Abstract
We have characterized the kinetic response of gene targets throughout the murine genome to transcriptional modulation by the glucocorticoid receptor (GR). In contrast to a model in which multiple genes are either repressed or activated during the GR response, the vast majority of responsive genes are subject to complex regulation profiles, frequently with alternate activation and repression phases. We also observe that GR binding at response elements does not always correlate with the target gene response profile. Thus, the cellular response to GR stimulation involves a highly orchestrated series of regulatory actions and not simply a binary response to hormone.
Glucocorticoid receptor target genes display a complex kinetic regulation profile wherein many genes are subject to alternate activation and repression phases.
The glucocorticoid receptor (GR) influences a large variety of important physiological processes, including rapid stress responses, tissue differentiation during organism development, programming of neuroendocrine function in adult animals, diurnal metabolic actions, and a number of other endocrine functions. Understanding the molecular basis of GR action on gene expression is derived almost exclusively from a limited set of model systems. Current paradigms developed from these models suggest that GR action is mediated by the selective activation or repression of key target genes. For example, it is widely reported that pro-inflammatory genes are subject to repression by glucocorticoids, either by direct binding of the receptor to negative response elements (1), or by indirect binding to other transcription factors (trans-repression) (2) (2,3,4,5).
Central to this thesis is the concept that the primary action for GR at a particular locus is either to repress or activate that gene. In contrast to this simple view, the response of some model systems to GR stimulation is known to be more complex. Rates of transcription for the mouse mammary tumor virus (MMTV) promoter are quickly elevated by GR, reaching a maximum at 20 min after hormone treatment, then decreasing with equal rapidity to much lower levels (6,7). Alternating levels of mature transcripts have also been reported for a number of GR-regulated genes in hippocampal cells (8). Furthermore, a cyclical transcriptional response to other steroid receptors has also been recently demonstrated in several model systems (9,10,11). To develop a more complete understanding of GR action, we have examined the genome-wide, time-dependent response of GR-regulated genes in two murine cell lines, a mammary cell line (3134) and a pituitary corticotroph (AtT-20). Microarray expression profiling of GR responsive genes was performed at several time points after hormone treatment, followed by quantitative-PCR (q-PCR) analysis of nascent transcripts for representative genes. We find that GR activation leads to a highly complex global response profile. The kinetic behavior of these genes can be classified in several patterns. Transcription rates for some genes are rapidly elevated and relatively quickly reach an induced level that remains stable over 24 h. A second class is rapidly induced, but rates of accumulation quickly return to low expression levels, in some cases not significantly higher than the uninduced rate of expression. For a third class, rates of induction develop more slowly but continue to increase throughout the induction period. Expression profiles for repressed genes are equally complex. One group of responsive genes is rapidly repressed relative to constitutive expression rates and remains suppressed for at least 24 h. In contrast, a second set of genes that is initially repressed returns to expression rates characteristic of untreated cells. Finally, a third group exhibits relatively slow but continuously decreasing levels of RNA over the 24-h period studied.
We conclude that the cellular response to GR activation cannot be modeled in simplistic terms of gene repression or activation but must be understood in terms of a complex kinetic pattern of gene activity. The majority of GR-regulated genes respond to hormone treatment with a complicated expression profile, with activity levels varying in multiple ways at various times after the initiation of hormone action. Thus, it is inappropriate to consider genes as susceptible to “repression” or “activation.” Rather, the GR response is characterized by a multifarious response of hundreds of genes throughout the induction cycle. We propose that the net effect of hormone on cellular activity is orchestrated through a complex web of induction and repression events.
Materials and Methods
Cell lines
The 3134 cell line was derived by transformation of C127, originally isolated from a mammary adenocarcinoma tumor of the RIII mouse (12). This line is a subclone of 904.13 (13). The AtT-20 cell line is an anterior pituitary corticotroph of murine origin (ATCC). Both cell lines were maintained in DMEM (Invitrogen Corp., Carlsbad, CA) supplemented with 10% fetal bovine serum (Gemini, Woodland, CA), 2 mm l-glutamine, 1 mm sodium pyruvate, 0.1 mm nonessential amino acids, 5 mg/ml penicillin-streptomycin (Invitrogen), and kept at 37 C incubator with 5% CO2. Cells were transferred to 10% charcoal-dextran-treated, heat-inactivated fetal bovine serum for 48 h before hormone treatment.
Microarray analysis of RNA expression
To analyze GR-responsive genes, RNA was extracted from cells either vehicle treated or treated with 100 nm dexamethasone (dex) for varying times. 3134 RNA for microarray analysis was prepared via standard manufacturer protocols (QIAGEN, Inc., Valencia, CA) using cells resuspended in TRIzol reagent (Invitrogen). Samples from dex-treated cells (1, 4, and 24 h) were labeled with Cy3, and samples from untreated cells labeled with Cy5. Labeled cDNA samples were hybridized onto spotted oligonucleotide microarrays as described in Ref. 14. Microarrays with 70-mer oligonucleotide probe sets directed to 31,769 murine loci have been described previously (http://www.ncbi.nlm.nih.gov/geo/). Cy3 and Cy5 fluorescence was scanned using a laser confocal scanner (Agilent Technologies, Inc., Palo Alto, CA). Images were analyzed using the ArraySuite 2.1 Extensions (Y. Chen, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD) on the IPLab program (Scanalytics Inc., Fairfax, VA) (15). Replicate microarrays were used for each time point: three, four, and three arrays for 1, 4, and 24 h, respectively. After preprocessing and normalization of log-transformed data, the zero-centered F test was applied to identify 965 differentially expressed genes with a P value less than 0.001. Similarly, RNA was harvested from the AtT-20 cell line after 0, 2, 4, or 24 h dex treatment. This RNA was subject to microarray analysis using Illumina bead microarrays (Illumina Inc., San Diego, CA). Replicate microarrays (two) were used for each time point. Raw bead array data were processed and normalized by the R packages “lumi” and “illuminaMousev1.”
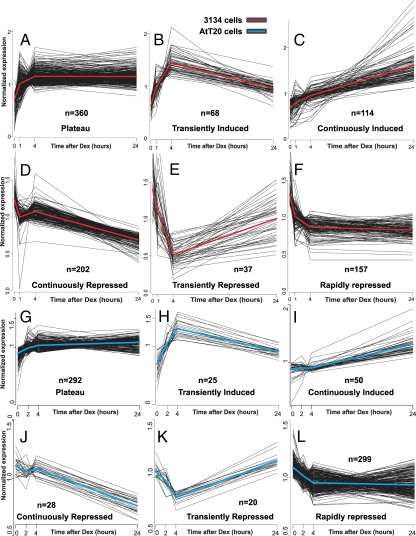
The class assignment of these responsive genes into the six kinetic categories was based on the following. Among the induced genes (mean log2 ratio of 3 time points >0), if the 24-h mean log2 ratio was within ±log2 (1.3) of the 4-h mean log2 ratio, then the gene was assigned to the plateau category. If the 24-h log2 ratio was more than or equal to (4 h log2 ratio) plus log2 (1.3), then the gene was considered continuously induced. If the 24-h log2 ratio was less than (4 h log2 ratio) minus log2 (1.3), then the gene was considered transiently induced. Classification was performed similarly for repressed genes (mean log2 ratio of 3 time points <0) with the signs reversed. All analyses were performed with the R-statistical computing environment. The plots in Fig. 1 show normalized expression levels obtained as follows. The log ratios were converted back to fold scale, where one was used for 3134 0-h values. The fold-change time series data were divided by the gene-specific time average value for each gene. The red and blue curves indicate the average expression within each kinetic profile for the 3134 and AtT-20 cells lines, respectively.
Figure 1.
Microarray profiling reveals a complex GR induction and repression profile in 3134 and AtT-20 cell lines. A–F, Mammary cells (3134) were treated with 100 nm dex for 1, 4, and 24 h. G–L, Pituitary corticotrophs (AtT-20) were treated with 100 nm dex for 0, 2, 4, and 24 h. Genes whose RNA levels are induced by dex treatment display complex induction profiles, either rapid initial induction followed by constant plateau levels (A and G) transient induction (B and H), or continuous induction (C and I). Genes whose RNA levels are repressed by dex treatment also show a time-dependent variation in the response pattern. One class of genes (D and J) is slowly and continuously repressed over the time period studied, a second class (E and K) is transiently repressed, and a third class (F and L) shows rapid and sustained repression. The plots show normalized expression levels (see Materials and Methods for a detailed description). The red or blue curves indicate the average expression within each kinetic profile for the 3134 and AtT-20 cell lines, respectively. Replicate arrays were used at each time point. Three, four, and three arrays were used for the 1, 4, and 24-h time points, respectively, for 3134. Two arrays were used for the 2-, 4-, and 24-h time points for AtT-20.
Chromatin immunoprecipitation (ChIP)
ChIPs were performed as per standard protocols (Upstate Biotechnology Inc., Lake Placid, NY). Briefly, cells were treated with either vehicle or 100 nm dex for the indicated times. Cells were cross-linked for 10 min at 37 C in 1% formaldehyde, followed by a quenching step for 10 min with 150 mm glycine. Sonicated, soluble chromatin was immunoprecipitated with 5 μg of an antibody to the GR (BuGR2, ABR) or pol II (Kevin Gardner, National Cancer Institute, National Institutes of Health, Bethesda, MD). DNA isolates from immunoprecipitates were used as templates for real-time q-PCR amplification. All ChIP experiments were performed at least two times.
Primer sequences for GR ChIP in 3134 are as follows: MMTV, forward TTTCCATACCAAGGAGGGGACAGTG and reverse CTTACTTAAGCCTTGGGAACCGCAA; Ccl2, forward TGGTAACCACCAAGTGGAGAGAATGand reverse TGGAAACACAGCCTAGCTTGCC; Mt2, forward CATAGCCAGGGCAGCCACAGAA and reverse GGCAATGCCTTCTTGACTCATTCC; Sgk Prox Prom, forward TCTAACTCGCCACCTCCTCA and reverse CCAACTAATCTCCGAGAACA; Sgk Distal, forward CTTCCCTTATCCAGCATGTCTTGTG and reverse TGCATCGTGCAATCTGTGGC; and Lcn2, forward TCACCCTGTGCCAGGACCAA and reverse TGGGGAAGGGTGAGCAAGCT. Primer sequences for GR ChIP in AtT-20 are as follows: Rgs4, forward TGTGAGCCTTAGACTCTATT and reverse TAACAATCGTGAACTCTCAC; Chst1, forward GTGGTTTTCTTTGTTTGCTT and reverse TACTGCTCTACTCCACAGAA; Ttr, forward GGGCAGAAACTGTTCTTGCT and reverse CATGCTGATCCCATTATGAT; and Lhfpl2, forward TATTTAACCAGACACAGTAGATCC and reverse TTGAGACAGGCTCTTCAGTA. Primer sequences used to detect pol II loading by ChIP are the same primers used to detect newly synthesized transcripts (see section below). All primer sequences are listed as 5′ to 3′.
q-PCR analysis of RNA
RNA was extracted from cell lines via standard methods using TRIzol reagent, and purified using RNeasy mini kits (QIAGEN). All RNA samples were treated with ribonuclease-free deoxyribonuclease (QIAGEN), and quality checked on 1% agarose gel. Reverse transcriptase of total RNA was performed with the Bio-Rad cDNA Synthesis Kit per the manufacturer's instructions and analyzed by q-PCR using SyBr green (Bio-Rad Laboratories, Inc., Hercules, CA). Primer sequences were designed to amplify only nascent RNA, using PCR amplicons that cross an exon/intron or untranslated region (UTR)/intron boundary. Primer sequences for q-PCR validation of expression microarray data are as follows.
Primer sequences for q-PCR characterization of nascent RNA in 3134 are as follows: Tgm2, forward TGTCACCAGGGATGAGAGACGG and reverse TCCAAATCACACCTCTCCAGGAG; Ampd3, forward AAGGAGCTTGCAGAGCAGAAGTC and reverse CAGCTCCCTCAGGTCTCACAACTAT; Mt2, forward GAACTCTTCAAACCGATCTCTCGTC and reverse TCCCAGAAATCCCGTCAGCA; SuOx, forward CTAATGAGGGAGAGGTGACTGACCA and reverse TGCAGAGCCTCAAGGGGGTT; Sgk, forward GGGAATGGTAGCGATTCTCATCG and reverse CGACGCCACACGCTAATCTG; Lcn2, forward ACCTCTCATTTCTTGCAGTTCCG and reverse CAGGATGGAGGTGACATTGTAGCT; Glul, forward GAGCAGAGTGTCTGAACAGCACG and reverse ACCCTCCGTGCGCTTACCAG; Edn1, forward CTTGCTCTGGACGCCTGAAGA and reverse GGGGCTCTGCACTCCATTCT; Ccl7, forward CCGCTGCTTTCAGCATCCAA and reverse GCAAAGCCAGCAAATGTGAGACTA; Dusp6, forward GTGTGTGTTCCCGCAGATGAAG and reverse TGCATGAGGTACGCCACTGT; and Ccl2, forward CTTTGAGTCCCCTTTTTCTACCTGC and reverse AGCACAGACCTCTCTCTTGAGCTTG. Primer sequences for q-PCR characterization of nascent RNA in AtT-20 are as follows: Chst1, forward ATGCAATGTTCTTGGAAGGCT and reverse CTCCTCACACAACCGCTCT; Tt2, forward CAGGATGGCTTCCCTTCGACTCTT and reverse GCCATGTCTGGATCGCTCACAG; and Rgs4, forward GAGTGCAAAGGACATGAAACATC and reverse TTTTCCAACGATTCAGCCCAT. All primer sequences are listed as 5′ to 3′.
Results
Genes sensitive to GR regulation
To identify genes that respond to GR stimulation, we performed microarray expression analysis with RNA isolated from the murine mammary adenocarcinoma cell line, 3134 (16). The 3134 cell line harbors an amplified array of MMTV reporter elements, allowing the direct visualization of GR binding to response elements in living cells (6,17). Thus, it is possible in this cell line to compare the action of receptor at genome-wide regulatory sites, monitored by static methodologies such as ChIP, with dynamic receptor interactions at the amplified MMTV loci measured in real time by photobleaching techniques, fluorescence recovery after photobleaching, or fluorescence loss in photobleaching (6). AtT-20 is a pituitary corticotroph-derived cell line that has been used extensively as a model system to study glucocorticoid-signaling mechanisms along the hypothalamus-pituitary-adrenal axis (18). RNA was prepared from 3134 or AtT-20 cells either vehicle treated, or treated for various times with 100 nm dex. RNA from treated and untreated cells was differentially labeled and subjected to hybridization on murine microarrays (see Materials and Methods) to determine relative induction levels at various time points after hormone stimulation. As seen in Fig. 1, approximately 950 genes in 3134 (Fig. 1, A–F) and 714 genes in AtT-20 (Fig. 1, G–L) were found to be modulated by treatment with dex (the complete data set is presented in supplemental Tables 1 and 2, which are published as supplemental data on The Endocrine Society's Journals Online web site at http://endo.endojournals.org, for 3134 and AtT-20, respectively). Surprisingly, the kinetic profiles for the responsive genes were quite complicated. A large set (n = 360 in 3134 and n = 292 in AtT-20) of loci is rapidly induced, and then remains at relatively constant levels of message accumulation over a 24-h period (Fig. 1, A and G). Although this was the expected pattern for GR induction, not all induced genes actually respond in this mode. A significant group of induced loci has rapidly elevated expression levels but then returns to relatively low levels of accumulated RNA (Fig. 1, B and H). A third group of genes is inefficiently induced, with RNA levels accumulating slowly but continuously over the 24-h induction period (Fig. 1, C and I).
The kinetic pattern for repressed genes is equally complex. One class of down-regulated genes is characterized by continuously decreasing levels of gene activity (Fig. 1, D and J). Two other types of repression patterns are evident in this data set. One group of loci includes genes that are rapidly repressed, then recover to levels equivalent to, or even greater than, expression levels in untreated cells (Fig. 1, E and K). A third group of genes is rapidly repressed, then remains at these repressed levels throughout the induction period (Fig. 1, F and L). Thus, the overall expression pattern for genes regulated by the GR is markedly complex, both for induced and repressed loci. A comparison of the GR-regulated genes between 3134 and AtT-20 underscores the cell type-specific nature of GR action (19). Despite the distinct sets of genes regulated in each cell line, the kinetic profiles of GR-regulated genes, as a whole, appear to be a conserved regulatory feature.
Kinetics of nascent RNA accumulation
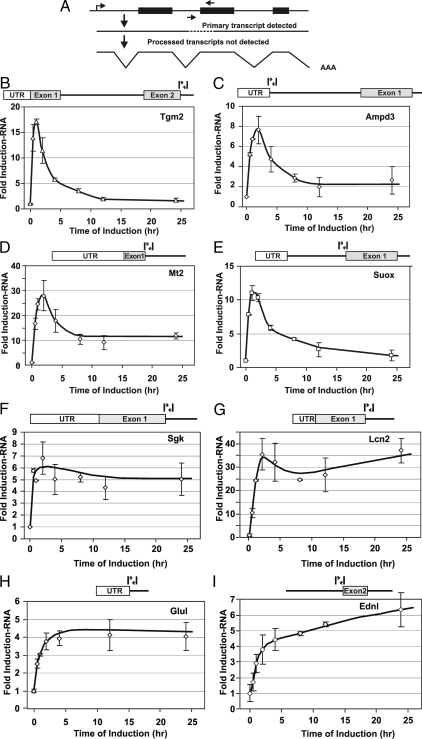
The expression levels monitored in the microarray experiments represent concentrations of accumulated RNA that would result from regulation at multiple steps, including posttranscriptional mechanisms. To evaluate more clearly the contribution of transcriptional regulation to the observed expression profiles, we characterized the concentration of nascent RNA transcripts, in the 3134 cell line, for a series of genes representative of each expression class. These studies were also performed at a greater time resolution, monitoring RNA levels for each locus at 0, 0.5, 1, 2, 4, 8, 12, and 24 h. In each case, nascent heterogeneous nuclear RNA (hnRNA) levels were measured by q-PCR, using primer pairs with one member internal to an exon and the second crossing the exon/intron boundary (20), thereby permitting the detection only of unprocessed, newly expressed transcripts (Fig. 2A).
Figure 2.
Nascent RNA analysis of GR-induced genes in 3134. To evaluate more closely the transcriptional component of the complex regulatory profile, nascent transcripts were characterized for representative genes from each induction class. Nascent RNA levels were measured at multiple time points after induction by q-PCR analysis, using primer pairs spanning intron/exon boundaries for each gene. A, Strategy for nascent transcript mapping. Nascent transcript levels were determined after hormone addition for members of the transiently induced class [Tgm2 (B), Ampd3 (C), Mt2 (D), and Suox (E)], the rapid induction class [Sgk (F), Lcn2 (G), and Glul (H)], and the continuously induced class [Ednl (I)]. Error bars show the sd from the mean. q-PCR experiments represent three independent experiments (biological replicates) performed in duplicate (technical replicates).
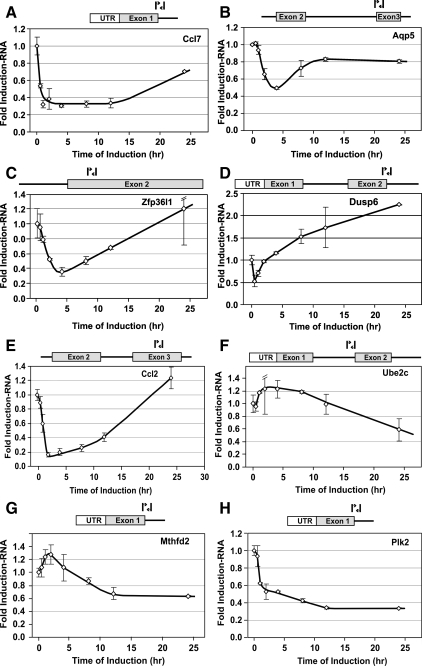
Results from these experiments confirm that transcription rates represent a primary mechanism in generating the diversity of responses described in Fig. 1. Tgm2, Ampd3, Mt2, and Suox are examples of GR-induced genes whose transcripts transiently accumulate, followed by a marked loss of RNA levels (transient induction class). For these genes the nascent RNA transcripts follow a similar pattern (Fig. 2, B–E). We note that the pattern of the appearance of nascent RNA for these genes is remarkably similar to that described for the MMTV promoter (6,7). The Sgk, Lcn2, and Glul genes are representative of the first induction class (rapid induction followed by plateau). For each of these loci, nascent RNA accumulates very rapidly, then remains at constant levels throughout the induction period (Fig. 2, F–H). In contrast, nascent transcripts for the Ednl locus are rapidly induced and continue to increase over the induction period tested. Thus, this locus is representative of the third induction class (slow and continuous accumulation). Again, the rates of nascent RNA production parallel the pattern seen for stable transcripts, implicating transcription as a major mechanism governing the behavior of this class. Similarly, for the repressed gene classes, transcription is a major contributor to the variation in repression behavior. Transcripts from the Ccl7, Aqp5, Zfp36L1, Ccl2, and Dusp6 genes are rapidly repressed but then recover to levels found in untreated cells (Fig. 3, A–D, transient repression class). In contrast, the Plk2 locus shows a rapid and sustained shutdown in levels of nascent RNA (Fig. 3H, rapid repression class), consistent with the findings observed for mature RNA levels (Fig. 1F). Finally, the Ube2c and Mthfd2 gene transcripts are slowly but continuously repressed (Fig. 3, F and G, gradual repression class), in agreement with the microarray expression data (Fig. 1D). We have also monitored nascent hnRNA levels for the Sgk, Glul, Tgm2, Sbp1, Ccl2, and Plk2 genes in the hepatocyte cell line, Hepa1c1c7 (supplemental Fig. S1). Consistent with the data obtained from the 3134 cell line, the rates of nascent RNA production in this cell line mirrors the expression patterns observed using microarrays.
Figure 3.
Analysis of RNA from representative genes repressed by GR in 3134. Transcript levels were determined after hormone addition for members of the transiently repressed class [Ccl7 (A), Aqp5 (B), Zfp36l1 (C), Dusp6 (D), and Ccl2 (E)], the continuously repressed class [Ube2c (F) and Mthfd2 (G)], and the rapidly repressed class [Plk2 (H)]. Error bars show the sd from the mean. q-PCR experiments represent three independent experiments (biological replicates) performed in duplicate (technical replicates).
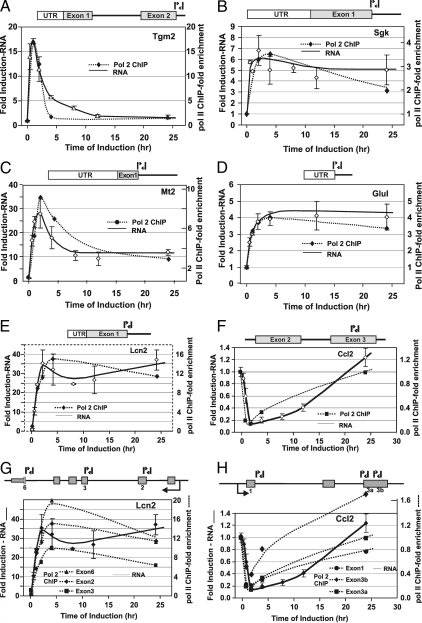
To further associate transcription as the main contributor to the various expression profiles, we have monitored RNA pol II loading (using ChIP) at several 3134 GR-regulated genes (Fig. 4). RNA and chromatin were harvested at several time points after hormone treatment (0, 0.5, 1, 2, 4, 8, 12, and 24 h). Expression and pol II ChIP profiling of six hormone induced or repressed genes [Tgm2 (A), Sgk (B), Mt2 (C), Glul (D), Lcn2 (E), and Ccl2 (F)], belonging to different kinetic categories, demonstrated that nascent RNA levels were coincident with levels of elongating polymerase. The primers used to detect RNA pol II amplify regions that span intron-exon junctions, thereby allowing for the capture of elongating RNA polymerase as opposed to promoter bound polymerase. Pol II occupancy at the Lcn2 and Ccl2 loci was additionally monitored using three different primer sets that spanned various intron-exon junctions across each transcribed locus (Fig. 4, G and H). Each primer set provided a pol II loading profile that tracked with nascent RNA levels. Therefore, the equilibrium association of RNA pol II appears to mirror largely the transcriptional state of the genes tested. We, therefore, conclude that transcriptional regulation is a primary mechanism in generating the wide variation in expression patterns observed for both glucocorticoid-induced and repressed genes. The observation that GR-regulated genes, in multiple cell lines, are subject to complex regulation profiles not only generalizes the biological relevance of this phenomenon but also implicates transcription as the key mechanism governing this behavior. However, it does remain a formal possibility that alternate mechanisms such as the differential processing of unspliced message (hnRNA) might be a contributor to the complex kinetic profiles observed.
Figure 4.
Time-dependent occupancy of RNA pol II at GR-regulated genes in 3134. Relative pol II occupancy at selected genes was determined by ChIP analysis and compared with levels of transcription. Values are presented as a loading ratio relative to the amount determined before hormone treatment. A, Tgm2: pol II occupancy (-♦-) over a 24-h period of hormone treatment is compared with nascent RNA (—) levels. B, Sgk: pol II binding (-♦-) over 24 h hormone treatment is compared with nascent RNA (—) levels. C, Mt2: pol II binding (-♦-) over 24 h hormone treatment is compared with nascent RNA (—) levels. D, Glul: pol II binding (-♦-) over 24 h hormone treatment is compared with nascent RNA (—) levels. E, Lcn2: pol II binding (-♦-) over 24 h hormone treatment is compared with nascent RNA (—) levels. F, Ccl2: pol II binding (-♦-) over 24 h hormone treatment is compared with nascent RNA (—) levels. G, Lcn2: pol II binding (-♦-, -▴-, -▪-) over 24 h hormone treatment is monitored using multiple intron-exon primers across the transcribed locus and is compared with nascent RNA (—) levels. H, Ccl2: pol II binding (-♦-, -•-, -▪-) over 24 h hormone treatment is monitored using multiple intron-exon primers across the transcribed locus and is compared with nascent RNA (—-) levels. Error bars show the sd from the mean. ChIP q-PCR experiments represent at least two independent experiments (biological replicates) performed in duplicate (technical replicates). Primer locations are highlighted in the genomic structure diagrams for each locus.
Glucocorticoid response element (GRE) site occupancy during hormone treatment
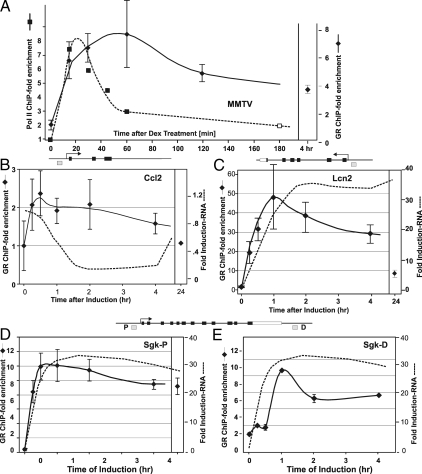
ChIP, coupled with high-density tiling of promoter regions, or with massive parallel sequencing (21,22), permits the localization of DNA-binding proteins at specific sites throughout large regions of a genome. Using these approaches we have identified GR binding sites associated with many of the GR-responsive genes described in Figs. 1–4 (19). We sought to characterize GR interaction with these sites during the time line under study, by performing a quantitative ChIP analysis of a representative set of binding elements in both the 3134 (Fig. 5) and AtT-20 (Fig. 6) cell lines. As seen in Figs. 5 and 6, GR occupancy of the sites examined follows a complex and variable pattern (see Ref. 19 for location of binding elements). For the MMTV locus (Fig. 5A), GR binding increases for the first hour, then decreases gradually over the following induction period. Because the MMTV locus is very well characterized, we know rigorously that the GRE at the MMTV locus represents the functional GRE. Importantly, the rate of transcription clearly does not rigorously correlate with the level of GR bound to the response element. Transcription rates have been measured for this locus by three independent methods, transcription run-on (23), level of GFP-Pol II on the promoter in living cells (6), and pol II ChIP analysis (7). These methods all show that transcription reaches a maximum at 20 min after induction, followed by a rapid down-regulation in transcription rate. Interestingly, this repression occurs despite high levels (see Discussion) of GR associated with the promoter, thereby suggesting that there is a disconnect between levels of GR occupancy at the MMTV promoter and rates of transcription, as measured by pol II ChIP or q-PCR assays measuring nascent transcripts.
Figure 5.
Nascent RNA analysis of GR-regulated genes and time-dependent occupancy of GR binding sites in 3134. Relative GRE occupancy at selected genes was determined by ChIP analysis and compared with levels of transcription. Values are presented as a loading ratio relative to the amount determined before hormone treatment. A, MMTV promoter: GR occupancy (-♦-) over a 4-h period of dex treatment at the nucleus. B/C GRE (31): the extent of Pol2 (-▪-) associated with MMTV reporter gene was assayed by ChIP analysis. B, Ccl2: GR binding (-♦-) at the GRE (GRE position indicated by the gray box; see diagram of the genomic structure) is shown relative to nascent RNA (—-) levels over 24 h hormone treatment. C, Lcn2: GR binding (-♦-) at the promoter proximal GRE is compared with nascent RNA (—-) levels over 24 h hormone treatment. D, Sgk-P: GR binding (-♦-) at the proximal GRE is shown relative to nascent RNA (—-) levels over 24 h hormone treatment. E, Sgk-D: GR binding (-♦-) at the distal GRE is compared with nascent RNA (—-) levels over 24 h hormone treatment. Error bars show the sd from the mean. ChIP q-PCR experiments represent at least two independent experiments (biological replicates) performed in duplicate (technical replicates).
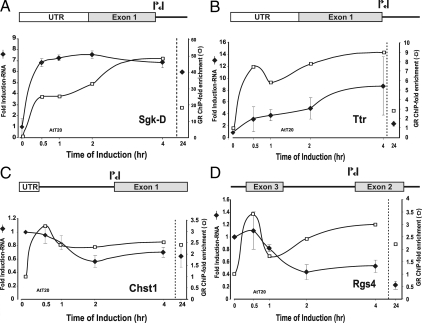
Figure 6.
Nascent RNA analysis of GR-regulated genes and time-dependent occupancy of GR binding sites in AtT-20. To evaluate more closely the transcriptional component of the complex regulatory profile, nascent transcripts were characterized for representative genes from different induction and repression classes. Nascent RNA levels were measured at multiple time points after induction by q-PCR analysis using primer pairs spanning intron/exon boundaries for each gene. Nascent transcript levels (-♦-) were determined after hormone addition for members of the rapid induction class [Sgk-D (A)], continuously induced class [Ttr (B)], rapidly repressed class [Chst1 (C), and continuously repressed class [Rgs4 (D)]. Error bars show the sd from the mean. q-PCR experiments represent three independent experiments (biological replicates) performed in duplicate (technical replicates). Relative GRE occupancy at selected genes was determined by ChIP analysis and compared with levels of transcription. Values are presented as a loading ratio relative to the amount determined before hormone treatment. A, Sgk-D: GR binding (□) at the distal GRE is shown relative to nascent RNA (-♦-) levels over 24 h hormone treatment. B, Ttr: GR binding (□) at the distal GRE is shown relative to nascent RNA (-♦-) levels over 24 h hormone treatment. C, Chst1: GR binding (□) at the distal GRE is shown relative to nascent RNA (-♦-) levels over 24 h hormone treatment. D, Rgs4: GR binding (□) at the distal GRE is shown relative to nascent RNA (-♦-) levels over 24 h hormone treatment. Error bars show the sd from the mean. ChIP q-PCR experiments represent at least two independent experiments (biological replicates) performed in duplicate (technical replicates).
For the other genes examined (Figs. 5, B–E, and 6, A–D), GR site occupancy is mostly, but not always, correlated with nascent transcript levels. In 3134, for the proximal Sgk site (Fig. 5D) or the GR binding site at the Mt2 locus (data not shown), GR binding tracks quite closely with nascent transcript levels. However, for the distal Sgk site (Fig. 5E), GR occupancy shows a pronounced decrease early after hormone addition (2 h in 3134). Finally, for one of the repressed genes, Ccl2, the level of bound GR at the promoter proximal site (Fig. 5B) is completely uncoupled from the apparent rate of transcription, as estimated by nascent RNA levels. Similarly in AtT-20, the disconnect between GR occupancy and transcription is most evident at the distal Sgk site (at the 24 h time point, Fig. 6A), whereas other genes show GR occupancy paralleling nascent RNA levels (Fig. 6, B and C). For some loci studied, there is a small decrease in bound GR levels after 2 h (similar to that shown for MMTV and Lcn2; Fig. 5, A and C) that corresponds to the well-characterized down-regulation of GR protein levels during hormone stimulation (24,25). However, this phenomenon cannot serve as the primary mechanism for GRE occupancy because the levels of GR associated with disparate sites vary in a pattern completely unlinked to overall intracellular levels of GR protein. Thus, the dramatic transitions that occur in primary transcription levels cannot be simply explained by variation in GR binding to promoter regulatory sites.
Discussion
Regulation of gene expression by the GR is a critical mechanism in mammalian homeostasis. This important signaling system serves to mediate a large variety of physiological changes, from rapid events such as the stress response, to longer-term processes such as circadian rhythm and organ development. In particular, glucocorticoids serve as major modulators of pro-inflammatory responses, and steroids designed to suppress these responses are among the most frequently prescribed medications available to the modern physician.
A large literature deals with the activation and suppression of responsive genes by GR, and detailed molecular models have evolved to account for selective action of the receptor. In general, these models describe mechanisms by which the receptor can either activate or repress a given gene locus. This unimodal view of GR regulation reflects the widely held concept that in a given tissue, and under a specific set of physiological circumstances, a set of genes can either be induced or repressed. In contrast to this view, a complex GR activation and repression profile was first reported for MMTV in 1994 (23). More recently, waves of GR-regulated gene expression have been described in hippocampal tissue slices (8).
Time-dependent responses for the genes described in this report can be classified in six categories (supplemental Fig. S2). The most abundant class for induced genes includes genes whose RNA levels are rapidly activated, and then remain at relatively unchanged levels for extended periods during the continuous presence of hormone (Fig. 1, A and G). The second most frequent class consists of genes that manifest continuously increasing levels of transcript (Fig. 1, C and I). Although each of these classes represents variations on the theme of transcriptional induction, the third class (transient induction; Fig. 1, B and H) is much more complex. The presence of a significant number of genes showing this type of regulation is quite unexpected. Importantly, variation in primary transcription rates must play a major role in the mechanisms governing these accumulation rates because the quantitative analysis of nascent transcripts associated with representative members of each class (Figs. 2, 3, 5, and supplemental Fig. S1) shows dramatic, time-dependent changes in the quantity of primary transcripts observed for a number of the genes.
The time-dependent behavior of repressed genes is also complex. This gene set can be arbitrarily organized in three groups (supplemental Fig. S2), rapid repression and plateau, slow continuous repression, and transient repression. These groups represent mirror images for the induced classes and further illustrate the complexity of the GR response.
A relatively straightforward interpretation of these response classes would invoke a time-dependent variation in receptor associated with critical response elements for a given gene. Under this model, GR levels found on the relevant GREs for a given gene should track roughly with the RNA output level for that locus. This model is apparently excluded for some of the gene responses described here. For the MMTV example (Fig. 5A), the amount of GR associated with the well-characterized GRE does not correlate well with the observed transcription rate. For this locus, actual rates of transcription have been measured both by run-on analysis (23) and by pol II levels found on the promoter (6), and these rates reach a maximum much earlier than promoter-associated GR levels. The behavior of this response class (represented by Tgm2, Ampd3, Mt2, Suox, and MMTV) is characterized by a rapid burst of transcription, followed by a “refractory” period of low transcription rates that are nonresponsive to further stimulation. Indeed, addition of fresh hormone (23,26), or even transfection into the cell of new receptor (27), cannot overcome this refractory period. The level of receptor detected by ChIP analysis on a given site does not represent a “static” association of the factor with the template but rather reflects the equilibrium distribution of rapidly exchanging receptor at that site (see Refs. 28 and 29 for a complete discussion). Thus, the promoter moves through a series of programmed events, which in turn leads to constant modulation of promoter activity. A given gene can be alternately activated, or repressed, depending on the program encoded in the regulatory elements associated with that gene. Under this model for GR action, hormone modulation of gene activity is viewed as an ongoing, time-dependent process, rather than simple activation or repression events. In the constant presence of activating ligand, promoters are subject to a wide variety of specific regulatory responses. Current models for GR function focus heavily on the concepts of “trans” repression (30), with receptor acting in a non-DNA binding mode, whereas promoter activation is driven primarily through “cis-acting,” direct DNA binding events. The results presented here show that receptor action on the global scale is much more complex. For the GR, we conclude that hormone action must be understood as the induction of a series of promoter-specific programs, which in turn produce the time-dependent accumulation of gene products that are appropriate for a given biological response.
Supplementary Material
Footnotes
Disclosure Summary: The authors have nothing to declare.
First Published Online January 8, 2009
Abbreviations: ChIP, Chromatin immunoprecipitation; dex, dexamethasone; GR, glucocorticoid receptor; GRE, glucocorticoid response element; hnRNA, heterogeneous nuclear RNA; MMTV, mouse mammary tumor virus; q-PCR, quantitative PCR; UTR, untranslated region.
References
- Dostert A, Heinzel T 2004 Negative glucocorticoid receptor response elements and their role in glucocorticoid action. Curr Pharm Des 10:2807–2816 [DOI] [PubMed] [Google Scholar]
- Reichardt HM, Tuckermann JP, Gottlicher M, Vujic M, Weih F, Angel P, Herrlich P, Schutz G 2001 Repression of inflammatory responses in the absence of DNA binding by the glucocorticoid receptor. EMBO J 20:7168–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bosscher K, Vanden Berghe W, Haegeman G 2006 Cross-talk between nuclear receptors and nuclear factor κB. Oncogene 25:6868–6886 [DOI] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR 2005 The glucocorticoid receptor blocks P-TEFb recruitment by NFκB to effect promoter-specific transcriptional repression. Genes Dev 19:1116–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens C, Bilodeau S, Maira M, Gauthier Y, Drouin J 2005 Protein-protein interactions and transcriptional antagonism between the subfamily of NGFI-B/Nur77 orphan nuclear receptors and glucocorticoid receptor. Mol Endocrinol 19:885–897 [DOI] [PubMed] [Google Scholar]
- Becker M, Baumann CT, John S, Walker D, Vigneron M, McNally JG, Hager GL 2002 Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep 3:1188–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Zhao Y, Becker M, John S, Parekh BS, Huang S, Hendarwanto A, Martinez ED, Chen Y, Lu H, Adkins NL, Stavreva DA, Wiench M, Georgel PT, Schiltz RL, Hager GL 2006 HDAC1 acetylation is linked to progressive modulation of steroid receptor induced gene transcription. Mol Cell 22:669–679 [DOI] [PubMed] [Google Scholar]
- Morsink MC, Steenbergen PJ, Vos JB, Karst H, Joels M, de Kloet ER, Datson NA 2006 Acute activation of hippocampal glucocorticoid receptors results in different waves of gene expression throughout time. J Neuroendocrinol 18:239–252 [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M 2000 Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843–852 [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F 2003 Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751–763 [DOI] [PubMed] [Google Scholar]
- Burakov D, Crofts LA, Chang CP, Freedman LP 2002 Reciprocal recruitment of DRIP/mediator and p160 coactivator complexes in vivo by estrogen receptor. J Biol Chem 277:14359–14362 [DOI] [PubMed] [Google Scholar]
- Lowy DR, Rands E, Scolnick EM 1978 Helper-independent transformation by unintegrated Harvey sarcoma virus DNA. J Virol 26:291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragoso G, Pennie WD, John S, Hager GL 1998 The position and length of the steroid-dependent hypersensitive region in the mouse mammary tumor virus long terminal repeat are invariant despite multiple nucleosome B frames. Mol Cell Biol 18:3633–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird K, Davis S, Antonescu CR, Harper UL, Walker RL, Chen Y, Glatfelter AA, Duray PH, Meltzer PS 2005 Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res 65:9226–9235 [DOI] [PubMed] [Google Scholar]
- Chen Y, Dougherty ER, Bittner ML 2007 Ratio-based decisions and the quantitative analysis of cDNA microarray images. J Biomed Opt 2:364–374 [DOI] [PubMed] [Google Scholar]
- Kramer P, Fragoso G, Pennie WD, Htun H, Hager GL, Sinden RR 1999 Transcriptional state of the mouse mammary tumor virus promoter can effect topological domain size in vivo. J Biol Chem 274:28590–28597 [DOI] [PubMed] [Google Scholar]
- McNally JG, Mueller WG, Walker D, Wolford RG, Hager GL 2000 The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262–1265 [DOI] [PubMed] [Google Scholar]
- Drouin J 2006 Molecular mechanisms of pituitary differentiation and regulation: implications for hormone deficiencies and hormone resistance syndromes. Front Horm Res 35:74–87 [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung MH, Biddie S, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL 2008 Interaction of the glucocorticoid receptor with the global chromatin landscape. Mol Cell 29:611–624 [DOI] [PubMed] [Google Scholar]
- Li GJ, Zhao Q, Zheng W 2005 Alteration at translational but not transcriptional level of transferrin receptor expression following manganese exposure at the blood-CSF barrier in vitro. Toxicol Appl Pharmacol 205:188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith OL, He A, Marra M, Snyder M, Jones S 2007 Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods 4:651–657 [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007 High-resolution profiling of histone methylations in the human genome. Cell 129:823–837 [DOI] [PubMed] [Google Scholar]
- Archer TK, Lee HL, Cordingley MG, Mymryk JS, Fragoso G, Berard DS, Hager GL 1994 Differential steroid hormone induction of transcription from the mouse mammary tumor virus promoter. Mol Endocrinol 8:568–576 [DOI] [PubMed] [Google Scholar]
- Wang X, DeFranco DB 2005 Alternative effects of the ubiquitin-proteasome pathway on glucocorticoid receptor down-regulation and transactivation are mediated by CHIP, an E3 ligase. Mol Endocrinol 19:1474–1482 [DOI] [PubMed] [Google Scholar]
- Wallace AD, Cidlowski JA 2001 Proteasome-mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J Biol Chem 276:42714–42721 [DOI] [PubMed] [Google Scholar]
- Lee HL, Archer TK 1998 Prolonged glucocorticoid exposure dephosphorylates histone H1 and inactivates the MMTV promoter. EMBO J 17:1454–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Hager GL 1997 Transcriptional regulation of mammalian genes in vivo: a tale of two templates. J Biol Chem 272:27493–27496 [DOI] [PubMed] [Google Scholar]
- Hager GL, Elbi C, Johnson TA, Voss TC, Nagaich AK, Schiltz RL, Qiu Y, John S 2006 Chromatin dynamics and the evolution of alternate promoter states. Chromosome Res 14:107–116 [DOI] [PubMed] [Google Scholar]
- Hager GL, Nagaich AK, Johnson TA, Walker DA, John S 2004 Dynamics of nuclear receptor movement and transcription. Biochim Biophys Acta 1677:46–51 [DOI] [PubMed] [Google Scholar]
- Schaaf MJ, Cidlowski JA 2002 Molecular mechanisms of glucocorticoid action and resistance. J Steroid Biochem Mol Biol 83:37–48 [DOI] [PubMed] [Google Scholar]
- Fletcher TM, Xiao N, Mautino G, Baumann CT, Wolford RG, Warren BS, Hager GL 2002 ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol Cell Biol 22:3255–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.