Abstract
The adaptor protein talin serves both to activate the integrin family of cell adhesion molecules and to couple integrins to the actin cytoskeleton. Integrin activation has been shown to involve binding of the talin FERM domain to membrane proximal sequences in the cytoplasmic domain of the integrin β-subunit. However, a second integrin-binding site (IBS2) has been identified near the C-terminal end of the talin rod. Here we report the crystal structure of IBS2 (residues 1974-2293), which comprises two five-helix bundles, “IBS2-A” (1974-2139) and “IBS2-B” (2140-2293), connected by a continuous helix with a distinct kink at its center that is stabilized by side-chain H-bonding. Solution studies using small angle x-ray scattering and NMR point to a fairly flexible quaternary organization. Using pull-down and enzyme-linked immunosorbent assays, we demonstrate that integrin binding requires both IBS2 domains, as does binding to acidic phospholipids and robust targeting to focal adhesions. We have defined the membrane proximal region of the integrin cytoplasmic domain as the major binding region, although more membrane distal regions are also required for strong binding. Alanine-scanning mutagenesis points to an important electrostatic component to binding. Thermal unfolding experiments show that integrin binding induces conformational changes in the IBS2 module, which we speculate are linked to vinculin and membrane binding.
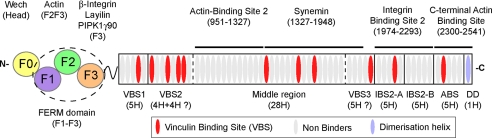
Talin (∼270 kDa) is one of a number of adaptor proteins (including α-actinin, filamin, tensin, ILK, skelemin, and melusin) that couple the integrin family of cell adhesion molecules to the actin cytoskeleton (1). However, it appears thus far to be unique in providing the necessary final step to integrin (“inside-out”) activation. Talin is composed of a head region (residues 1-400) containing an extended FERM domain, a linker region (residues 401-481) of unknown structure, and finally a long helical rod (residues 482-2541), in which ∼62 α-helices are organized into a tandem series of ∼12-13 mostly 5-helix bundles (2, 3). The C-terminal helix is a principal mediator of talin dimerization, forming an antiparallel 2-helix coiled-coil (Fig. 1).
FIGURE 1.
Domain structure and binding partners of talin. Schematic diagram of the talin molecule indicating the regions involved in binding to various ligands. The talin head (residues 1-400) contains a FERM domain (comprising F1, F2, and F3 subdomains) preceded by a domain referred to here as F0. The rod domain contains 62 predicted α-helices (ovals) organized into a series of amphipathic helical bundles. Domain boundaries based on structural determination are indicated by solid lines. Dashed lines indicate boundaries that are tentative. The ∼11 vinculin-binding sites (VBS) are shown in red. The last α-helix contains the dimerization domain (DD).
The FERM subdomain F3 has a phosphotyrosine-binding domain-like fold (4, 5) that binds to and sequesters the cytoplasmic tail of the integrin β-subunit, activating integrins in a two-step process that requires interaction with acidic membrane phospholipids. In the first step of activation, F3 makes critical interactions with the “mid-section” of the integrin tail, comprising a WXXXXNPLYXXA motif (residues 739-752 in β3). Trp-739 (it is Phe in integrin β2) inserts its side chain into a well defined hydrophobic pocket made up of residues Arg-358, Ala-360, and Tyr-377 near to the membrane-proximal surface of F3, whereas the NPXY motif forms a helical turn that nestles into a shallow groove at the membrane distal end of the F3 subdomain; the intervening residues form β-sheet interactions with the edge of the β6-strand of F3. In the second step, F3 engages the membrane-proximal helix of the β-integrin as well as the membrane itself. This is believed to cause the separation of the α- and β-integrins tails, which releases the quaternary constraints that hold the integrin in its low affinity conformation. This sets in motion or potentiates conformational changes that are transduced across the plasma membrane to the extracellular domains, promoting high affinity binding to matrix proteins or counter-receptors on other cells (6, 7). Consistent with this two-step model, talin mutants that bind normally to the integrin mid-section of the β-integrin tail but are defective in binding either to the membrane-proximal helix, or the membrane itself, have a dominant-negative phenotype. Other cell adhesion molecules bind competitively to the region in F3 where it engages the mid-section of integrin tails, utilizing variants on the integrin motif. Such molecules include the phosphatidylinositol phosphate kinase-type 1γ, which engages via a C-terminal motif (WVYSPLH) (8, 9) (in which Ser rather than Asn serves as the N-cap to the helical turn), as well as two sequences in the cytoplasmic domain of layilin (a hyaluronan receptor), one observed experimentally (WVENEIY) (10) and another predicted (FVTNDIY) (11). In these non-integrin cases, the sequence binds in a canonical phosphotyrosine-binding domain-peptide mode, and the shorter intervening sequence allows for tighter packing of the NPXY motif or its homolog against F3; however, none of these molecules activates integrin (12, 13).
The talin rod contains at least two binding sites for F-actin (14), the best characterized of which is at the C terminus (15, 16), as well as numerous potential binding sites for the cytoskeletal protein, vinculin (3, 17), which is recruited by talin to stabilize nascent focal adhesions (18, 19). Interestingly, the talin rod contains a binding site for the muscle-specific intermediate filament protein α-synemin (20), thus providing a potential link between integrin-talin-actin complexes and the intermediate filament network. Evidence has also slowly accumulated for the existence of an additional integrin-binding site, IBS2,2 in the talin rod. Initial indications that the rod contained such a site came from gel filtration studies (21). More recently, Xing et al. (22) showed that purified rod captured in microtiter wells bound αIIbβ3 integrin in a dose-dependent manner and that an antibody to the talin rod blocked αIIbβ3 binding to intact talin by only ∼50%. Moreover, αIIbβ3 integrin bound to a recombinant talin fragment from the C-terminal region of the rod (residues 1984-2541) but not to an N-terminal rod fragment (434-1975). Surface plasmon resonance studies also showed that the talin rod bound to β3-integrin tails, although the affinity was ∼40-fold weaker than that of the talin head (23). Tremuth et al. (24) further localized the binding site in the rod to residues 1984-2113, using a combination of pulldown and surface plasmon resonance assays. They also reported that binding was inhibited by a mutation of the integrin NPXY motif (Y → A), as observed for the talin head. Subsequently, Moes et al. (25) identified a 42-residue talin fragment (residues 2072-2113) that co-localized with integrin in focal adhesions, and a 23-residue peptide (residues 2077-2099) that bound GST-β3 integrin tails in a blot assay. However, none of these rod fragments was able to activate integrin when transfected into Chinese hamster ovary cells.
Further evidence in support of a second integrin-binding site in talin has come from elegant studies in Drosophila (26). As predicted, an Arg-367 → Ala mutation in the Drosophila talin FERM F3 domain (equivalent to mouse Arg-358) abrogated recruitment of the talin head to integrin-containing muscle attachment sites; furthermore, a full-length talin R367A mutant was unable to support the development of talin-null embryos to adulthood. However, the R367A mutant was able to partially rescue the talin-null phenotype in adult flies. Similarly, in embryos, the R367A mutant rescued the talin-null phenotype in various tissues, including muscle, and was recruited to integrin-containing junctions. However, close analysis showed that the muscle ends had pulled away from their matrix attachment sites, indicating a reduction in adhesion strength. It is well established that both the affinity of individual integrins and the avidity of clustered integrins for matrix proteins contribute to the overall strength of adhesion. The observations can therefore be rationalized by postulating that, although the Drosophila R367A mutant is unable to induce affinity changes, it retains the ability to support integrin clustering at cell-matrix junctions. The authors of this work suggested a model in which the talin head and rod serve distinct functions: the head converts integrins to the high affinity state, while the rod contributes to integrin clustering via its IBS2 function (26).
We have previously determined the structures of the two domains flanking IBS2, the “VBS3” domain, residues 1843-1973 (27), and the C-terminal actin-binding module, residues 2300-2482 (16). Here we describe the structure of the intervening fragment (1974-2293) comprising IBS2. The structure reveals a tandem pair of five-helix bundles forming a functional module. The N-terminal bundle has been implicated in integrin binding (24, 25), but we show that both domains are required for high affinity binding. Moreover, both domains of the module are required for focal adhesion localization and for binding to acidic phospholipids. We map the regions of the β-integrin tail critical for IBS2 binding and show that both membrane-proximal and -distal interactions are required for high affinity binding. Together, these results suggest that the two major integrin-binding sites on talin share many common features but have distinct functions.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—The cDNAs encoding murine talin residues 1974-2293, 1974-2140, and 2137-2293 were synthesized by PCR using a mouse talin1 cDNA as template and cloned into expression vector pET-151/d-TOPO (Invitrogen). Constructs were expressed in Escherichia coli BL21 Star (DE3), cultured either in LB or, for preparation of 15N-labeled samples for NMR, in minimal media containing 1 g of 15N-ammonium chloride per liter. Recombinant His-tagged talin 1974-2293 was expressed in E. coli B834 strain for selenomethionine (SeMet) incorporation, and cultured in appropriate minimal media. Recombinant His-tagged talin polypeptides were purified by nickel-affinity chromatography following standard procedures. The His tag was removed by cleavage with AcTEV protease (Invitrogen), and the protein was further purified by anion-exchange chromatography. Recombinant His-tagged chicken vinculin domain 1 (residues 1-258) was expressed using a pET-15b expression plasmid and purified as described previously (28). The concentration of purified proteins was determined using the CB-Protein Assay (Calbiochem).
X-ray Crystallography—Crystals of talin residues 1974-2293 were obtained at 19 °C by vapor diffusion equilibration against 10% (w/v) polyethylene glycol 8000, 100 mm HEPES, 1% (w/v) polyethylene glycol 3350, 10 mm sodium thiocyanate at pH 7.5. Protein at 5.0 mg/ml in 0.2 m NaCl, 2 mm dithiothreitol, and 20 mm Tris-HCl, pH 8.0, was mixed with an equal volume of precipitant. Crystals adopt space group P21, but two distinct forms were observed. Native protein yielded Form 1 crystals, with 1 molecule per asymmetric unit, whereas SeMet crystals yielded Form 2 crystals containing 2 molecules per asymmetric unit (see Table 1), with solvent contents of 47% and 45%, respectively. The two forms are closely related in their crystal packing, but there is a near doubling of the c-axis in Form 2 to accommodate the second molecule.
TABLE 1.
Summary of crystallographic analysis and refinement statistics for talin residues 1974-2293
Rsym = S|I - <I>|SI, where I is the observed intensity and <I> is the average intensity of the multiple observations of symmetry-related reflections. R = S||Fo| - |Fc||/S|Fo|; Rfree is calculated for a randomly selected 5% number of the reflections; Rfactor is calculated for the remaining 95% of the reflections used in refinement. Values in parentheses represent the outer resolution shell.
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions | a = 59.4 | a = 58.7 Å |
| b = 57.1 Å | b = 57.6 Å | |
| c = 92.1 Å | c = 92.6 Å | |
| β = 102.8° | β = 103.0° | |
| No. of molecules in asymmetric unit | 2 | 2 |
| Data set | Peak | High resolution |
| Wavelength (Å) | 0.9791 | 0.9757 |
| Resolution (Å) | 20-2.5 | 20-1.85 |
| Measured reflections | 154011 | 559438 |
| Unique reflections | 40327 | 52439 |
| Completeness (%) | 99.8 (99.5) | 98.1 (87.5) |
| Rsym | 9.0 (30.6) | 8.5 (44.9) |
| I/σI | 12.8 (5.7) | 16.8 (1.7) |
| Refinement statistics | ||
| Resolution range (Å) | 20-1.85 | |
| Unique reflections (free) | 49457 | |
| Rwork (%) | 21.4 (29.0) | |
| Rfree (%) | 26.0 (41.0) | |
| Number of residues/atoms | 632/5010 | |
| Number of solvent molecules | 332 | |
| Average B value (Å2) | 31 | |
| r.m.s.d. bond length (Å) | 0.016 | |
| r.m.s.d. bond angles (Å) | 1.45 |
Diffraction data were collected from native crystals (Form 1) at European Synchrotron Radiation Facility beamline ID23-1, and from SeMet crystal (Form 2) at beamline 14-4, recorded on ADSC Q315R charge-coupled device detectors. Data were processed with DENZO and SCALEPACK (29). Phases were determined from the anomalous data collected at the selenium absorption peak (λ = 0.9791) from SeMet Form 2 crystals. 10 of the 12 possible selenium atoms were located (6 per molecule), and a map was constructed using these preliminary phases at 2.5-Å resolution. An initial atomic model was built using SOLVE/RESOLVE (30), and, following phase improvement with DM (31), the model was rebuilt manually with Coot (32) and refined using maximum likelihood refinement in Refmac5 (33). Subsequently, the structure was refined against a new 1.85-Å data set collected from Form 2 SeMet crystals.
The final model converged to an RWORK of 21.4% for all data between 20 and 1.85 Å, and an Rfree of 26.0%. The final Ramachandran plot shows 96.9% of residues in favored regions, 2.8% in additional favored regions, and 0.2% in generously allowed regions, as defined by PROCHECK (34). The structure has been submitted to the Protein Data Bank with the accession number 3dyj (www.rcsb.org). The figures were generated with CCP4mg (35). The Form 1 native crystals did not diffract as well as the SeMet derivatives, and the data were partially refined to an Rfree of 23.7%.
Gel Filtration and Proteolysis—Analytical gel filtration chromatography of recombinant talin fragments 1974-2293, 1974-2140, and 2137-2293, as well as vinculin Vd1-(1-258), was performed using Superdex-75 (10/300) GL (Amersham Biosciences) at room temperature. The proteins were mixed and incubated at various temperatures for 30 min prior to loading onto the column, which was pre-equilibrated with and run with 20 mm Tris, pH 8.0, 150 mm NaCl, and 2 mm dithiothreitol at a flow rate of 0.8 ml/min. All proteolysis experiments were carried out at 20 °C for 1 h using a 1:50 (w/w) trypsin:protein ratio. The buffer was 150 mm NaCl, 20 mm Tris, pH 8.0.
SAXS—Small angle x-ray scattering (SAXS) experiments were carried out at station 2.1 of the U.K. Synchrotron Radiation Source at Daresbury, using a multiwire gas detector covering a momentum-transfer range of 0.02 Å-1 < q < 0.70 Å-1, where q = 4π sin Θ/λ (2Θ is the scattering angle and λ the x-ray wavelength, 1.54 Å). Measurements on talin 1974-2293 were performed at 4 °C at concentrations of 2 and 10 mg/ml in a buffer comprising 20 mm sodium phosphate, pH 6.5, 50 mm NaCl, and 2 mm dithiothreitol. Experimental data were accumulated in 60-s frames, and, before averaging, frames were inspected for x-ray-induced damage or aggregation. The background was subtracted using the scattering from the buffer solution alone. No protein aggregation was detected, and the linearity of the Guinier plot (supplemental Fig. S1) indicated that the protein solutions were homogeneous. Data reduction was carried out with software provided at the Daresbury facility, and subsequent analysis was done with the ATSAS program package (36). The theoretical Rg for the crystal structure of the IBS2 domain was calculated using Crysol (37).
NMR Spectroscopy—NMR spectra were collected using a 0.2 mm protein solution in 20 mm sodium phosphate buffer (pH 6.5), 50 mm NaCl, and 2 mm dithiothreitol at 298 K on a Bruker AVANCE DRX600 spectrometer equipped with a cryoprobe. Spectra were processed and analyzed using TopSpin software (Bruker).
Binding of Integrin Tails to Talin Rod Using a Pulldown Assay—Purified talin rod fragments were diluted to 250 nm in PN buffer (10 mm PIPES, 50 mm NaCl, 150 mm sucrose, 50 mm NaF, 40 mm sodium phosphate, pH 6.8). To assay for integrin binding, 700 μl of protein solution was mixed (incubated for 2 h at room temperature) with 10 μg of His-Avi-tagged integrin tails immobilized on NeutrAvidin-coated beads. Unbound proteins were removed by three washes in PN buffer containing 5% Triton X-100, and bound proteins were solubilized in Laemmli sample buffer and detected by Western blotting (38). Mouse anti-V5 antibody (Invitrogen) was used to detect talin rod domains, and a rabbit Anti-His antibody (Santa Cruz Biotechnology) was used to detect the talin head.
Binding of Integrin Tails to Talin Rod by ELISA—Microtiter wells (ELISA high binding plate, white, Fisher) were coated with a solution of 10 μg/ml NeutrAvidin (Pierce) and incubated with 150 μl of blocking buffer (1% heat inactivated-bovine serum albumin in PBS). Wells were then incubated with purified recombinant His-Avi-tagged integrin tails (2 μg/ml) diluted in PBS containing 1% bovine serum albumin and 0.2% Tween 20 (sample buffer). Talin rod fragments (25-2500 nm) in sample buffer were added to the wells, and bound talin was detected with primary mouse Anti-V5 antibody (Invitrogen, ratio 1:5000) and a secondary goat anti-mouse horseradish peroxidase-conjugated antibody (BIOSOURCE) using luminescence with ECL (Amersham Biosciences). All incubations were for 1 h at 37 °C in 50 μl of buffer unless otherwise stated, and plates were washed three times with PBS containing 0.2% Tween 20 after each step. Controls included wells without NeutrAvidin, NeutrAvidin without integrin, and wells coated with αIIb-integrin. Integrin loading onto NeutrAvidin plates was quantitated using mouse 7H8 anti-helix monoclonal antibody, and some wells were coated with the talin fragments to verify equal loading of the various constructs.
Expression of GFP-tagged Talin IBS2 Polypeptides in Vinculin-null Cells—Vinculin-null mouse embryonic fibroblasts (39) cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum with 2 mm l-glutamine were plated onto glass coverslips, and 24 h later transfected with cDNAs encoding various pEGFP-C2-tagged mouse IBS2 polypeptides using FuGENE 6 (Roche Applied Science), according to the manufacturer's instructions. Cells were fixed with pre-warmed para-formaldehyde (4% (w/v) in PBS), permeabilized with 0.5% Triton X-100 in PBS, and stained using a mouse monoclonal antibody to paxillin (clone 349, BD Biosciences, diluted 1:300 in 0.05% Triton X-100 in PBS) followed by a goat anti-mouse conjugated to AlexaFluor 568 fluorescent dye. Cells were imaged using an inverted Zeiss Axiovert 200M microscope equipped with a 63× oil immersion lens (numerical aperture = 1.3). Digital images were processed in Adobe Photoshop CS2.
Phospholipid Binding—Phosphatidylinositol phosphate strips (Invitrogen) were treated at room temperature for 5 h with 3% ovalbumin in TBS-T (10 mm Tris, pH 8.0, 150 mm NaCl, 0.1% Tween 20) to eliminate nonspecific binding, and incubated overnight at 4 °C with 1 μg/ml talin fragments in TBS-T containing 3% ovalbumin. After incubation, the strips were washed three times at room temperature in TBS-T containing 0.1% ovalbumin, and talin binding was detected with a mouse anti-His horseradish peroxidase-conjugated antibody (ratio 1:6000, 1 h at room temperature, Alpha Diagnostics), followed by three washes in TBS-T. The signals were detected by enhanced chemiluminescence (Pierce).
SPOT Synthesis—Peptides (25- and 36-mers) based on the mouse β-integrin sequences were synthesized on a fully automated SPOT synthesizer Multipep (Intavis AG, Germany). The derivatization of hydroxyl groups of cellulose sheets (Schleicher & Schuell, Germany) was carried out with Fmoc-alanine, 1-methylimidazole, and dicyclohexylcarbodiimide in dimethyl formamide overnight. The peptides were then synthesized by repeated deposition of pre-activated amino acids onto derivatized cellulose sheets via Fmoc chemistry, using dicyclohexylcarbodiimide/1-hydroxybenzotriazole activation of amino acids in N-methyl-2-pyrrolidone and Fmoc deprotection with 20% (v) piperidine in dimethyl formamide. During the first three cycles of synthesis, residual amino groups and the final N-terminal amino groups were blocked with a mixture of 80% acetic anhydride/10% N,N-diisopropylethylamine/10% dimethyl formamide (v/v) for 30 min. Finally, cleavage of side-chain protection groups was carried out in 95% trifluoroacetic acid/5% dichloromethane (v/v) for 45 min. Typically, the loading of peptides was ∼100 nmol per spot.
Talin IBS2 Binding to β-Integrin SPOT-peptide Arrays—Membranes were treated overnight with 10% fetal bovine serum in Tris-buffered saline (50 mm Tris-HCl, pH 7.0, 137 mm NaCl, 2.7 mm KCl). Murine talin fragment C (residues 1975-2541), N-terminally tagged with T7 & GFP, and C-terminally with His-7, was expressed using pET23A-T7 (40) and purified on nickel-nitrilotriacetic acid-Sepharose (Qiagen) according to the manufacturer's protocol. Eluted protein was subjected to Mono Q ion-exchange chromatography (Amersham Biosciences), and purified T7/GFP-talin C was transferred into PBS. Membranes were overlaid for 2 h with T7/GFP-talinC (1 μm) in Tris-buffered saline with 1% bovine serum albumin at room temperature. Bound T7/GFP-talin fragment C was detected using a monoclonal T7 antibody (Novagen) and alkaline phosphatase-coupled anti-mouse Ig (Jackson Laboratories), as described previously (3).
Differential Scanning Calorimetry—DSC experiments were carried out using an NDSC II calorimeter (CSC) at a scanning rate of 1 K/min under 3.0 atm of pressure. Protein samples were exchanged into DSC buffer comprising 20 mm PIPES (pH 7.5) and 100 mm NaCl. β1A-integrin peptides (47- and 25-mers) were synthesized by Dr. Sven Rothemund (Interdisziplinäres Zentrum für Klinische Forschung Leipzig), and also dissolved in DSC buffer. Protein samples were analyzed at 0.7 mg/ml with β-integrin peptide at 100 or 200 μm.
RESULTS
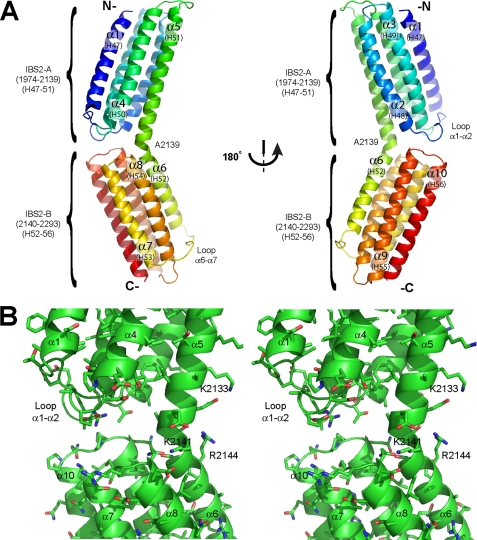
Crystal Structure of Talin IBS2 (Residues 1974-2293)—To determine the structure of IBS2, we expressed recombinant talin residues 1974-2293 in E. coli, and obtained crystals from the purified polypeptide that diffracted x-rays. We determined initial phases using anomalous scattering from a selenomethionine derivative from Form 2 crystals (see “Experimental Procedures”) at 2.5-Å resolution, and subsequently collected a diffraction set to 1.85-Å resolution for high resolution refinement (Table 1). The final high resolution model includes two copies of IBS2 (residues 1975-2291) within the asymmetric unit. IBS2 comprises a tandem pair of five-helix bundles with the same topology, comprising five anti-parallel α-helices (ranging from 24 to 30 residues in length). Helices α2-α5 and α7-α10 are folded into left-handed up-down-up-down 4-helix bundles. In both cases, a long 10-residue linker connects the first and second helices (i.e. α1-α2 and α6-α7); otherwise, the helices are connected by short loops (Fig. 2A and supplemental Fig. S2). The two IBS2 five-helix bundles can be superposed using Coot (32) with an r.m.s.d. of 2.4 Å on backbone atoms (supplemental Fig. S3A). This bundle topology has only previously been seen (DALI (41)) in the five-helix bundle at the N terminus of the talin rod (residues 482-655) (28), and both IBS2 bundles superpose with this domain with an r.m.s.d. of ∼2.5 Å for backbone atoms (supplemental Fig. S3B).
FIGURE 2.
Structure of IBS2 in the talin rod. A, schematic representation of the talin 1974-2293 crystal structure. The upper five-helix bundle is called IBS2-A, and the lower one IBS2-B. The helix numbers shown in brackets are for full-length talin. B, stereo representation of the area located between the two domains in the crystal structure; there is no evidence of hydrophobic or electrostatic interactions between the two domains.
The two bundles are linked by an almost continuous helix, but with a distinct kink between the bundles. Several intrahelical main-chain H-bonds are lost between residues Glu-2138 and Thr-2143, dividing the helix into two segments (α5 and α6), which are assigned to the N- and C-terminal bundles, respectively. The kink is, however, stabilized by several intra- and interhelical H-bonds that replace the lost main-chain H-bonds (Fig. 2B). Of particular note, three interactions with the beginning of helix α8 stabilize the kink: the amide side chain of Gln-2198 makes simultaneous H-bonds with the “orphan” amides of Gly-2142 and Thr-2143, whereas the side chains of Thr-2140 and Arg-2144 act as C-terminal caps to helix α5. In addition, the side chain of Glu-2199 makes a salt bridge with Lys-2141, and the side chain of Ile-2202 packs against Gly-2142. These features are conserved in all known sequences and are structurally conserved in all three copies of the two crystal forms, suggesting that the helical kink is a biological feature of the two-domain module. Other contacts between the bundles are limited and vary in different crystal environments (see below).
The two molecules (A and B) in the Form 2 asymmetric unit have similar tertiary and quaternary organizations, but there are some significant differences. Thus, the individual bundles overlay with r.m.s.d. values of <0.5 Å for most main-chain residues. However, the large α1-α2 loop in the first bundle adopts two distinct conformations, beginning at residue Asn-2005 and propagating down helix α2 as far as Lys-2024. At the apex of the α1-α2 turn, residue Ala-2009 in molecule A shifts by 5.5 Å toward the second bundle compared with molecule B so that it makes hydrophobic contacts with the α9-α10 loop from the second bundle; there are also several water-mediated polar interaction; nevertheless, the interface is limited. In molecule B, the only significant contacts between the two domains are two long range hydrogen bonds (Arg-2006 H-bonds to the C=O of Gln-2259, and Lys-2260 H-bonds to Gly-2008 C=O). The interfacial difference is linked to a significant alteration in the quaternary organization of the module, involving a 2- to 3-Å translation of the second bundle with respect to the first in the two molecules. This is accommodated by a gradual bend in helix α5 with little change in the kink angle (∼40°). In Form 1 crystals (see “Experimental Procedures”), the intrabundle contacts closely resemble those of Form 2 molecule A. Furthermore, analysis of crystal contacts shows that this region in Form 2 Molecule B forms several lattice contacts, whereas molecule A does not. These observations point to a significant degree of flexibility within this interfacial region.
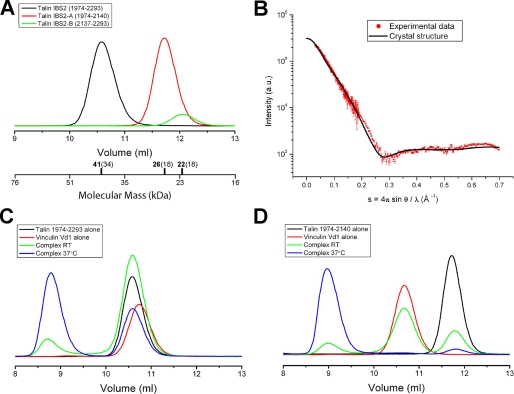
Solution studies support the crystallographic studies, pointing to an extended conformation with some flexibility, as judged by analytical gel filtration, SAXS, and NMR (Fig. 3 (A and B) and supplemental Fig. S4). Likewise, NMR line-widths point to a module of intermediate flexibility: the increase in line width of the IBS2 module compared with the individual bundles (supplemental Fig. S4, A-C) is greater than that expected for a pair of domains tumbling independently, but it is also greater than that expected for a rigid domain pair. Additionally, the positions of the resolved resonances change very little compared with the two domains in isolation, consistent with the small interfacial area demonstrated crystallographically.
FIGURE 3.
Biochemical characterization of the talin IBS2 polypeptide. A, talin polypeptides spanning residues 1974-2293 (IBS2), 1974-2140 (IBS2-A), and 2137-2293 (IBS2-B) were analyzed on a Superdex-75 (10/300) GL gel filtration column. The apparent molecular mass for each domain is indicated with their theoretical molecular mass in brackets. The talin IBS2 polypeptides showed an anomalous elution profile indicative of an extended conformation. B, SAXS of the talin IBS2 polypeptide indicates a different domain organization from that in the crystal structure. Experimental scattering profile of talin IBS2 (red) compared with the simulated scattering profile based on the crystal structure (black line) (goodness-of-fitχ = 8.9). C and D, binding of the vinculin Vd1 domain to talin IBS2 (C) or IBS2-A (D) was analyzed on a Superdex-75 (10/300) GL gel filtration column at room temperature (RT). Incubation of either IBS2 or IBS2-A with Vd1 at room temperature resulted in rather little complex formation, and most of the talin and vinculin polypeptides remained in the free form. However, preincubation of the proteins at 37 °C resulted in formation of a talin-Vd1 complex.
The IBS2 module is relatively resistant to trypsin digestion, despite its high Arg and Lys content (11%). The most abundant cleavage site is at Lys-2133 between the two five-helix bundles, whereas Lys-2141 and Arg-2144, which are also surface-exposed in the crystal structure, are resistant to cleavage. The crystal structure rationalizes these data: thus Lys-2133 is fully exposed and makes only a weak ionic interaction with Asp-2137 (Fig. 2B); by contrast, Lys-2141 is sandwiched between two glutamates, Glu-2138 and Glu-2139, whereas Arg-2144 forms multiple intramolecular interactions contributing to the α5 helix cap, as noted above.
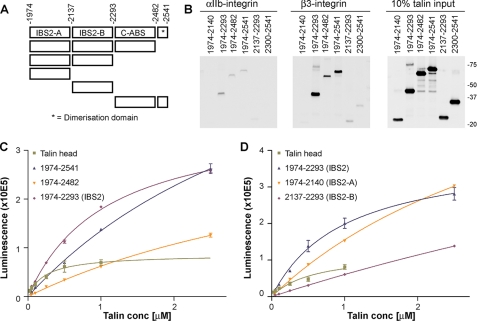
IBS2 Binds the β1 and β3 Integrin Cytoplasmic Domains—To confirm that the C-terminal region of the talin rod interacts with integrin cytoplasmic tails, we first performed pulldown experiments using biotinylated integrin tails immobilized on NeutrAvidin beads. IBS2, as well as two longer constructs, 1974-2482 and 1974-2541 (the latter includes the C-terminal dimerization domain), all bound strongly to integrin β3, whereas the C-terminal domain alone (residues 2300-2541) bound weakly (Fig. 4, A and B). These data are consistent with the integrin-binding site being located within IBS2. In contrast to previous reports, we found that individual IBS2A and IBS2B bundles bound integrin weakly or not at all. IBS2 also bound weakly to the αIIb-integrin tail, although its significance is unclear.
FIGURE 4.
The talin 1974-2293 IBS2 polypeptide binds to β3-integrin tails in pulldown and ELISA-type assays. A, schematic of talin rod polypeptides used where each box represents a five-helix bundle with the exception of the C-terminal dimerization domain, which is composed of a single α-helix that forms an anti-parallel dimer (16). All constructs include an N-terminal His tag followed by a V5 epitope. B, pulldown assays using αIIb- and β3-integrin tails immobilized on NeutrAvidin beads with purified recombinant talin rod polypeptides. Binding of talin rod polypeptides was detected using an anti-V5 antibody, while binding of the talin head (used as a positive control) was detected using an anti-His antibody (data not shown). C and D, binding of talin polypeptides to microtiter wells coated with β3-integrins using ELISA. The talin head polypeptide (residues 1-405) was used as a positive control. Binding to wells coated with αIIb-integrin or not coated with integrins were used as negative controls. The talin IBS2 polypeptide, which contains both the IBS2-A and IBS2-B five-helix bundles, binds to β3-integrin with much higher affinity than the individual IBS2-A and IBS2-B five-helix bundles.
To further characterize these interactions, we used an ELISA-type assay in which biotinylated integrin tails were immobilized on NeutrAvidin-coated microtiter wells. Binding of V5-tagged talin rod polypeptides was quantified using an anti-V5 monoclonal antibody. Talin head was used as a positive control, and uncoated NeutrAvidin-treated wells as negative controls. αIIb-coated wells were also tested. Talin head (residues 1-405) bound in a dose-dependent manner to β3-integrin tail (Fig. 4C) with an EC50 of 0.4 ± 0.2 μm, consistent with published surface plasmon resonance studies (23). The long construct, talin 1974-2541, bound with lower affinity (EC50 >2.5 μm), consistent with published data on the talin rod (23). However, the IBS2 module alone bound with significantly higher affinity (EC50 0.9 ± 0.2 μm), comparable to that of the head, suggesting that elements C-terminal to the IBS2 module may be autoinhibitory. A very similar EC50 was found for binding of IBS2 to β1A-integrin (0.9 ± 0.1 μm). The individual bundles of IBS2 did show dose-dependent integrin binding (Fig. 4D), but it was weak and not saturable under the conditions employed, with estimated EC50 values of >2.5 μm. These results confirm that strong integrin binding requires the intact IBS2 module.
The IBS2-A Five-helix Bundle Contains a Cryptic Vinculin Binding Site—IBS2-A contains a potential vinculin-binding site in helix α4 (talin rod helix 50) (3), but, as with all such sites, the vinculin-binding residues are buried in the hydrophobic core of the bundle. Very little binding between the IBS2 module and vinculin d1 domain (Vd1) was observed at room temperature, but incubation at 37 °C led to substantial complex formation as detected by gel filtration (Fig. 3C). We have previously shown a similar temperature dependence for the vinculin-binding site in talin 482-655 (28, 42). IBS2 has a Tm of 58 °C, and we conclude that increasing the temperature to the more physiologic 37 °C has a destabilizing effect on bundle integrity that facilitates vinculin binding. As expected, only the IBS2-A bundle bound Vd1 (Fig. 3D and data not shown).
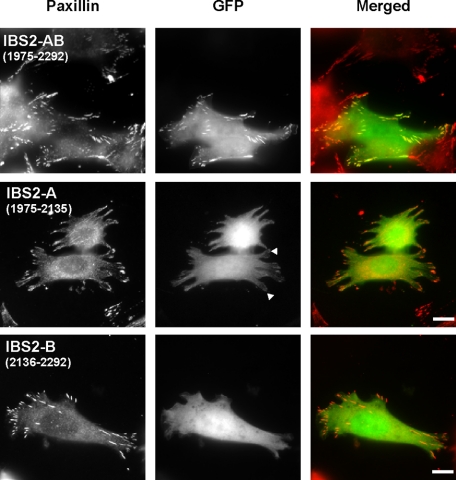
IBS2 Localization to Focal Adhesions Does Not Require Vinculin—Talin fragments from the IBS2 region have been shown to localize to focal adhesions (FAs) (25, 43), but because IBS2A contains a vinculin-binding site (3), it was unclear whether localization reflected binding to integrin or vinculin. To test this, we expressed GFP-tagged IBS2 in vinculin-null mouse embryo fibroblasts and found that it co-localized efficiently to FAs (Fig. 5). Thus, vinculin binding is not required for IBS2 localization to FAs. In contrast, GFP-IBS2-A and GFP-IBS2-B displayed only a diffuse cytoplasmic fluorescence. The data mirrors that on integrin binding and suggest that FA localization of IBS2 reflects its ability to bind integrins.
FIGURE 5.
GFP-talin IBS2 localizes to FAs in vinculin null cells. Mouse embryonic fibroblasts derived from vinculin knockout mice (39) were transfected with cDNAs encoding EGFP-tagged IBS2 fragments. FAs were visualized by paxillin staining. The IBS2 double domain construct clearly localized to FAs, whereas the individual five-helix bundles IBS2-A and IBS2-B showed little or no targeting.
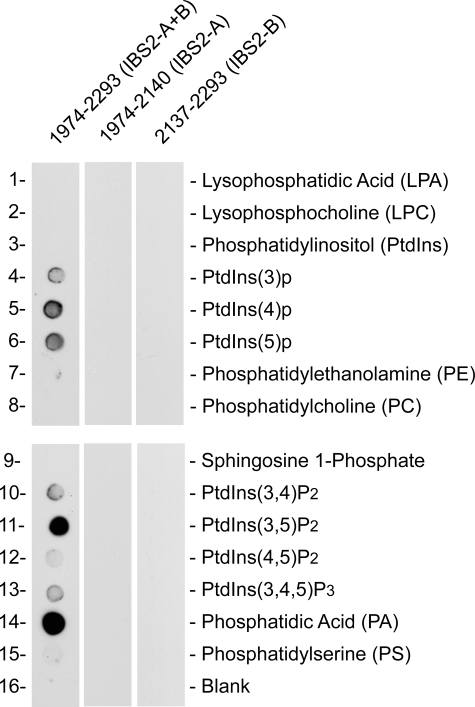
Because activation of integrins by the talin FERM domain is dependent on interactions between basic residues on the membrane-proximal surface of the FERM domain and acidic phosphoinositides in the plasma membrane (5), we explored the binding of IBS2 to a range of phospholipids spotted on a nitrocellulose membrane. We found that IBS2 bound strongly to certain acidic lipids, including phosphatidylinositol 3,5-bisphosphate and phosphatidic acid (Fig. 6), and that, as in the case of integrin-binding and FA targeting, the two-domain IBS2 module is required. Interestingly, binding to phosphatidylinositol 4,5-bisphosphate, which is up-regulated at focal adhesions, was significantly weaker.
FIGURE 6.
Talin IBS2 but not IBS2-A or IBS2-B binds to acidic phospholipids. Binding of His-tagged talin polypeptides to phosphatidylinositol phosphate strips containing an array of acidic phospholipids (Invitrogen) was detected using an anti-His antibody. Each spot contains 100 pmol of phospholipid, and the membrane was challenged with 1 μg/ml protein. Talin IBS2 (residues 1974-2293) binds to several phospholipids, whereas the individual five-helix bundles that make up IBS2, i.e. residues 1974-2140 (IBS2-A) and residues 2137-2293 (IBS2-B), did not bind to any of the phospholipids tested.
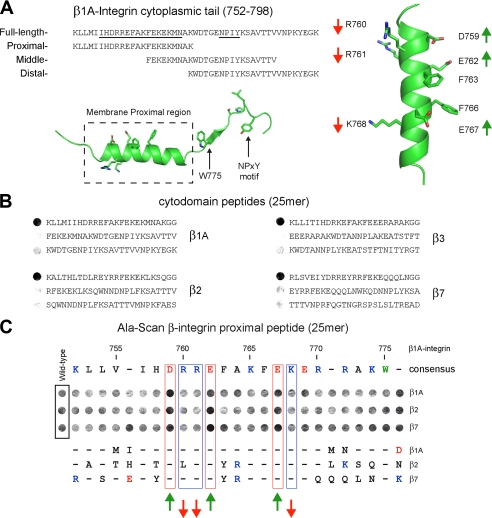
Fine Mapping of the IBS2-binding Determinants on Integrin—We first probed filters containing three overlapping SPOT-synthesized 25-mer peptides spanning the cytoplasmic tails of β1A, β2, β3, and β7 integrins (Fig. 7, A and B) with an IBS2 polypeptide spanning residues 1974-2541. The data locate the major IBS2-binding site within the 23 membrane-proximal residues of the β-integrin tails (Fig. 7B) and show that all the β subunits tested bind equally well. We next carried out alanine point mutagenesis (Fig. 7C) on this region. Replacement of most acidic residues led to a substantial increase in IBS2 binding, whereas substitution of basic residues caused a small but consistent reduction, indicating an important role for charged residues in the interaction, and suggesting that the binding site on IBS2 is acidic.
FIGURE 7.
Talin IBS2 binds to membrane proximal β-integrin tail peptides. A, alignment of the full-length β1A-integrin cytoplasmic domain peptide with the membrane proximal, middle, and distal tail peptides used in this study. The membrane proximal helical region and NPXY motif are underlined. Two schematic representations of the β-integrin cytoplasmic domain highlight key positions. The amino acid numbering is for β1A-integrin. B and C, analysis of the binding of a talin 1975-2541 polypeptide to a series of immobilized β-integrin cytoplasmic domain peptides. B, binding to membrane proximal (top), middle, and distal tail peptides equivalent to β1A-, β2-, β3-, and β7-integrins (25-mers). Binding to membrane proximal peptides from all β-integrin tails was observed (including β5 and β6; data not shown). C, analysis of binding to a series of membrane proximal β-integrin peptides (25-mers) in which each residue in turn was substituted by alanine. Amino acid substitutions that consistently affected talin binding are highlighted. Amino acid numbering for β1A-integrin is shown. Mutation of β-integrin membrane proximal peptides identify important charged residues required for optimal talin 1975-2541 binding (see A).
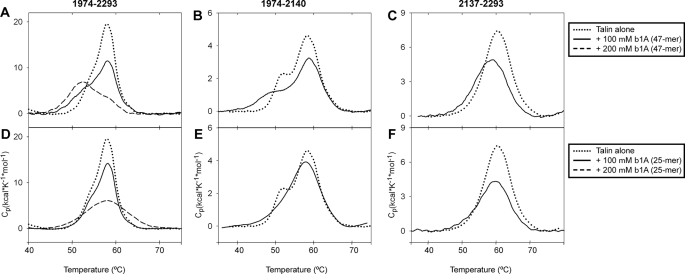
Effects of β-Integrin Peptides on the Stability of the Talin IBS2 Module—To further characterize integrin binding to IBS2, we used DSC to monitor the effect of integrin binding on the melting temperature (Tm) and unfolding enthalpy of IBS2. Binding of the full-length β1A-integrin tail peptide (47-mer) to IBS2 (Fig. 8A and Table 2) led to a decrease in ΔH at 100 μm integrin. At 200 μm integrin, ΔH was reduced further, and there was also a large reduction in Tm, suggestive of a conformational change and perhaps partial unfolding of IBS2. Two peaks with different Tm values are evident in the IBS2 module, and integrin binding promotes the peak with the lower melting temperature. The melting curves for IBS2-A and IBS2-B (Fig. 8, B and C) show that the lower peak was entirely attributable to IBS2-A. IBS2-B did bind integrin, as judged by a significant reduction in Tm and ΔH, but still melted as a single peak. Binding of the shorter membrane-proximal peptide (25-mer) caused a similar decrease in ΔH but did not lead to the formation of the peak at lower Tm (Fig. 8, D-F) suggesting that membrane-distal portions of the integrin contribute significantly to binding and conformational change, especially in IBS2-A.
FIGURE 8.
DSC analysis shows binding of various talin rod domains to β-integrin tail peptides in solution. DSC analysis of talin (A and D) 1974-2293, (B and E) 1974-2140 (IBS2-A), and (C and F) 2137-2293 (IBS2-B) in the presence of 47-mer β1A-integrin peptide (A-C) and 25-mer β1A-integrin membrane proximal peptide (D-F). The concentration of the talin constructs was 0.7 mg/ml.
TABLE 2.
Thermodynamic parameters obtained from DSC scans for talin 1974-2293, 1974-2140, and 2137-2293 upon binding β1A-integrin peptides
The absolute error in Tm values did not exceed ±0.2 °C; the relative error in ΔHcal values did not exceed ± 10%.
|
1974-2293
|
1974-2140
|
2137-2293
|
||||
|---|---|---|---|---|---|---|
| Tm | ΔH | Tm | ΔH | Tm | ΔH | |
| °C | kcal/mol | °C | kcal/mol | °C | kcal/mol | |
| Talin alone | 57.9 | 119 | 58.4 | 44 | 60.3 | 76 |
| Talin + 100 μm β1A (47-mer) | 58.0 | 94 | 58.7 | 36 | 58.7 | 63 |
| Talin + 200 μm β1A (47-mer) | 52.4 | 67 | ||||
| Talin + 100 μm β1A (25-mer) | 58.1 | 90 | 58.2 | 42 | 59.1 | 59 |
| Talin + 200 μm β1A (25-mer) | 58.2 | 72 | ||||
DISCUSSION
We have determined the structure of talin residues 1974-2293 (IBS2), which consists of two five-helix bundles connected by a long helix with a pronounced kink or flexible region at its center. The topology of the two bundles is identical and so far unique to talin. The modest inter-bundle interface (∼600 Å2) comprises mostly charged residues. Solution studies by SAXS and NMR are consistent with an extended conformation and with a relatively small interface with limited influence of one bundle on the other. Weak interactions between domains in the talin rod are not unexpected, because talin is thought to switch between a globular inactive and a more extended active conformation, which must involve changes in the interactions between at least some of the bundles. A relatively flexible interface and end-to-end packing between the helical bundles seems to be typical of the C-terminal region of the talin rod but contrasts sharply with the staggered arrangement of the two bundle module at the N terminus of the rod, which is stabilized by an extensive hydrophobic interface (28).
Integrin binding to the IBS2 region has been reported previously, and we have confirmed this by pulldown and ELISA assays. We also show for the first time that IBS2 binds to all β-integrin tails with comparable affinity, and moreover an affinity that is only ∼2-fold weaker than binding by the talin FERM domain. However, our studies contrast with earlier reports (24, 25, 44) by showing that tight binding to integrins requires both five-helix bundles. Moreover, only the two-domain IBS2 polypeptide localized strongly to FAs in vinculin-null cells, whereas the individual five-helix bundles showed a diffuse cytoplasmic distribution (vinculin-null cells were used because IBS2-A contains a vinculin-binding site). Because the structures of the individual bundles appear not to be significantly influenced by the presence of the other bundle (as judged by NMR), this suggests that both bundles directly contribute to integrin binding. We showed that this was indeed the case by using DSC, which clearly indicated binding of integrin tails to both bundles. We considered the possibility that integrin binds to residues at the interface between the two domains, but as these are not conserved this seems unlikely.
Using blot overlays and DSC experiments, we mapped the major determinants of IBS2 binding to the membrane proximal (∼23 residue) region of the β-integrin tails, although more membrane-distal regions also contribute to strong binding. We further found that alanine substitution of acidic residues in β-integrin tails consistently and uniquely increased IBS2 binding, while substitution of basic residues had the opposite effect, suggesting that the interaction has a strong electrostatic component. Rodius et al. (44) has also presented evidence that electrostatic interactions are important in IBS2 binding to β-integrin tails but concluded that two acidic residues in the β3-integrin tail, Glu-726 and Glu-733 were involved in binding to Lys-2085 and Lys-2089 on the surface of talin helix 50 in IBS2-A. It should be noted, however, that their experiments employed a fragment of talin (1843-2108) that includes part of IBS2-A but lacks the last α-helix.
Our studies also differ in significant respects from those of Tremuth et al. (24), who mapped the integrin-binding site to talin residues 1984-2113. Our crystal structure shows that this fragment comprises part of IBS2-A only and lacks part of the first helix and the whole of helix 5, raising significant concerns over its structural integrity and utility in binding studies. However, our DSC experiments show that integrin-binding destabilizes both IBS2-A and IBS2-B bundles, raising the possibility that the 1984-2113 construct favors integrin binding precisely, because the structure is destabilized. If so, this would be strongly reminiscent of our studies of vinculin binding, where the talin bundles must unfold to engage the vinculin head or F-actin, a process that is favored by truncated or mutant variants of the talin bundle with altered stability (16, 42). Indeed, Moes et al. (25) showed that GST-β3-integrin tails bound to an immobilized talin peptide corresponding to a single helix (helix 50, residues 2077-2099) in the talin IBS2-A bundle. The significance of these studies employing subdomain fragments remains to be determined.
In an attempt to characterize the interaction between talin IBS2 and β-integrin tails in more detail, we performed NMR experiments using unlabeled β3-integrin peptides and 15N-labeled talin 1974-2293 (in collaboration with S. Lorenz, N. Anthis, and I. D. Campbell). Unfortunately, the complex precipitated (both the integrin peptide and talin polypeptide could be detected in the precipitate by SDS-PAGE). Precipitation was seen with the high affinity fragments, full-length β3-integrin tail (residues 716-762) and the membrane proximal peptide (residues 716-740), but not with weakly binding membrane-distal peptides (residues 736-749 and 744-762). Precipitation at the high concentrations employed for NMR studies is consistent with the DSC experiments suggesting that IBS2 partially unfolds upon binding integrin. In vivo, it is conceivable that conformational changes induced by integrin binding to IBS2 are stabilized by association with the plasma membrane, which is in close proximity, and our demonstration that IBS2 binds acidic phospholipids in vitro may be relevant here. Although speculative at this point, destabilization of the IBS2 bundle might also lead to exposure of the vinculin-binding site present in helix 50 (or vice versa), resulting in vinculin binding and stabilization of the focal adhesion complex.
Talin is thought to exist in an inactive form in which the integrin-binding sites are masked, and the recent studies of Goksoy et al. (45) have provided the first insights into the structural basis for such autoinhibition. Thus, they have shown that the talin F3 FERM subdomain binds to residues 1654-2344 in the talin rod, and that this interaction masks the binding site for the membrane proximal helix of the β-integrin cytoplasmic domain in F3. The above region of the talin rod overlaps with IBS2, and it will be interesting to see whether binding of F3 to the rod also masks IBS2, i.e. the two integrin-binding sites may be co-regulated.
Supplementary Material
Acknowledgments
We thank Peter Moody (Leicester) and Bog Stec (Burnham Institute, La Jolla) for their help with the crystallography and Sonja Lorenz, Nick Anthis, and Iain Campbell for NMR titrations with the integrin peptides.
The atomic coordinates and structure factors (code 3dyj) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported, in whole or in part, by National Institutes of Health Cell Migration Consortium Grant U54 GM64346 from NIGMS. This work was also supported by the Wellcome Trust and Cancer Research UK. The work in Leipzig was supported by the Deutsche Forschungsgemeinschaft. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Figs. S1-S4, and additional references.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: IBS2, integrin-binding site 2; GST, glutathione S-transferase; SeMet, selenomethionine; SAXS, small angle x-ray scattering; ELISA, enzyme-linked immunosorbent assay; PBS, phosphate-buffered saline; GFP, green fluorescent protein; DSC, differential scanning calorimetry; r.m.s.d., root mean square deviation; Vd1, vinculin d1 domain; FA, focal adhesion; Fmoc, 9-fluorenylmethyloxycarbonyl.
References
- 1.Critchley, D. R., and Gingras, A. R. (2008) J. Cell Sci. 121 1345-1347 [DOI] [PubMed] [Google Scholar]
- 2.McLachlan, A. D., Stewart, M., Hynes, R. O., and Rees, D. J. (1994) J. Mol. Biol. 235 1278-1290 [DOI] [PubMed] [Google Scholar]
- 3.Gingras, A. R., Ziegler, W. H., Frank, R., Barsukov, I. L., Roberts, G. C., Critchley, D. R., and Emsley, J. (2005) J. Biol. Chem. 280 37217-37224 [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Alvarez, B., de Pereda, J. M., Calderwood, D. A., Ulmer, T. S., Critchley, D., Campbell, I. D., Ginsberg, M. H., and Liddington, R. C. (2003) Mol. Cell 11 49-58 [DOI] [PubMed] [Google Scholar]
- 5.Wegener, K. L., Partridge, A. W., Han, J., Pickford, A. R., Liddington, R. C., Ginsberg, M. H., and Campbell, I. D. (2007) Cell 128 171-182 [DOI] [PubMed] [Google Scholar]
- 6.Calderwood, D. A., Zent, R., Grant, R., Rees, D. J., Hynes, R. O., and Ginsberg, M. H. (1999) J. Biol. Chem. 274 28071-28074 [DOI] [PubMed] [Google Scholar]
- 7.Calderwood, D. A., Yan, B., de Pereda, J. M., Alvarez, B. G., Fujioka, Y., Liddington, R. C., and Ginsberg, M. H. (2002) J. Biol. Chem. 277 21749-21758 [DOI] [PubMed] [Google Scholar]
- 8.Barsukov, I. L., Prescot, A., Bate, N., Patel, B., Floyd, D. N., Bhanji, N., Bagshaw, C. R., Letinic, K., Di Paolo, G., De Camilli, P., Roberts, G. C., and Critchley, D. R. (2003) J. Biol. Chem. 278 31202-31209 [DOI] [PubMed] [Google Scholar]
- 9.de Pereda, J. M., Wegener, K. L., Santelli, E., Bate, N., Ginsberg, M. H., Critchley, D. R., Campbell, I. D., and Liddington, R. C. (2005) J. Biol. Chem. 280 8381-8386 [DOI] [PubMed] [Google Scholar]
- 10.Wegener, K. L., Basran, J., Bagshaw, C. R., Campbell, I. D., Roberts, G. C., Critchley, D. R., and Barsukov, I. L. (2008) J. Mol. Biol. 382 112-126 [DOI] [PubMed] [Google Scholar]
- 11.Liddington, R. C., Bankston, L. A., and de Pereda, J. M. (2003) Curr. Biol. 13 R94-R95 [DOI] [PubMed] [Google Scholar]
- 12.Loer, B., Bauer, R., Bornheim, R., Grell, J., Kremmer, E., Kolanus, W., and Hoch, M. (2008) Nat. Cell Biol. 10 422-428 [DOI] [PubMed] [Google Scholar]
- 13.Legate, K. R., Montanez, E., Kudlacek, O., and Fassler, R. (2006) Nat. Rev. Mol. Cell Biol. 7 20-31 [DOI] [PubMed] [Google Scholar]
- 14.Hemmings, L., Rees, D. J., Ohanian, V., Bolton, S. J., Gilmore, A. P., Patel, B., Priddle, H., Trevithick, J. E., Hynes, R. O., and Critchley, D. R. (1996) J. Cell Sci. 109 2715-2726 [DOI] [PubMed] [Google Scholar]
- 15.McCann, R. O., and Craig, S. W. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 5679-5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gingras, A. R., Bate, N., Goult, B. T., Hazelwood, L., Canestrelli, I., Grossmann, J. G., Liu, H., Putz, N. S., Roberts, G. C., Volkmann, N., Hanein, D., Barsukov, I. L., and Critchley, D. R. (2008) EMBO J. 27 458-469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bass, M. D., Smith, B. J., Prigent, S. A., and Critchley, D. R. (1999) Biochem. J. 341 257-263 [PMC free article] [PubMed] [Google Scholar]
- 18.Saunders, R. M., Holt, M. R., Jennings, L., Sutton, D. H., Barsukov, I. L., Bobkov, A., Liddington, R. C., Adamson, E. A., Dunn, G. A., and Critchley, D. R. (2006) Eur. J. Cell Biol. 85 487-500 [DOI] [PubMed] [Google Scholar]
- 19.Chandrasekar, I., Stradal, T. E., Holt, M. R., Entschladen, F., Jockusch, B. M., and Ziegler, W. H. (2005) J. Cell Sci. 118 1461-1472 [DOI] [PubMed] [Google Scholar]
- 20.Sun, N., Critchley, D. R., Paulin, D., Li, Z., and Robson, R. M. (2008) Exp. Cell Res. 314 1839-1849 [DOI] [PubMed] [Google Scholar]
- 21.Horwitz, A., Duggan, K., Buck, C., Beckerle, M. C., and Burridge, K. (1986) Nature 320 531-533 [DOI] [PubMed] [Google Scholar]
- 22.Xing, B., Jedsadayanmata, A., and Lam, S. C. (2001) J. Biol. Chem. 276 44373-44378 [DOI] [PubMed] [Google Scholar]
- 23.Yan, B., Calderwood, D. A., Yaspan, B., and Ginsberg, M. H. (2001) J. Biol. Chem. 276 28164-28170 [DOI] [PubMed] [Google Scholar]
- 24.Tremuth, L., Kreis, S., Melchior, C., Hoebeke, J., Ronde, P., Plancon, S., Takeda, K., and Kieffer, N. (2004) J. Biol. Chem. 279 22258-22266 [DOI] [PubMed] [Google Scholar]
- 25.Moes, M., Rodius, S., Coleman, S. J., Monkley, S. J., Goormaghtigh, E., Tremuth, L., Kox, C., van der Holst, P. P., Critchley, D. R., and Kieffer, N. (2007) J. Biol. Chem. 282 17280-17288 [DOI] [PubMed] [Google Scholar]
- 26.Tanentzapf, G., and Brown, N. H. (2006) Nat. Cell Biol. 8 601-606 [DOI] [PubMed] [Google Scholar]
- 27.Gingras, A. R., Vogel, K. P., Steinhoff, H. J., Ziegler, W. H., Patel, B., Emsley, J., Critchley, D. R., Roberts, G. C., and Barsukov, I. L. (2006) Biochemistry 45 1805-1817 [DOI] [PubMed] [Google Scholar]
- 28.Papagrigoriou, E., Gingras, A. R., Barsukov, I. L., Bate, N., Fillingham, I. J., Patel, B., Frank, R., Ziegler, W. H., Roberts, G. C., Critchley, D. R., and Emsley, J. (2004) EMBO J. 23 2942-2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otwinowski, Z., and Minor, W. (1997) in Methods in Enzymology (Carter, C. W. J., and Sweet, R. M., eds) pp. 849-861, Academic Press, New York
- 30.Terwilliger, T. C., and Berendzen, J. (1999) Acta Crystallogr. D. Biol. Crystallogr. 55 849-861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowtan, K. (1994) Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31 34-38 [Google Scholar]
- 32.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 33.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 34.Morris, A. L., MacArthur, M. W., Hutchinson, E. G., and Thornton, J. M. (1992) Proteins 12 345-364 [DOI] [PubMed] [Google Scholar]
- 35.Potterton, E., McNicholas, S., Krissinel, E., Cowtan, K., and Noble, M. (2002) Acta Crystallogr. D. Biol. Crystallogr. 58 1955-1957 [DOI] [PubMed] [Google Scholar]
- 36.Konarev, P. V., Petoukhov, M. V., Volkov, V. V., and Svergun, D. I. (2006) J. Appl. Crystallogr. 39 277-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svergun, D. I., Petoukhov, M. V., and Koch, M. H. (2001) Biophys. J. 80 2946-2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim, C. J., Han, J., Yousefi, N., Ma, Y., Amieux, P. S., McKnight, G. S., Taylor, S. S., and Ginsberg, M. H. (2007) Nat. Cell Biol. 9 415-421 [DOI] [PubMed] [Google Scholar]
- 39.Xu, W., Baribault, H., and Adamson, E. D. (1998) Development 125 327-337 [DOI] [PubMed] [Google Scholar]
- 40.van der Ven, P. F., Wiesner, S., Salmikangas, P., Auerbach, D., Himmel, M., Kempa, S., Hayess, K., Pacholsky, D., Taivainen, A., Schroder, R., Carpen, O., and Furst, D. O. (2000) J. Cell Biol. 151 235-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm, L., and Sander, C. (1995) Trends Biochem. Sci. 20 478-480 [DOI] [PubMed] [Google Scholar]
- 42.Patel, B., Gingras, A. R., Bobkov, A. A., Fujimoto, L. M., Zhang, M., Liddington, R. C., Mazzeo, D., Emsley, J., Roberts, G. C., Barsukov, I. L., and Critchley, D. R. (2006) J. Biol. Chem. 281 7458-7467 [DOI] [PubMed] [Google Scholar]
- 43.Gilmore, A. P., Wood, C., Ohanian, V., Jackson, P., Patel, B., Rees, D. J., Hynes, R. O., and Critchley, D. R. (1993) J. Cell Biol. 122 337-347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodius, S., Chaloin, O., Moes, M., Schaffner-Reckinger, E., Landrieu, I., Lippens, G., Lin, M., Zhang, J., and Kieffer, N. (2008) J. Biol. Chem. 283 24212-24223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goksoy, E., Ma, Y. Q., Wang, X., Kong, X., Perera, D., Plow, E. F., and Qin, J. (2008) Mol. Cell 31 124-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.