85 versus Phe86 and Leu98 versus Ile99 for HdmX versus Hdm2 modify the shape of the Trp pocket. In particular, the bottom of the Trp pocket is not yet completely filled by the 6-chlorine for HdmX. The important van der Waals interactions made by the 6-chlorine with Phe86 for Hdm2 are only partially substituted by hydrophobic interactions with Leu98 for HdmX. The diagram is programmed for stereo viewing.
A comparison of HdmX-compound 1 and HdmX-compound 2 with zebrafish MdmX (mutations L46V and V95L; PDB entry code 2Z5T) yields r.m.s.d. values of 0.47 and 0.51 Å (sequence identity is 71.1%), respectively. The Cα atom of Pro95 from HdmX-compound 2 moves by 1.7 Å with respect to Cα of Pro92 from the mutated zebrafish MdmX. In addition, several important side chain differences (in particular for Tyr99 versus Tyr96) are found for the latter structures.
Finally, a comparison of HdmX-compound 1 with a HdmX-p53 peptide complex (PDB entry code 3DAB) yields an r.m.s.d. value of 0.43 Å, with the largest backbone change occurring for Val62 with 1.4 Å. Overall these structures are similar, but in the binding pocket there are side chain differences for Leu56, Val92, and Tyr99. A comparison of HdmX-compound 2 with this HdmX-p53 peptide complex (PDB entry code 3DAB) yields an r.m.s.d. value of 0.53 Å. A large backbone change again occurs around the Cα atom of Pro95 from HdmX-compound 2, which moves by 1.8 Å with respect to the HdmX-p53 peptide complex. In addition, several crucial side chain reorientations (Leu98 and Tyr99) are found for the latter structures (cf. below).
Taken together, these comparisons show that HdmX has in all structures α2′-shifted (toward its C terminus) and tilted (N terminus toward α1′) in comparison with Hdm2. This overall structural change is likely to be caused by the sequence differences Pro95 versus His96 and Pro97 versus Lys98 for HdmX versus Hdm2. In addition, HdmX displays a surprising backbone adaptability (not yet observed for Hdm2) at the N terminus of α2′ and in the loop β2′-α2′ as seen in the two complexes with compounds 1 and 2.
Compounds 1 and 2 Make Conserved Hydrophobic Interactions as Well as Two Direct and Several Water-mediated Hydrogen-bond Interactions with HdmX—The following amino acids of HdmX have a non-hydrogen atom closer than 4 Å to the ligand in the HdmX-compound 1 complex: Met53, Leu56, Gly57, Gln58, Ile60, Met61, Tyr66, Gln71, His72, Val74, Val92, Lys93, Pro95, Leu98, and Tyr99. The average B-factor for the ligand (13.6 Å2) is lower than the average B-factor for the protein (15.8 Å2), consistent with the fact that excellent electron density for all non-hydrogen atoms of compound 1 is visible. The most flexible regions of compound 1 with B-factors > 25 Å2 are the carboxylate group of the glutamate side chain and the phosphono group of phosphonomethylphenyl alanine (Pmp). In the HdmX-compound 2 complex the same amino acids as for compound 1 make contacts closer than 4 Å to the ligand in the p53 binding-pocket, with the exception of Tyr99 (which is flipped away from the Leu pocket, cf. below). The most flexible regions of compound 2, with B factors > 25 Å2, are the same as for compound 1 The overall conformations of the peptide analogues 1 and 2 (Figs. 1, B and C) bound to HdmX are similar. They form short α-helical turns and bind to the same hydrophobic crevice as the Phe19-Leu26 α-helical part of p53 via their Phe-, Trp (6-Cl-Trp), and Leu side chains. Importantly, the Leu side chain of compound 2 protruded 1 1.5 Å more deeply into the Leu pocket than for compound 1 (enabled by structural changes of HdmX, cf. below), whereas the Trp and Phe side chains of the ligands are practically superposable The side chain of the Pmp residue makes van der Waals contacts with Gln71, His72, and Val92 (similarly as for the complex with Hdm2 (20)). Its phosphono group points away from the protein, i.e. does not make a direct salt bridge with Lys93 but only a water-mediated one (Fig. 1A). The N1-H of the Trp-(6-Cl-Trp) residue establishes an important hydrogen-bond interaction (2.9 Å) with CO-Met53, and the main chain NH of the Phe residue is involved in a hydrogen-bond contact (2.7 Å) with OE1-Gln71 (Fig. 1A). In addition, there is a network of water-mediated interactions with HdmX (e.g. with NE2-Gln58, ND1-His54).
A Chlorine Substituent at the Bottom of the Trp Pocket of HdmX Can Change the Shape of the Leu Pocket; Leu98 Mediates Cross-talk between These Pockets—An inspection of the compound 1 complex might lead to the conclusion that the Trp pocket of HdmX could not accept a 6-chlorine substituent (Fig. 2B) because of steric clashes with CD1-Leu98 (2.8 Å) and CD2-Leu98 (3.1 Å). For Hdm2 the corresponding Ile99 on α2′ does not obstruct the entry to the bottom of the Trp pocket because it is farther away (Fig. 3A) due to a displacement of α2′. Surprisingly, HdmX nevertheless accepts a 6-chlorine group as shown in the compound 2 complex. This is enabled by an unexpected conformational change of Leu98. In particular, χ1 = -174° and χ2 = 80° for Leu98 in the compound 1 complex changed to χ1 = -49° and χ2 = 173° for the compound 2 complex. As a consequence of this new side chain conformation of Leu98, Pro95 had to move away from the Leu pocket (Cα moves by 1.8 Å) (Fig. 2B). The new position of Pro95 induces a drastic side chain conformational change of Tyr99, i.e. a switch from the closed conformation to the unexpected open conformation. In particular, Tyr99 changes its side chain conformation from χ1 = -64° and χ2 = -23° in the compound 1 complex to χ1 = 177° and χ2 = 66° for compound 2 (Fig. 2B). This shows importantly that Tyr99 is not rigidly blocking a part of the p53 binding pocket, as thought previously (21,22) but, rather, can move away and adopt an open conformation. This cascade of conformational changes can be viewed as cross-talk between the Trp-and Leu pockets, mediated by Leu98, and highlight the conformational adaptability of this part of the protein.
The Leu and Trp Pockets of HdmX and Hdm2 Have Important Differences—The amino acid sequence differences Pro95 versus His96, Met53 versus Leu54, and Leu98 versus Ile99 for HdmX versus Hdm2 modify the shape and characteristics of the Leu pocket. In particular, His96 of Hdm2 allows an aromatic interaction with a group in the Leu pocket, whereas Pro95 of HdmX does not allow such an interaction. In the HdmX-compound 1 complex, also the position of CB-Pro95 restricts the size of the Leu pocket (Fig. 3A). As a consequence, the Leu side chain of compound 1 cannot penetrate deeply into the Leu pocket of HdmX (e.g. compared with the Hdm2 complex with PDB entry code 1T4F) (Fig. 3A). The presence of the longer Met53 for HdmX compared with Leu54 for Hdm2 also slightly restricts the Leu pocket. This difference, however, does not hinder a deeper penetration of a Leu side chain into the Leu pocket of HdmX, as demonstrated by the complex with compound 2 (Fig. 2B). In the latter HdmX complex the presence of a 6-chlorine substituent in the Trp pocket leads to a widening of the Leu pocket and, thus, enables a deeper penetration of the Leu side chain of the ligand. This is accompanied by a flip of Tyr99 from a closed conformation to an open conformation. In the widened Leu pocket, CO-Val92 is exposed and could act as a potential hydrogen-bond acceptor. This shows that, surprisingly, the Leu pocket of HdmX can be, for suitable ligands, wider (and deeper) than for Hdm2.
The amino acid sequence differences Leu85 versus Phe86 and Leu98 versus Ile99 for HdmX versus Hdm2 modify the shape and characteristics of the Trp pocket (Fig. 3C), in particular of its bottom part. For Hdm2, the edge of Phe86 forms the bottom, and interactions with it can favorably be made by a polarizable 6-chlorine substituent. By contrast, Leu85 of HdmX is shorter and, thus, leaves room for a longer substituent. In the HdmX-compound 2 complex, the 6-chlorine substituent makes contacts (distance <4.2 Å) at the bottom of the Trp pocket with the protein atoms CB-Leu56, CD1-Ile60, CD1-Leu98, and CD2-Leu98. By contrast, in the Hdm2-compound 2 complex (PDB access code 2GV2, (20)) the 6-chlorine makes contacts (distance <4.2 Å) with CB-Leu57, CG-Leu57, CD1 Ile61, CG2-Ile99, and importantly with CE2-Phe86 and CZ-Phe86. Because of the shift of α2′ toward its C terminus for HdmX, Cβ-Leu98 is closer to the Trp pocket than Cβ-Ile99 for Hdm2. Leu98 can thus, depending on its side chain conformation, either block or open the bottom of the Trp pocket for HdmX, which increases its flexibility for adaptation to different ligands.
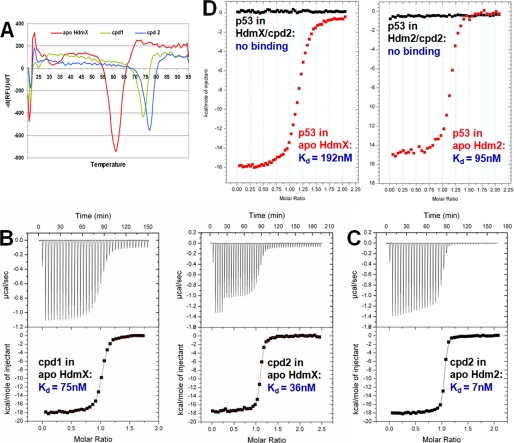
DSF and ITC Measurements Reveal That Compound 2 Binds with a Higher Binding Affinity to HdmX Than Compound 1 and That It Is a Potent Dual Inhibitor of HdmX and Hdm2—DSF measurements (Fig. 4A) comparing compound 1 and 2 binding to HdmX resulted in Δ Tm values of 12.6 and 15.7 °C, respectively. This indicates that the presence of a 6-chlorine substituent on the indole in the Trp pocket contributes additional favorable interactions (compared with 6-H). This was confirmed by ITC measurements (Fig. 4B), which revealed Kd values of 75 ± 3 and 36 ± 2 nm for compounds 1 and 2, respectively. Both methods show that the 6-chlorine substituent of compound 2 enhances the affinity for HdmX but by a factor that is much less pronounced than the 60-fold increase seen for Hdm2 (23). This difference can be explained by the favorable interactions made by the 6-chlorine with the edge of Phe86 in the case of Hdm2 and a possible energy penalty associated with the conformational rearrangement of Leu98 seen in the complex of HdmX with compound 2. ITC measurements for the interaction of compound 2 with Hdm2 (Fig 4C) revealed a Kd value of 7 ± 2 nm. This shows that compound 2 is an unexpected potent dual binder to both Hdm2 and HdmX. ITC measurements for the binding of an 8-mer p53 peptide to HdmX-compound 2 and Hdm2-compound 2 complexes indicated no significant binding under the conditions used (Fig. 4D). By contrast, the p53 peptide bound with Kd values of 192 ± 8 and 95 ± 8 nm to apoHdmX and apoHdm2, respectively (Fig. 4D). This indicates that compound 2 is indeed a potent dual inhibitor of the HdmX/p53 and Hdm2/p53 interactions and not only a dual binder.
FIGURE 4.
DSF and ITC measurements of ligand binding to HdmX and Hdm2. A, DSF-scans (20-95 °C, 1 °C/min) for apoHdmX (red) and the complexes with compounds 1 (green) and 2 (blue). The resulting ΔTm values (ligand-induced changes in protein thermal stability) are 12.6 and 15.7 °C for compounds 1 and 2, respectively. RFU, relative fluorescence units. B, calorimetric titration of compounds 1 and 2 into a sample cell containing HdmX. The titrations were performed at 25 °C in 50 mm Tris, pH 7.5, 200 mm NaCl, 1 mm TCEP, 1 mm EDTA, and 10% glycerol (same buffer as used for DSF; A) as described under “Experimental Procedures.” The data were fit with a one-site binding model to obtain Kd values of 75 ± 3 and 36 ± 2 nm for compounds 1 and 2, respectively. DSF and ITC show that the 6-chlorine substituent of compound 2 enhances the affinity for HdmX, but by a factor that is much less pronounced than the 60× for Hdm2 (23). C, calorimetric titration of compound 2 into a sample cell containing Hdm2. The titrations were performed at 25 °C in 50 mm Tris, pH 8.0, 200 mm NaCl, 1 mm TCEP, 1 mm EDTA, and 10% glycerol as described under “Experimental Procedures.” The data were fit with a one-site binding model to obtain a Kd value of 7 ± 2 nm for compound 1, showing that it is a potent dual binder to HdmX and Hdm2. D, calorimetric titrations of an 8-mer p53 peptide into sample cells containing HdmX and Hdm2, respectively (red), compared with calorimetric titrations of the peptide into sample cells containing HdmX-compound 2 and Hdm2-compound 2 complexes, respectively (black). The raw data are shown without base-line adjustment to zero. The p53 peptide bound with Kd values of 192 ± 8 and 95 ± 8 nm to apoHdmX and apoHdm2, respectively, whereas titration into the compound 2 complexes indicated no significant binding anymore.
The modest gain in potency obtained by the 6-chlorine substitution in the case of HdmX may explain the poor affinity of the current classes of potent non-peptide Hdm2-p53 antagonists (nutlins (9) and spiro-oxindoles (10)) for HdmX (11, 12). As testified by the dramatic 60-fold increase in potency obtained with compound 2, the 6-chlorine interaction is a key determinant of high affinity for the Hdm2 pocket. This structural feature is present in all known potent inhibitors of the Hdm2-p53 interaction. A similar key interaction imparting high affinity has yet to be discovered for low molecular mass inhibitors in the HdmX pocket.
Summary and Conclusion—In this paper we report the first crystal structures of human HdmX complexed with 8-mer p53-derived peptide analogues at 1.35 and 1.33 Å resolution. The synthetic peptidomimetic ligands are Ac-Phe-Met-Aib-Pmp-Trp-Glu-Ac3c-Leu-NH2 (compound 1) and Ac-Phe-Met-Aib-Pmp-6-Cl-Trp-Glu-Ac3c-Leu-NH2 (compound 2). Our data reveal for HdmX the unexpected conformational changes possible in the p53 binding cleft. In particular, we have found that a 6-chlorine substituent in the Trp pocket leads to conformational changes for Pro95, Leu98, and Tyr99 via a cross-talk between the Trp and Leu pockets. These changes drastically modify the shape of the p53 binding cleft of HdmX, thus providing new opportunities for structure-based drug design. The previous hypothesis that HdmX has an inherently narrower binding cleft (e.g. with Tyr99 always in the closed conformation) is, thus, contradicted, and our results also emphasize the inherent conformational adaptability of HdmX. Taken together, our data would, therefore, support an emerging view in the field of structural biology that intrinsic protein disorder (as in the case of the p53 N terminus) and flexibility (as in the cases of Hdm2 and HdmX) provide the basis for efficient signaling networks (35). Indeed, it is the flexibility of relatively structured proteins such as Hdm2 or HdmX that allows them to interact with various partners in a many-to-one interaction format. Additional interaction partners binding with the N-terminal domain of Hdm2 have been already proposed (36). It remains to be determined what other partners exist for HdmX. Our observations, therefore, provide the molecular basis to better understand and predict the interactions of HdmX with its potentially diverse biological targets (35, 37), which should ultimately lead to a better understanding of the biological roles of HdmX.
In contrast with previous hypotheses, we show that high affinity binding to Hdm2 can be compatible with high affinity binding to HdmX. First, our observation that a p53 wild-type peptide binds with similar affinity to both Hdm2 and HdmX (Kd values of 95 and 192 nm, respectively) fits well with the genetic and molecular evidence pointing to an equally important role of both proteins in regulating p53 (1, 2). Based on this finding, it is tempting to speculate that the biological roles of HdmX are more important than previously assumed. Second, ITC and DSF measurements have shown that compound 2 is a potent dual inhibitor of HdmX and Hdm2 (Kd values of 36 and 7 nm, respectively). This is supported by the observation that wild-type p53 peptide was unable to compete away the compound from either protein under the utilized conditions, emphasizing the stability of the interactions. Third, the observed conformational changes of HdmX when bound to various ligands suggests that our previous view of HdmX being a rigid structure to which the disordered p53 peptide binds is limited. In this sense, low molecular mass antagonists would be expected to not only have lower required binding energy (37) but could also bind to HdmX in a manner that may mimic an induced fit (38). This would suggest the association rate for these compounds to be low. However, the detailed analysis of the binding kinetics remains to be investigated. Currently existing p53-Hdm2 interaction inhibitors (such as nutlins (9) or the spiro-oxindole series (10)) do not show dual inhibitory activity, although accumulating evidence would suggest p53-HdmX interaction is as relevant a clinical target as p53 Hdm2. This is most strikingly seen in retinoblastoma, where HdmX and not Hdm2 is preferentially amplified to inhibit p53 activity (11). Hence, our work should also contribute to expanding potential clinical applications for this important class of protein-protein interaction inhibitors.
Acknowledgments
We thank R. Cebe, A. Saenger, P. Graff, E. Koch, E. Bourgier, G. Rummel, K. Yeung, and R. Wille for excellent technical assistance and P. Floersheimer for contributions. The experimental assistance from the Swiss Light Source (PX Beam Line X10SA), C. Schulze-Briese, and E. Pohl is gratefully acknowledged. We thank H. Widmer, S. Jacob, C. Garcia-Echeverria, and M. Lang for interest and support and A. Dietrich, M. Kroemer, and A. Widmer for computing support. We also thank C. Garcia-Echeverria for careful reading of the manuscript.
The atomic coordinates and structure factors (codes 3FE7 and 3FEA) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Hdm2, human Mdm2; HdmX, human MdmX; Aib, α-aminoisobutyric acid; Ac3c,1-amino-cyclopropanecarboxylic acid; ITC, isothermal titration calorimetry; DSF, differential scanning fluorimetry; r.m.s.d., root mean square deviation; Pmp, phosphonomethylphenylalanine; TCEP, Tris (2-carboxyethyl) phosphine; MES, 4-morpholineethanesulfonic acid.
References
- 1.Toledo, F., and Wahl, G. M. (2006) Nat. Rev. Cancer 6 909-923 [DOI] [PubMed] [Google Scholar]
- 2.Marine, J. C., Dyer, M. A., and Jochemsen, A. G. (2007) J. Cell Sci. 120 371-378 [DOI] [PubMed] [Google Scholar]
- 3.Finch, R. A., Donoviel, D. B., Potter, D., Shi, M., Fan, A., Freed, D. D., Wang, C. Y., Zambrowicz, B. P., Ramirez-Solis, R., Sands, A. T., and Zhang, N. (2002) Cancer Res. 62 3221-3225 [PubMed] [Google Scholar]
- 4.Jones, S. N., Roe, A. E., Donehower, L. A., and Bradley, A. (1995) Nature 378 206-208 [DOI] [PubMed] [Google Scholar]
- 5.Migliorini, D., Lazzerini Denchi, E., Danovi, D., Jochemsen, A., Capillo, M., Gobbi, A., Helin, K., Pelicci, P. G., and Marine, J. C. (2002) Mol. Cell. Biol. 22 5527-5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montes de Oca Luna, R., Wagner, D. S., and Lozano, G. (1995) Nature 378 203-206 [DOI] [PubMed] [Google Scholar]
- 7.Parant, J., Chavez-Reyes, A., Little, N. A., Yan, W., Reinke, V., Jochemsen, A. G., and Lozano, G. (2001) Nat. Genet. 29 92-95 [DOI] [PubMed] [Google Scholar]
- 8.Dudkina, A. S., and Lindsley, C. W. (2007) Curr. Top. Med. Chem. 7 952-960 [DOI] [PubMed] [Google Scholar]
- 9.Vassilev, L. T., Vu, B. T., Graves, B., Carvajal, D., Podlaski, F., Filipovic, Z., Kong, N., Kammlott, U., Lukacs, C., Klein, C., Fotouhi, N., and Liu, E. A. (2004) Science 303 844-848 [DOI] [PubMed] [Google Scholar]
- 10.Ding, K., Lu, Y., Nikolovska-Coleska, Z., Wang, G., Qiu, S., Shangary, S., Gao, W., Qin, D., Stuckey, J., Krajewski, K., Roller, P. P., and Wang, S. (2006) J. Med. Chem. 49 3432-3435 [DOI] [PubMed] [Google Scholar]
- 11.Laurie, N. A., Donovan, S. L., Shih, C. S., Zhang, J., Mills, N., Fuller, C., Teunisse, A., Lam, S., Ramos, Y., Mohan, A., Johnson, D., Wilson, M., Rodriguez-Galindo, C., Quarto, M., Francoz, S., Mendrysa, S. M., Guy, R. K., Marine, J. C., Jochemsen, A. G., and Dyer, M. A. (2006) Nature 444 61-66 [DOI] [PubMed] [Google Scholar]
- 12.Shangary, S., Qin, D., McEachern, D., Liu, M., Miller, R. S., Qiu, S., Nikolovska-Coleska, Z., Ding, K., Wang, G., Chen, J., Bernard, D., Zhang, J., Lu, Y., Gu, Q., Shah, R. B., Pienta, K. J., Ling, X., Kang, S., Guo, M., Sun, Y., Yang, D., and Wang, S. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 3933-3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu, B., Gilkes, D. M., Farooqi, B., Sebti, S. M., and Chen, J. (2006) J. Biol. Chem. 281 33030-33035 [DOI] [PubMed] [Google Scholar]
- 14.Patton, J. T., Mayo, L. D., Singhi, A. D., Gudkov, A. V., Stark, G. R., and Jackson, M. W. (2006) Cancer Res. 66 3169-3176 [DOI] [PubMed] [Google Scholar]
- 15.Wade, M., Wong, E. T., Tang, M., Stommel, J. M., and Wahl, G. M. (2006) J. Biol. Chem. 281 33036-33044 [DOI] [PubMed] [Google Scholar]
- 16.Bottger, V., Bottger, A., Garcia-Echeverria, C., Ramos, Y. F., van der Eb, A. J., Jochemsen, A. G., and Lane, D. P. (1999) Oncogene 18 189-199 [DOI] [PubMed] [Google Scholar]
- 17.Hu, B., Gilkes, D. M., and Chen, J. (2007) Cancer Res. 67 8810-8817 [DOI] [PubMed] [Google Scholar]
- 18.Macchiarulo, A., Giacche, N., Carotti, A., Baroni, M., Cruciani, G., and Pellicciari, R. (2008) J. Chem. Inf. Model. 48 1999-2009 [DOI] [PubMed] [Google Scholar]
- 19.Kussie, P. H., Gorina, S., Marechal, V., Elenbaas, B., Moreau, J., Levine, A. J., and Pavletich, N. P. (1996) Science 274 948-953 [DOI] [PubMed] [Google Scholar]
- 20.Sakurai, K., Schubert, C., and Kahne, D. (2006) J. Am. Chem. Soc. 128 11000-11001 [DOI] [PubMed] [Google Scholar]
- 21.Popowicz, G. M., Czarna, A., Rothweiler, U., Szwagierczak, A., Krajewski, M., Weber, L., and Holak, T. A. (2007) Cell Cycle 6 2386-2392 [DOI] [PubMed] [Google Scholar]
- 22.Popowicz, G. M., Czarna, A., and Holak, T. A. (2008) Cell Cycle 7 2441-2443 [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Echeverria, C., Chene, P., Blommers, M. J., and Furet, P. (2000) J. Med. Chem. 43 3205-3208 [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 25.Collaborative Computational Project, N. (1994) Acta Crystallogr. D Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 26.Vagin, A., and Teplyakov, A. (1997) J. Appl. Crystallogr. 30 1022-1025 [Google Scholar]
- 27.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 28.Lamzin, V. S., Perrakis, A., and Wilson, K. S. (2006) International Tables for Crystallography F, pp. 720-722, Kluwer Academic Publishers Group, Dordrecht, Netherlands
- 29.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Crystallogr. Sect. A 47 110-119 [DOI] [PubMed] [Google Scholar]
- 30.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 31.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 32.Cummings, M. D., Farnum, M. A., and Nelen, M. I. (2006) J. Biomol. Screen. 11 854-863 [DOI] [PubMed] [Google Scholar]
- 33.Niesen, F. H., Berglund, H., and Vedadi, M. (2007) Nat. Protoc. 2 2212-2221 [DOI] [PubMed] [Google Scholar]
- 34.Grasberger, B. L., Lu, T., Schubert, C., Parks, D. J., Carver, T. E., Koblish, H. K., Cummings, M. D., LaFrance, L. V., Milkiewicz, K. L., Calvo, R. R., Maguire, D., Lattanze, J., Franks, C. F., Zhao, S., Ramachandren, K., Bylebyl, G. R., Zhang, M., Manthey, C. L., Petrella, E. C., Pantoliano, M. W., Deckman, I. C., Spurlino, J. C., Maroney, A. C., Tomczuk, B. E., Molloy, C. J., and Bone, R. F. (2005) J. Med. Chem. 48 909-912 [DOI] [PubMed] [Google Scholar]
- 35.Dunker, A. K., Oldfield, C. J., Meng, J., Romero, P., Yang, J. Y., Chen, J. W., Vacic, V., Obradovic, Z., and Uversky, V. N. (2008) BMC Genomics 9 Suppl. 2, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, Z., and Zhang, R. (2005) Curr. Cancer Drug Targets 5 9-20 [DOI] [PubMed] [Google Scholar]
- 37.Cheng, Y., LeGall, T., Oldfield, C. J., Mueller, J. P., Van, Y. Y., Romero, P., Cortese, M. S., Uversky, V. N., and Dunker, A. K. (2006) Trends Biotechnol. 24 435-442 [DOI] [PubMed] [Google Scholar]
- 38.Koshland, D. E., Ray, W. J., and Erwin, M. J. (1958) Fed. Proc. 17 1145-1150 [PubMed] [Google Scholar]
- 39.DeLano, W. L. (2008) DeLano Scientific LLC, Palo Alto, CA