Abstract
Background
FGF signaling has multiple roles in regulating processes in animal development, including the specification and patterning of the mesoderm. In addition, FGF signaling supports self renewal of human embryonic stem cells and is required for differentiation of murine embryonic stem cells into a number of lineages.
Methodology/Principal Findings
Given the importance of FGF signaling in regulating development and stem cell behaviour, we aimed to identify the transcriptional targets of FGF signalling during early development in the vertebrate model Xenopus laevis. We analysed the effects on gene expression in embryos in which FGF signaling was inhibited by dominant negative FGF receptors. 67 genes positively regulated by FGF signaling and 16 genes negatively regulated by FGF signaling were identified. FGF target genes are expressed in distinct waves during the late blastula to early gastrula phase. Many of these genes are expressed in the early mesoderm and dorsal ectoderm. A widespread requirement for FGF in regulating genes expressed in the Spemann organizer is revealed. The FGF targets MKP1 and DUSP5 are shown to be negative regulators of FGF signaling in early Xenopus tissues. FoxD3 and Lin28, which are involved in regulating pluripotency in ES cells are shown to be down regulated when FGF signaling is blocked.
Conclusions
We have undertaken a detailed analysis of FGF target genes which has generated a robust, well validated data set. We have found a widespread role for FGF signaling in regulating the expression of genes mediating the function of the Spemann organizer. In addition, we have found that the FGF targets MKP1 and DUSP5 are likely to contribute to the complex feedback loops involved in modulating responses to FGF signaling. We also find a link between FGF signaling and the expression of known regulators of pluripotency.
Introduction
Fibroblast growth factors (FGFs) are small polypeptides that have multiple functions in early development and homeostasis of the adult organism. FGFs are present in all animal groups and are one of relatively few families of extracellular signaling molecules that are involved in regulating animal development. 22 FGFs have been identified in higher vertebrates [1].
FGF signaling has a key role in specifying the primary germ layers that give rise to all the tissues of the adult organism. Experiments initially carried out in amphibians, and later supported by studies in mammals, birds and fish, demonstrated that FGF signaling is required to regulate gene expression within the early vertebrate mesoderm, which is the germ layer giving rise to muscle, skeleton, connective tissue, blood and organs such as the kidney [2]–[6]. As well as regulating mesodermal gene expression, FGF signaling is involved in regulating the complex morphogenetic activity exhibited by mesoderm cells during vertebrate gastrulation [3], [7]. FGF signals produced by the mesoderm, acting on the adjacent ectoderm, are also required for induction and patterning of the vertebrate nervous system [8]–[11].
More recently it has been shown that FGF signaling plays a critical role in the commitment of mouse embryonic stem (ES) cells to mesodermal, as well as both neural and non-neural ectodermal lineages [12], [13]. FGF signaling is also important for maintaining self renewal in human ES and induced pluripotential stem (iPS) cells in culture [14]–[16].
Given the importance of FGF signaling in adult and embryonic life, the downstream transcriptional targets involved in mediating the activities of the FGFs are of great interest. In order to identify genes that respond to FGF signaling in early development we have compared gene expression in normal embryos with embryos in which FGF signaling has been inhibited. Our analysis identifies 67 genes which are significantly down regulated and 16 genes which are up regulated in response to FGF inhibition. A high proportion of the putative FGF target genes have predicted functions associated with cell signaling and transcriptional regulation.
We show that many of the targets are expressed in known regions of FGF activity during development. Our analysis reveals some interesting features of the FGF dependent transcriptome. We find that FGF signaling is required for the normal expression of multiple genes in the Spemann organizer, a structure orthologous to the node of higher vertebrates and which is required for the establishment of the main body axis.
Intrestingly, we find that inhibition of FGF signaling down regulates expression of Lin28 and FoxD3, two genes which have been implicated in regulating the pluripotential state of embryonic stem (ES) cells [14], [17]–[19].
Cluster analysis, based upon temporal expression, identified a number of distinct waves of expression from FGF targets following the initial activation of FGF signaling in the amphibian embryo. Following the activation of endogenous FGF signalling in blastula stages, we show that two of the earliest expressed target genes are the MAP kinase phosphatase 1 (MKP1) gene and a novel Xenopus gene related to human Dual Specificity Phosphatase 5 (DUSP5). We show that both DUSP5 and MKP1 inhibit FGF dependent ERK/MAP kinase phosphorylation and mesoderm formation induced by FGF in Xenopus tissues. Our analysis indicates that DUSP5 and MKP1 are members of the FGF synexpression group and are components of the negative feedback network required to limit the extent of FGF signaling in the early embryo.
Results
Timing of FGF signaling in the Xenopus embryo
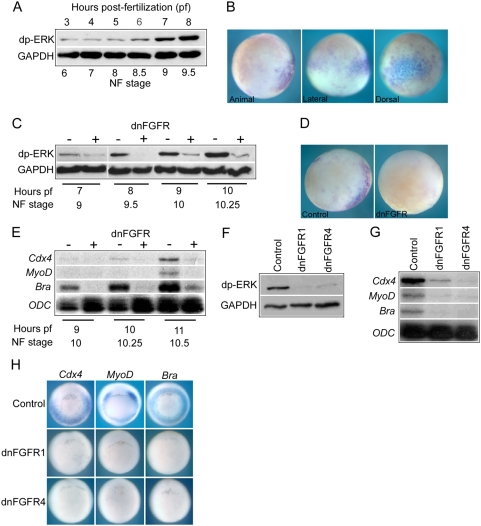
The aim of this study was to identify FGF targets that are induced shortly after the initial activation of zygotic FGF signaling in the embryo. It was therefore necessary to accurately determine when FGF signaling is activated in the embryo. We examined the temporal profile and spatial distribution of activated diphospho-ERK (dp-ERK), which is a key signal transduction effector of FGF signaling in the Xenopus embryo [20]–[22]. Figure 1A is a Western blot showing levels of dp-ERK in embryos from early cleavage to late blastula stages (NF stage 3 to 9.5). We detect constant low levels of activated ERK up to stage 8.5. The initial rise in the level of dp-ERK is detected at mid-blastula stage 8.5 and there is robust increase by stage 9 (7 hours post-fertilization (pf) at 23°C), corresponding to the onset of major zygotic transcription at the mid-blastula transition (MBT) [23].
Figure 1. FGF signaling in early development.
(A) is a Western blot showing levels of diphospo-ERK (dp-ERK) in whole embryos from cleavage stage 6 to late blastula stage 9.5 (3 to 8 hours pf at 23°C). GAPDH is a ubiquously expressed loading control. (B) shows whole mount immunohistochemistry for dp-ERK in blastula stage 9 embryos. In animal hemisphere view dorsal side is to the right. In lateral view the animal hemisphere is to the top and the dorsal side is to the right. In dorsal view the animal hemisphere is to the top. (C) is a Western blot comparing dp-ERK levels in control uninjected embryos and embryos injected with 4 ng dnFGFR4 mRNA from blastula stage 9 to early gastrula stage 10.25. (D) shows whole mount immunohistochemistry for dp-ERK at blastula stage 9 in a control uninjected embryo and an embryo injected with 4 ng dnFGFR4 embryo. Embryos are viewed from the animal hemisphere with dorsal side to the right. (E) is an RNAase protection analysis (RPA) showing the expression of Cdx4, MyoD, brachyury and ODC in control uninjected embryos and embryos injected with 4 ng dnFGFR4 mRNA from early gastrula stage 10 until stage 10.5. ODC is a ubiquioulsy expressed loading control. 10 µg of total RNA were used per hybridization. (F) is a Western blot showing dp-ERK levels in control uninjected embryos, embryos injected with 4 ng dnFGFR1 mRNA and embryos injected with 4 ng dnFGFR4 mRNA at early gastrula stage 10.5. The embryos are siblings to one set of the three biological replicates used for the microarray analysis. (G) is an RPA showing expression of Cdx4, MyoD and brachyury in control uninjected embryos, embryos injected with 4 ng dnFGFR1 mRNA and embryos injected with 4 ng dnFGFR4 mRNA at early gastrula stage 10.5. The embryos are siblings to one set of the three biological replicates used for the microarray analysis. (H) shows whole mount in situ hybridizations for Cdx4, MyoD and brachyury in control uninjected embryos , embryos injected with 4 ng dnFGFR1 mRNA and embryos injected with 4 ng dnFGFR4 mRNA at early gastrula stage 10.5. The embryos are siblings to one set of the three biological replicates used for the microarray analysis.
Our data show that the initial activation of ERK is in a dorsal to ventral gradient within a broad belt of tissue around the equator of the embryo at late blastula stage 9 (Figure 1B). Initial dp-ERK activation is not limited to the presumptive mesoderm of the marginal zone but extends a considerable distance into the animal hemisphere on the dorsal side of the embryo. Figures 1C and D show that this early zygotic ERK activity is blocked by over expression of a dominant negative FGF receptor (dnFGFR). The dnFGFRs used in this study are carboxy-terminal truncations of the receptors lacking tyrosine kinase activity and block FGF signaling by associating with endogenous receptors to form non-functional dimers [24], [25]. We conclude that zygotic activation of the FGF signaling pathway commences at mid-blastula stage 8.5 (6 hours pf at 23°C).
Timing of transcriptional responses to FGF signaling in the Xenopus embryo
The genes coding for the brachyury, MyoD and Cdx4 transcription factors have previously been identified as targets of the FGF signaling pathway. These genes are activated by FGF even in the presence of the translation inhibitor cycloheximide and are defined as immediate early responses to FGF signaling [26]–[28]. We have used expression of these genes to indicate when the initial transcriptional responses to FGF signaling occur in the Xenopus embryo. Figure 1E is an RNAase protection analysis (RPA) showing that Brachyury expression is detected by early gastrula stage 10 and Cdx4 by stage 10.25. However, robust expression of all three immediate early FGF response genes is not detected until stage 10.5 (11 hours pf at 23°C). Furthermore, we show that the initial expression of all three genes is blocked by over expression of a dnFGFR. Based on the timing of ERK activation and transcriptional activation of known FGF target genes, early gastrula stage 10.5 (11 hours of culture at 23°C) was chosen as the stage for the analysis of FGF targets.
Identifying transcriptional responses to FGF signaling
In order to identify transcriptional targets of FGF signaling gene expression in control embryos was compared to sibling embryos in which FGF signaling was inhibited by over expression of dominant negative versions of FGFR1 (dnFGFR1) or FGFR4 (dnFGFR4) [24], [25].
In order to undertake statistical analysis of the microarray data three biological replicates were carried out. Each replicate set consisted of control embryos and embryos injected with dnFGFR1 or dnFGFR4 collected at stage 10.5 (11 hours pf at 23°C). Before proceeding to microarray analysis each replicate set was checked for effective FGF inhibition. Sibling embryos from each replicate set were analysed for dp-ERK levels and expression of Cdx4, Brachyury and MyoD by both RPA and in situ hybridization. Figures 1F–1H are analyses of a representative biological replicate set showing that, with the experimental conditions used, overexpression of dnFGFR1 or dnFGFR4 results in potent inhibition of ERK activation (Figure 1F) and down regulation of transcription from known FGF target genes relative to sibling controls (Figures 1G and 1H).
Changes in gene expression resulting from dnFGFR1 and dnFGFR4 overexpression
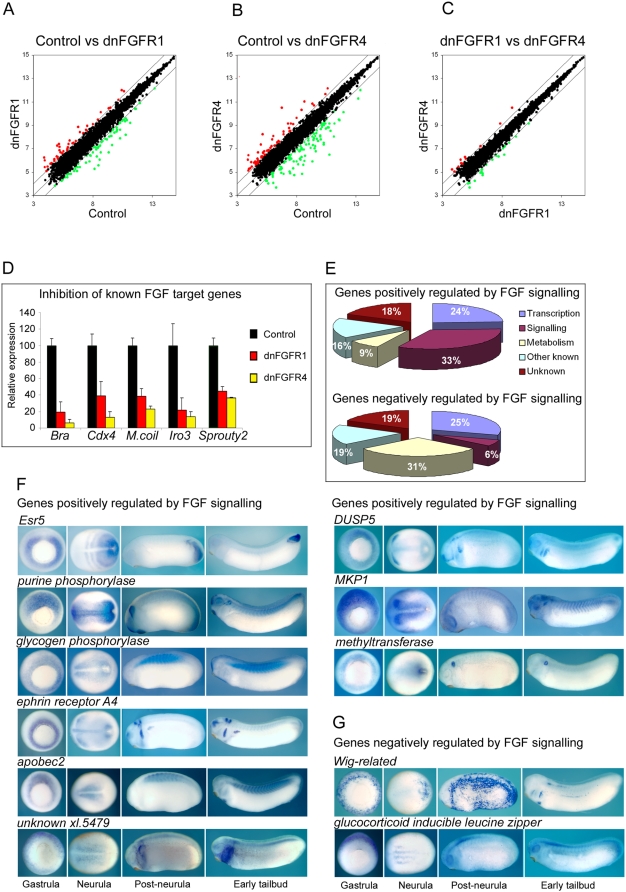
After these quality control checks, the RNA samples from each of the three biological replicates were analysed using Affymetrix GeneChip Xenopus laevis Genome Arrays, which allow the analysis of more than 14,400 transcipts expressed in early development. Figures 2A and 2B are scatterplots of log2 gene expression values in controls versus dnFGFR1 and dnFGFR4 injected embryos. Probe sets showing greater than 2-fold changes of expression in control versus experimental groups are indicated by red and green points. These data show that the expression of a considerable number of genes is altered in response to FGF inhibition by both dnFGFR1 and dnFGFR4.
Figure 2. Identification of FGF target genes.
(A, B and C) are scatterplots of log2 probeset expression values from the Affymetrix microanalysis undertaken on early gastrula stage 10.5 control embryos, dnFGFR1 injected embryos and dnFGFR4 injected embryos . Values for each point are the average of three biological replicates. The centre line represents a ratio of 1∶1 between the two groups indicating no difference in expression. The outlying lines represent two fold differences in expression. Points representing probe sets showing ≥2 reduction in expression are indicated in green. Points representing probesets showing ≥2 increase in expression are indicated in red. (D) is a chart showing the expression of brachyury (bra), cdx4, marginal coil (M.coil), Iro3 and sprouty2 in control embryos and embryos injected with 4 ng dnFGFR1 mRNA or 4 ng dnFGFR4 mRNA. Microarray derived expression values are based on the average of three biological replicates. Relative expression is calculated as a percentage of the expression in control embryos. Standard deviation bars are indicated. (E) shows pie charts of genes positively and negatively regulated by FGF signaling. Percentages of each group classified according to their putative cellular function are indicated. Details of the up regulated and down regulated genes are contained Tables 1, 2 and Tables S2 to S11. (F) shows the expression patterns of genes positively regulated by FGF signaling at determined by in situ hybridization at early gastrula stage 10.5, early neurula stage 14, post-neurula stage 22 and early tailbud stage 30. Gastrula embryos are vegetal views with dorsal to the top. Neurula embryos are dorsal views with anterior to the left. Post-neurula and tailbud embryos are lateral view with dorsal to the top and anterior to the left. (G) shows the expression patterns of genes negatively regulated by FGF signaling.
In contrast to Figure 2A and 2B, in dnFGFR1 versus dnFGFR4 injected embryos we see that expression levels of relatively few probe sets are greater than two-fold different between the two groups (Figure 2C). Our analysis shows that only four genes, using the criteria of ≥2-fold change in expression and a significance level of p≤0.01, exhibit different expression levels in dnFGFR1 versus dnFGFR4 injected embryos (Table S1). This indicates that at this stage of development the genes affected by inhibition with the different dominant negative receptors are largely the same. The differences in gene expression profiles of dnFGFR1 and dnFGFR4 injected embryos are quantitative differences in the levels of expression from the same set of target genes. This conclusion is supported by Figure 2D, which shows that both dnFGFR1 and dnFGFR4 down regulate the expression of several known FGF targets. However, in all cases dnFGFR4 has a more potent effect on gene expression. The data in Figure 2A and 2B follow a similar trend, in which there are greater fold changes in gene expression following dnFGF4 injection than with dnFGFR1. We conclude that inhibition with dnFGFR1 and dnFGFR4 affects the same sets of genes but that on a per mass of injected mRNA basis, dnFGFR4 is more potent.
Classification of putative FGF target genes
Lists of genes affected by FGF inhibition were compiled by comparing gene expression changes in dnFGFR4 injected embryos versus control embryos. The criteria of at least a 2-fold change in expression and a significance level of p≤0.01 were used to compile the gene lists. After the elimination of multiple probe sets representing the same gene, using the stated criteria, we find that 67 genes are significantly down-regulated by FGF inhibition, indicating that in normal development these genes are positively regulated by FGF signaling. Table 1 shows these genes in order of mean fold inhibition in dnFGFR injected embryos relative to controls. The T-box gene brachyury shows the highest fold inhibition (>19-fold). We find that only 16 genes are significantly up-regulated by FGF inhibition, indicating that in normal development these genes are negatively regulated by FGF signaling (Table 2).
Table 1. Genes positively regulated by FGF signaling.
| Gene | Fold inhibition | GenBank accession | Affymetrix probe set |
| Brachyury | 19.2 | M77243 | Xl.514.1.S1_at |
| Egr1 | 12.4 | AF250345 | Xl.637.1.A1_at |
| FoxD5A | 10.8 | AF162782 | Xl.642.1.S1_at |
| SIP1 | 9.8 | AB038353 | Xl.958.1.S2_at |
| Cdx4 | 8.6 | UO2034 | Xl.10269.1.S1_at |
| Esr5 | 8.5 | BJ057112 | Xl.14524.1.S1_at |
| Purine phosphorylase | 7.9 | BM172525 | Xl.16206.1.A1_at |
| Marginal coil | 7.7 | BJ044312 | Xl.5454.1.S1_at |
| Paraxial protocadherin | 7.3 | AW782445 | Xl.6173.1.A1_at |
| Glycogen phosphorylase | 7.0 | BJ056085 | Xl.7815.1.A1_at |
| NADH dehydrogenase sub-unit | 6.5 | BJ051675 | Xl.12993.1.A1_at |
| FoxD3A | 6.0 | AB014611 | Xl.525.1.S1_at |
| G-coupled receptor P2Y5 | 5.7 | BQ401062 | Xl.19933.1.S1_at |
| Related to DC-STAMP domain receptor | 5.6 | BI447679 | Xl.15270.1.A1_at |
| Meso05 | 5.1 | BF615090 | Xl.7720.1.A1_at |
| Uncharacterised protein C2orf32 | 5.1 | CB756627 | Xl.25136.1.A1_at |
| Frzb1 | 5.1 | U78598 | Xl.212.2.S1_a_at |
| XPO | 5.0 | BJ051206 | Xl.5908.1.S1_s_at |
| Ephrin receptor A4 | 4.8 | BJ080037 | Xl.13.2.A1_at |
| XSpr2 | 4.5 | BJ049843 | Xl.2755.1.S1_a_at |
| Zic3a | 4.5 | AB005292 | Xl.7969.1.S1_at |
| Xiro3 | 4.4 | AF027175 | Xl.4522.1.S1_at |
| Gravin-like | 4.4 | AF308810 | Xl.3468.1.S1_at |
| Alkaline phosphatase | 4.3 | BC043760 | Xl.1299.1.S1_at |
| Apobec2 | 4.2 | AW766385 | Xl.5876.1.A1_a |
| p75-like fullback receptor | 4.2 | AF131890 | Xl.3540.1.S1_at |
| Wnt8 | 4.0 | X57234 | Xl.49.1.S1_at |
| Fructokinase-related protein | 3.9 | CB756273 | Xl.15623.1.A1_at |
| Crescent | 3.9 | AF260729 | Xl.619.1.S1_at |
| Pinhead | 3.8 | BJ056268 | Xl.3529.1.A1_at |
| Wnt5b | 3.6 | AW148258 | Xl.11619.1.S1_at |
| Unknown | 3.6 | BJ092401 | Xl.5479.1.A1_at |
| Retrotransposon protein 1a11 | 3.6 | L11263 | Xl.3352.1.S1_at |
| FoxA4 | 3.4 | S93559 | Xl.1082.1.S1_at |
| Mitotic phosphoprotein 67 | 3.2 | BJ077239 | Xl.20772.1.A1_at |
| Cdx1 | 3.2 | CB564190 | Xl.23739.1.A1_at |
| Sprouty2 | 3.1 | AF331825 | Xl.11965.1.S1_at |
| DUSP5 | 3.0 | BJ077463 | Xl.15374.1.A1_at |
| Chordin | 2.8 | BF610870 | Xl.3549.1.S1_at |
| MKP1 | 2.7 | AJ320159 | Xl.2803.1.S1_at |
| Unknown | 2.7 | BI312705 | Xl.18179.1.S1_at |
| Xom | 2.7 | X98454 | Xl.37.1.S1_at |
| Putative nucleolar GTP binding protein | 2.7 | BM179370 | Xl.14776.1.A1_at |
| Lin28a homologue | 2.7 | BJ047699 | Xl.3418.1.A1_at |
| Glut1 transporter | 2.7 | BJ049047 | Xl.24121.1.A1_at |
| Unknown | 2.6 | BJ056692 | Xl.15382.1.A1_at |
| Dkk1 | 2.6 | AF030434 | Xl.251.1.S1_at |
| Unknown | 2.5 | AW460550 | Xl.11594.1.A1_at |
| RALDH2 | 2.5 | BI449483 | Xl.18999.1.A1_at |
| Prickle | 2.5 | AF387815 | Xl.7556.1.S1_at |
| ADMP | 2.4 | BF231842 | Xl.3809.1.A1_at |
| Unknown | 2.3 | BJ085271 | Xl.1521.1.A1_at |
| Cytochrome B561 | 2.3 | U16364 | Xl.11917.1.S1_at |
| Goosecoid | 2.3 | BJ056432 | Xl.801.1.S1_at |
| FoxC1 | 2.3 | AF116844 | Xl.180.1.S1_at |
| Noggin | 2.2 | M98807 | Xl.834.1.S1_at |
| Sprouty1 | 2.2 | BG022481 | Xl.10087.1.A1_Fat |
| Oct1 | 2.2 | BG022051 | Xl.1265.1.S1_at |
| Rexp52 | 2.1 | BG555868 | Xl.3023.1.A1_at |
| Grb10 interacting protein2 | 2.1 | BJ098841 | Xl.14208.1.A1_at |
| Putative methyltransferase | 2.1 | BJ100128 | Xl.20056.1.S1_a_at |
| Connexin 29 | 2.1 | BJ076720 | Xl.8924.1.A1_at |
| SMCT | 2.1 | BJ047968 | Xl.6392.1.A1_at |
| Weakly similar to Rab1 | 2.1 | BJ079872 | Xl.3365.1.A1_at |
| Unknown | 2.1 | CA972457 | Xl.19961.1.S1_at |
| Moderately similar to Brain protein 44 | 2.1 | BJ088835 | Xl.15887.1.S1_x_at |
| Ephrin receptor A2 | 2.0 | BF025525 | Xl.14496.1.A1_at |
Table 2. Genes negatively regulated by FGF signaling.
| Gene | Fold activation | GenBank accession | Affymetrix probe set |
| XIRG protein | 13.5 | AJ278067 | Xl.4965.1.S1_at |
| PDGF A chain | 5.6 | M23238 | Xl.841.3.S1_a_at |
| WIG-related | 5.3 | BJ044287 | Xl.23988.1.S1_at |
| CP2-like transcription factor | 4.3 | BJ046394 | Xl.16094.1.A1_at |
| Glucocorticoid inducible leucine zipper | 4.1 | BC043841 | Xl.12378.1.S1_at |
| Unknown | 3.8 | AW147865 | Xl.2077.1.A1_at |
| WIG | 3.7 | AF310008 | Xl.736.1.S1_at |
| XANF1 | 3.1 | X60099 | Xl.131.1.S1_at |
| HES-related 1B | 3.0 | AB071434 | Xl.12126.1.S1_at |
| Darmin | 2.9 | CD324819 | Xl.6024.1.S1_at |
| ODC2 | 2.8 | AF217544 | Xl.8949.1.S1_at |
| Unknown | 2.4 | AW460608 | Xl.11598.1.A1_at |
| Thioredoxin binding protein 2 | 2.3 | BQ399899 | Xl.24749.1.A1_at |
| Unknown | 2.2 | BM192746 | Xl.25985.1.A1_at |
| Selenophosphate synthetase 1 | 2.1 | BJ091471 | Xl.6522.1.A1_at |
| Adenosine deaminase | 2.1 | BJ090126 | Xl.24155.1.A1_at |
Where possible we have classified FGF target genes based on cellular function. We find that a large proportion of genes positively regulated by FGF signaling are either involved in transcriptional regulation (24%) or cell signaling (18%), with a relatively small number of genes (9%) involved in various aspects of metabolism. The corresponding figures for genes negatively regulated by FGF signaling are 29% involved in transcriptional regulation, 6% in cell signaling and 29% in metabolism. These data are represented as pie charts in Figure 2E and the detailed breakdown of FGF target classification, together with relevant references are presented in Tables S2, S3, S4, S5, S6, S7, S8, S9, S10 and S11. Tables S12 and S13 show the Gene Ontology (GO) term classifications for FGF targets derived from the available Affymetrix annotation files.
Expression of FGF target genes
Consistent with the pattern of FGF activity in the late blastula and early gastrula stage embryo (Figure 1) many of the genes identified as being down regulated following FGF inhibition are expressed in the mesoderm or dorsal ectoderm at the start of gastrulation. Expression data for previously characterised genes, along with new data from this study are summarized in Table S14.
We have undertaken a more detailed expression analysis of a number of these positively regulated FGF targets at early gastrula, early neurula, post-neurula and early tailbud stages. (Figure 2F) We find that all of these genes are expressed in the mesoderm around the mesoderm. In post-gastrula stages the expression patterns of these genes diversify; however, some common patterns are detected. For example, the posterior mesoderm, the paraxial mesoderm and the tailbud are common sites of expression. Other sites of expression include the anterior CNS and branchial arches. In the case of a putative methyltransferase the post-gastrula expression is remarkably restricted, being limited to a very tight domain around the closed blastopore and later in the developing otic vesicle.
The expression patterns were also determined for two of the genes that are up regulated in response to FGF inhibition, which we predict will be negatively regulated by FGF signaling in normal development (Figure 2G).
Wig-related codes for a protein similar to Xwig1, which is a putative endoplasmic reticulum resident protein [29]. GILZ (glucocorticoid inducible leucine zipper), is a member of the Tsc-22 family of transcription factors related to Drosophila bunched [30]. In contrast with genes positively regulated by FGF, the early expression of these genes is excluded from the circum-blastoporal region. The Wig-related gastrula and post-gastrula expression pattern is highly dynamic, before resolving to a stable pattern of expression in the CNS, branchial arches and lateral mesoderm. After gastrulation GILZ is expressed in the neurogenic region of the open neural plate and subsequently in the neural tube.
Validation of FGF targets
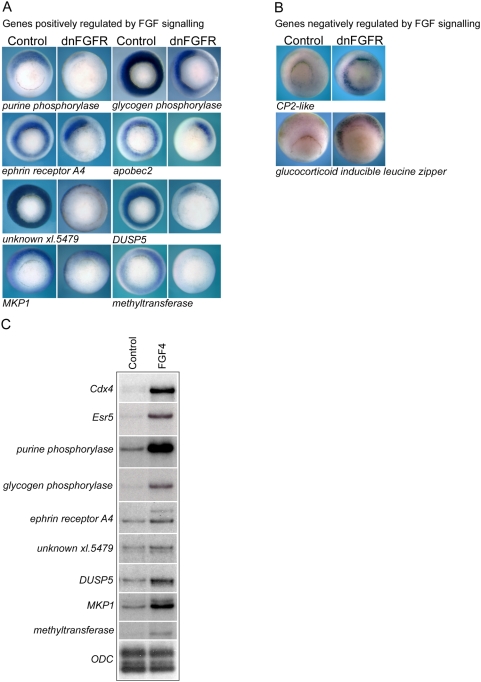
It is generally accepted that gene lists identified by microarray analysis should be validated by independent methodology. For a number of the putative target genes independent validation of their response to FGF signaling was undertaken by in situ hybridization. Figure 3A shows the effects of FGF inhibition on the spatial expression of genes identified as being down regulated in the microarray-based screen. Injection of dnFGFR leads to dramatic inhibition of the circum-blastoporal expression of these genes in gastrula stage embryos.
Figure 3. Validation of FGF target genes.
(A) shows whole mount in situ hybridizations for genes positively regulated by FGF signaling at gastrula stage 10.5 in control embryos and embryos injected with 2 ng of dnFGFR1 mRNA. (B) shows the expression gene negatively regulated by FGF signaling in control embryos and embryos injected with 2 ng dnFGFR1 mRNA. All embryos are vegetal view with dorsal to the top. Non-uniform down regulation around the circumference of the blastopore is apparent in some embryos and is likely due to variability in the diffusion of injected dnFGFR mRNA. (C) is an RPA showing the expression at gastrula stage 10.5 of a number of genes in control animal cap explants and explants treated with FGF4 protein. 5 µg total RNA was used per hybridization. ODC is a loading control.
Conversely, the size of the expression domains of two putative targets negatively regulated by FGF signaling are dramatically increased in embryos injected with dnFGFR mRNA (Figure 3B). In the case of CP2-like and GILZ, inhibition of FGF signaling leads to elevated expression in the circum-blastoral region indicating that in normal development FGF signaling is required to exclude their expression from this region of the embryo.
Further validation of the FGF target genes was undertaken by showing that FGF signaling is not only necessary but is also sufficient for their expression. Figure 3C shows an RNAase protection assay (RPA) on control animal hemisphere tissue explants (animal caps) and animal caps treated with recombinant FGF4 protein. With all genes tested, FGF treatment leads to marked increase in transcription as compared to control explants. Taken together, our analyses indicate that the microarray based screen has provided a well supported list of candidate FGF target genes.
FGF signaling and dorsal gene expression
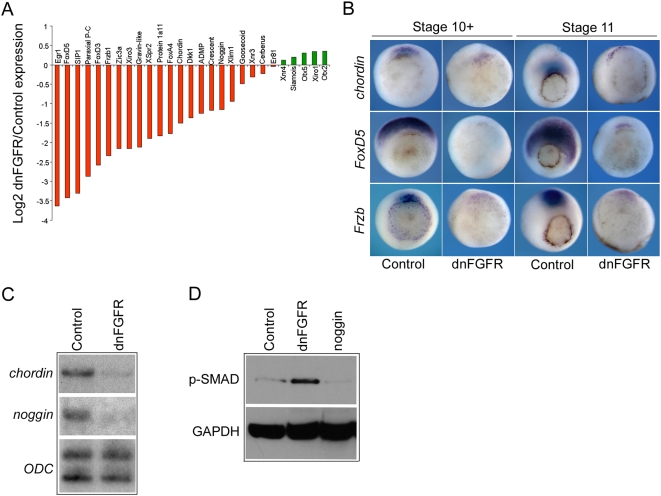
The identified FGF target genes included several genes, including chordin and noggin, which are required for the function of the Spemann organizer in the dorsal mesoderm [31], [32]. This region of the embryo plays a key role in regulating the formation of the main body axes. We investigated if there is a general role for FGF in regulation of dorsal gene expression. The data in Data in Figure 4A and Table 3 show that many dorsally biased genes, including Egr1 and FoxD5 are highly down regulated when FGF signaling is inhibited (>12-fold and >10-fold respectively). Many of the dorsally expressed FGF target genes have been shown to be directly involved in mediating organizer function, including Frzb1, chordin, noggin, FoxD3, FoxD5 and goosecoid. Other dorsally expressed genes, such as Xnr3 and cerberus, are not significantly down regulated, and others such as Otx2 show small increases in expression (not significant at the p = 0.01 level).
Figure 4. FGF signaling and regulation of dorsal gene expression.
(A) is a bar chart showing the log2 of the ratio of expression dnFGFR4 injected embryos versus control embryos for a number of dorsally expressed genes at gastrula stage 10.5. Microarray derived expression values are based on the average of three biological replicates. Bars in red below the centre line represents genes down regulated in response to FGF inhibition. Bars in green represent genes up regulated in response to FGF inhibition. (B) shows whole mount in situ hybridizations for chordin, FoxD5 and Frzb in control embryos and embryos injected with 4 ng dnFGF4 mRNA at very early gastrula stage 10+ and mid-gastrula stage 11. All embryos are vegetal views with dorsal to the top. (C) is an RPA showing the expression of chordin and noggin in control embryos and embryos injected with 4 ng dnFGFR1 mRNA at gastrula stage 10.5. (D) is a Western blot showing levels of phospho-SMAD1/5/8 (p-SMAD) at gastrula stage 10.5 in control embryos, embryos injected with 4 ng dnFGFR1 mRNA or 1 ng mRNA coding for secreted the BMP inhibitor noggin. GAPDH is a loading control.
Table 3. Effects of FGF inhibition on organizer gene expression.
| Gene | Ratio of expression in dnFGFR and control groups |
| Egr1 | 12.4 |
| FoxD5A | 10.8 |
| SIP | 9.8 |
| Paraxial P-C | 7.3 |
| FoxD3A | 6 |
| Frzb1 | 5.1 |
| Zic3A | 4.5 |
| Xiro3 | 4.5 |
| Gravin-like | 4.4 |
| Crescent | 3.9 |
| Xspr2 | 3.7 |
| Protein 1a11 | 3.6 |
| FoxA4 | 3.4 |
| Chordin | 2.9 |
| Dkk1 | 2.6 |
| ADMP | 2.4 |
| Goosecoid | 2.3 |
| Noggin | 2.2 |
| Xlim1 | 1.9 |
| Xnr3 | 1.2 |
| Cerberus | 1.2 |
| Er81 | 1.0 |
| Xnr4 | 0.9 |
| Siamois | 0.9 |
| Otx5 | 0.8 |
| Xiro1 | 0.8 |
| Otx2 | 0.8 |
The in situ hybridizations in Figure 4B show that FGF inhibition dramatically reduces the spatial extend of chordin, FoxD5 and Frzb expression through early gastrula stages. We also show by RPA that chordin and noggin are strongly down regulated in early gastrula stage embryos following FGF inhibition (Figure 4C), indicating a role for FGF in regulating the expression of secreted BMP inhibitors.
Stimulation of BMP signaling leads to the phosphorylation and activation of SMAD1. Figure 4D is a Western blot for phospho-SMAD1/5/8 (p-SMAD). In keeping with a role for FGF signaling in regulating dorsally expressed secreted BMP inhibitors, such chordin and noggin, levels of p-SMAD1/5/8 are elevated in response to FGF inhibition. Our data indicate a widespread but not ubiquitous requirement for FGF signaling in the regulation of organizer gene expression during gastrula stages.
Expression profiling and cluster analysis of FGF target genes
In order to generate temporal expression profiles for individual FGF target genes from pre-MBT stages until early neurula stages we carried out Affymetrix microarray analysis on normally developing sibling embryos at hourly time points from 5 hours pf (stage 8) to 16 hours pf (stage 14) at 23°C.
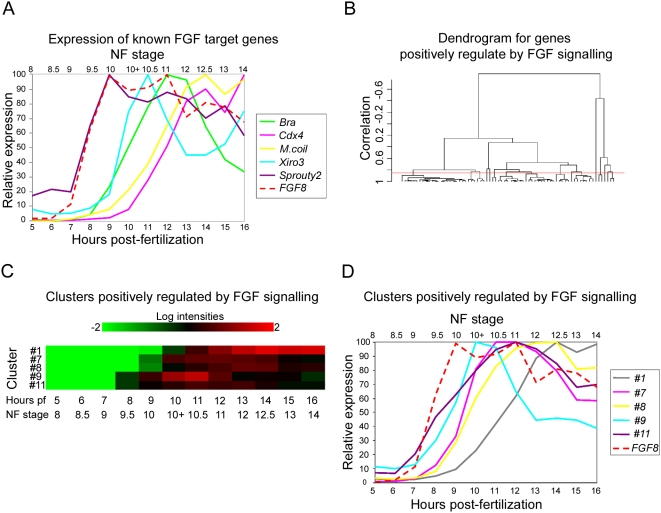
Figure 5A shows the relative expression profiles of FGF8 and 5 known FGF target genes. The initial rise in FGF8 expression is first detected at stage 9 (7 hours pf), indicating that FGF8 expression is activated very rapidly post-MBT. As mentioned earlier, this increase in FGF8 expression corresponds closely with the detected rapid elevation of dp-ERK levels in the embryo between stage 8.5 and stage 9 (Figure 1A). We note that although normal expression of these genes requires FGF signaling, the timing of gene activation relative to the initiation of FGF signaling in the embryo can be quite different. For example, sprouty2 is expressed at low levels maternally and the initial rise in levels of zygotic expression is detected at stage 9.5 which is 1 hour at 23°C after the activation of FGF8 expression. Subsequently, sprouty2 expression continues to closely follow that of FGF8 during late blastula stages (stage 9 to 10). The initiation of brachyury and Iro3 expression is somewhat delayed relative to initiation of FGF signaling, with a slight rise in expression by stage 9.5 and a more significant increase in expression by stage 10. Expression from Cdx4 and marginal coil are even more delayed and their expression levels only begin to rise steeply from stage 10 onwards.
Figure 5. Cluster analysis of genes down regulated in response to FGF inhibition.
(A) shows the temporal expression profiles of FGF8 (dashed line) and several known FGF target genes from blastula stage 8 until early neurula stage 14 (5 to 16 hours pf at 23°C). Profiles are derived from normalised microarray expression levels. Relative expression values are represented as percentages of the maximum expression value for each gene. (B) is a cluster dendrogram generated within BRB-ArrayTools for genes that are significantly down regulated in response to FGF inhibition i.e. positively regulated by FGF signaling. The red line indicates the level at which the dendrogram was cut, corresponding to a correlation coefficient of 0.85. (C) is a heat map of temporal expression for gene clusters positively regulated by FGF signalling. Only clusters containing ≥5 members are presented. Values at each time point from blastula stage 8 to early neurula stage 14 are derived from the mean of the expression levels for all the genes in each cluster. (D) shows the temporal expression profiles of FGF8 and gene clusters positively regulated by FGF signalling based upon the mean of the expression levels for all the genes in each cluster.
The differing dynamics of expression from known target genes indicate that there are different classes of FGF response genes. We investigated this further by undertaking a cluster analysis of FGF dependent genes based upon their temporal expression profiles from early blastula to early neurula stages. For the generation of clusters of genes positively regulated by FGF signaling a correlation value of p≥0.85 was used. The cluster analysis of genes negatively regulated by FGF is not presented because this group contains considerably fewer genes leading to multiple clusters containing single genes.
The genes in each of the clusters used for further analysis are shown in Table 4. The dendrogram generated during cluster analysis is shown in Figure 5B. Figure 5C shows a heat map of the relative expression of each of the generated clusters from stage 8 to stage 14. The expression profiles of the clusters positively regulated by FGF signaling, together with that of FGF8, are shown in Figure 5D. In keeping with our findings for individual known FGF targets, the initiation of expression from each of the clusters relative to the activation of FGF signaling varies considerably. For example, activation of expression from genes in clusters #11 and #9 rapidly follows the activation of FGF8 expression. However, at 23°C activation of expression from genes in cluster #1 is delayed 2–3 hours relative to that of FGF8. Activation of expression from genes in clusters #7 and #8 occurs at an intermediate time point with a 1–2 hour lag relative FGF8 and the activation of FGF signaling.
Table 4. Gene clusters positively regulated by FGF signaling.
| Cluster | Gene |
| #1 | Alkaline phosphatase |
| Cdx4 | |
| Cytochrome B561 | |
| FoxC1 | |
| Glycogen phosphorylase | |
| Gravin-like | |
| Lin28a homologue | |
| Marginal coil | |
| mitotic phosphoprotein 67 | |
| p75-lke fullback receptor | |
| Purine phosphorylase | |
| Putative methyltransferase | |
| Unknown | |
| SIP1 | |
| SMCT | |
| Uncharacterised protein C2orf32- xl.25136 | |
| Unknown-xl.15382 | |
| #7 | Dkk1 |
| FoxA4 | |
| FoxD5A | |
| Frzb1 | |
| NADH dehydrogenase sub-unit | |
| Pinhead | |
| Unknown-xl.3023 | |
| XSpr2 | |
| Zic3A | |
| #8 | Brachyury |
| Chordin | |
| Ephrin receptor A4 | |
| FoxD3A | |
| Glut1 transporter | |
| Paraxial protocadherin | |
| Retrotransposon protein 1a11 | |
| Unkown-Xl.18179 | |
| unkown-xl.5479 | |
| #9 | Egr1 |
| FoxA4 | |
| Goosecoid | |
| Prickle | |
| Related to DC-STAMP domain receptor | |
| #11 | ADMP |
| DUSP5 | |
| Esr5 | |
| Noggin | |
| Sprouty2 | |
| Wnt8 | |
| Xom |
Identification of a novel negative regulator of FGF signaling
The earliest activation of zygotic transcription from putative FGF target genes occurs between blastula stage 8.5 to 9. During this period a number of genes undergo >10-fold increase in expression. Amongst these rapid responders Dual specificity phosphatase 5 (DUSP5) has the highest fold increase during this period (>35). Xenopus laevis DUSP5 codes for a putative MAP kinase phosphatase with 61% peptide sequence identity to human DUSP5 (Figure S1).
Expression of another MAP kinase phosphatase, MKP1/XCL100 [33], is also significantly down regulated in response to FGF inhibition (Table 1 and Figure 3). Interestingly, another Xenopus MAP kinase phosphatase, MKP3, has been shown to inhibit FGF dependent mesoderm induction [21] and is implicated as a component of a negative feedback loop regulating FGF activity in the embryo [34]. Given the critical role that ERK/MAP kinase activity has in mediating responses to FGF signaling in the early Xenopus embryo we investigated the potential role of DUSP5 and MKP1 as negative regulators of FGF mediated MAP kinase signaling in early development.
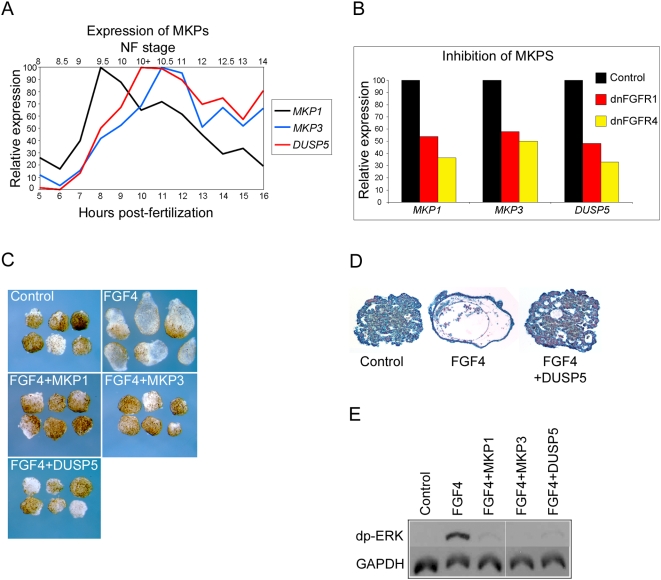
Figure 6A is a chart showing the temporal expression profiles of MKP1, MKP3 and DUSP5 as determined by microarray analysis. Expression of all three MKPs rises rapidly during late blastula stages, reaching maxima in early gastrula stages. Our data show that both MKP1 and DUSP5 are expressed in the circum-blastoporal region of the embryo during gastrula stages (Figures 2F). MKP3 is also expressed in this region in the gastrula [34]. In post-gastrula stages MKP1 is expressed in much of the open neural plate and subsequently in the anterior CNS and somites (Figure 2F). In contrast, at neurula stages DUSP5 is expressed in the posterior mesoderm around the closed blastopore and in a restricted domain in the anterior open neural plate. Later in development it is expressed in distinct domains in the anterior CNS, the tailbud and branchial arches.
Figure 6. MKPs and FGF signalling.
(A) shows the temporal expression profiles of MKP1, MKP3 and DUSP5 from blastula stage 8 until early neurula stage 14 (5 to 16 hours pf at 23°C). Profiles are derived from normalised microarray expression levels. Relative expression values are represented as percentages of the maximum expression value for each gene. (B) is a chart showing the expression of MKP1. MKP3 and DUSP5 in control embryos and embryos injected with 4 ng dnFGFR1 mRNA or 4 ng dnFGFR4 mRNA. Microarray derived expression values are based on the average of three biological replicates. Relative expression is calculated a percentage of the expression in control embryos. Experiments in (C, D and E) were carried out on animal cap explants removed from blastula stage 8 embryos. In all cases control explants are from uninjected embryos, FGF4 treatment was with 10 units of recombinant protein and mRNA injections were with 10 ng MKP1, MKP3 or DUSP5. (C) shows the morphology of animal cap explants at tailbud stage 41. (D) shows 10 µm histological sections of animal cap explants at stage 41. (E) is a Western blot showing levels of dp-ERK and the loading control GAPDH in animal cap explants at stage 10.5.
Figure 6B shows the degree to which expression of the three MKPs in the early gastrula is down regulated in response to FGF inhibition with dnFGFR1 and dnFGFR4. Consistent with this, expression of MKP1 and DUSP5 in the circum-blastoporal region during gastrula stages is dependent on FGF signaling (Figure 3A). Similar FGF dependence has been reported for MKP3 [34]. We also show that FGF signaling is sufficient for MKP1 and DUSP5 expression; treatment of animal cap explants with FGF proteins leads to marked up regulation of both genes (Figure 3C).
Treatment of animal cap explants from blastula stage embryos with FGF protein results elongation of the explant and development of vesicles containing a range of mesodermal tissues. Figure 6C shows that overexpression of MKP1, MKP3 or DUSP5 inhibits the formation of vesicles, indicating that like MKP3, MKP1 and DUSP5 are able to block FGF mediated formation of mesodermal tissues. We have confirmed this by examining histology of the differentiated tissues in FGF and DUSP5 treated explants (Figure 6D). Sections of untreated, control explants show the presence of a mass of atypical epidermis, whereas, FGF treated explants contain copious mesodermal tissue types, including a layer of smooth muscle (mesothelium) and loosely packed mesenchyme. Histology reveals the absence of mesodermal differentiation in response to FGF treatment when explants are over expressing DUSP5. Figure 6E shows that treatment of animal cap explants with FGF4 protein leads to phosphorylation and activation of ERK/MAP kinase and that over expression of either MKP1, MKP3 or DUSP5 in animal hemisphere explants dramatically inhibits FGF induced ERK phosphorylation.
Our data indicate that normal expression of MKP1 and DUSP5 in the early embryo requires a functional FGF pathway and that MKP3, MPK1 and DUSP5 have similar abilities to negatively regulate FGF signaling.
Discussion
FGF signaling in the early embryo
There is a wealth of evidence indicating that FGF signalling is involved in regulating multiple developmental processes before and during amphibian gastrulation [1], [35]. FGF dependent regulatory pathways have been shown to operate at different levels within the cell. For example, FGF signal transduction involving PKC and Ca++ modulates the planar cell polarity pathway necessary for the morphogenetic movements of gastrulation [36]. Another key function of FGF signalling during gastrula stages is as a regulator of gene transcription and it is this latter function which is the focus of the present study.
Identification of transcriptional targets of FGF signaling
Our study was designed to identify early transcriptional responses to FGF signalling. The reagents that we used to inhibit FGF signaling were dominant negative mutants of FGF receptor 1 (dnFGFR1) and FGF receptor 4a (dnFGFR4a) [24], [25]. Over expression of either dnFGFR1 or dnFGFR4a leads to very similar effects on gene expression at the start of gastrulation.
Using the strict criteria outlined, we have identified 67 genes which are down regulated and 16 genes which are up regulated in response to FGF signaling. The target validation undertaken indicates that these FGF targets lists are well supported and should provide the basis for further studies into FGF dependent transcriptional regulation.
Different waves of FGF dependent gene regulation
As part of this study we carried out a time course analysis of gene expression from mid-blastula to early neurula stages. Our cluster analysis based on these expression profiles reveals that activation of expression from FGF dependent genes occurs in a number of waves following the mid-blastula transition (MBT). Some genes are activated very rapidly following the MBT and closely follow the expression profile of FGF8. Expression of other genes, including Brachyury and Cdx4, which are known immediate early response genes, activated by FGF signaling in the absence of protein synthesis, occurs in later waves [26], [37]. At present it is unclear why some immediate early responses are more rapid than others. However, we speculate that the presence of identifiable clusters of FGF response genes indicates that similar upstream mechanisms are involved in regulating the expression of genes within the same cluster. The identification of such clusters of co-expressed transcriptional targets of FGF signaling will allow the analysis of these genes for shared regulatory elements required to drive their common modes of expression.
It is important to note that our analysis does not rule out the involvement of other signaling pathways in the regulation of the identified FGF target genes. Indeed this is to be expected, given that FGF signaling has been shown to interact with a number of pathways regulating gene expression in early development, including the activin/nodal and Wnt signaling pathways [38]–[41].
Patterns of FGF target gene expression
The initial zygotic expression of several FGF ligand genes, including FGF3, FGF4, FGF8 [42]–[44] and FGF20 (Figure S2) is restricted to the early mesoderm during late blastula stages. In keeping with this we find that many target genes positively regulated by FGF signaling are also expressed in the mesoderm. Our analysis of FGF dependent gene expression in later development shows that there is diversity in their later expression. However, we note that the posterior mesoderm, the paraxial mesoderm, the branchial arches and tailbud are common sites of expression for the FGF target genes. These are all known sites of FGF activity and ligand expression including FGF3, FGF4 and FGF8 [22], [42]–[44]. We show that FGF7 and FGF10 are also expressed in the posterior of the embryo and in the branchial arch region (Figure S2). An interesting example of the correspondence of target gene expression with sites of FGF signaling in later development is seen with a putative methyltransferase gene, which in tailbud stages is expressed exclusively in the developing otic vesicle in close proximity to FGF10 expression (Figure S2).
FGF regulation of organizer gene expression
Early studies of FGF function in early amphibian development focused on their potential role as regulators of gene expression in the ventro-lateral mesoderm [35], [45], [46]. However, more recent studies have provided evidence that FGF signaling is also required for the expression of genes within the dorsal organiser region of the amphibian embryo [47]–[49]. The large scale analysis of gene expression provided by our microarray experiments show that FGF signaling is required for the normal expression of a large number of organizer genes, including goosecoid, chordin, noggin, dkk1 and Frzb, and a number of genes, such as Sip1 and FoxD5, which are expressed in the dorsal neuroectoderm (See Table S14). Such a role is very much in keeping with the observed activity of MAP kinase signalling in the dorsal marginal zone and dorsal neuroectoderm, and supports the view that FGF signaling is required during late blastula and early gastrula stages for the establishment of both the Spemann organizer and the presumptive neuroectoderm.
There are also indications that FGF signaling is required to negatively regulate and therefore restrict gene expression in the dorsal region of the embryo. For example, the transcription factors Hes1b is up regulated in response to FGF inhibition. We note that Hes1b is expressed in the dorsal region of the gastrula but at some distance from the highest levels of FGF activity in the blastopore region [50]. It is also seems likely that negative regulation by FGF restricts XANF1 expression to the deeper layers of the organizer region in the early gastrula [51].
Gene function downstream of FGF signaling
A detailed description of the putative function for each of the identified target genes is beyond the scope of this discussion and we refer the reader to the extensive annotation and literature resources provided in Tables S2, S3, S4, S5, S6, S7, S8, S9, S10 and S11.
A number of the genes identified in the screen are of unknown function either because they are orthologs of genes with poorly characterized function or because we were unable to identify orthologous genes in the databases and may therefore represent novel Xenopus genes. However, analysis of the annotated genes identified in the screen reveals that a large proportion of the genes regulated by FGF signaling, are themselves also involved in gene regulation, either directly, as in the case of transcription factors, or via involvement in other signaling pathways. This indicates the key position of FGFs as upstream regulators of genetic pathways leading to germ layer specification during the late blastula to early gastrula stage of amphibian development. In addition, FGF signaling is required for the normal expression of genes such as Prickle, marginal coil and Ephrin receptor A4 (pagliaccio) which are involved in regulating cell movements and adhesion during gastrulation [52]–[54].
We also note that the purine phosphorylase and glycogen phosphorylase genes, which code for enzymes involved in nucleotide and carbohydrate metabolism respectively, are dependent on FGF signaling and are expressed in highly restricted, FGF associated domains in the early mesoderm. Further studies will be required to determine if these genes have previously unsuspected roles in the regulation of developmental mechanisms.
Previous studies have identified a number of FGF inducible inhibitors of the FGF signaling pathway, including the Sprouty genes, which we find are significantly down regulated in our screen [36], [55]. In the present study we identify the MAP kinase phosphatase genes MKP1 and DUSP5 as FGF targets which are activated rapidly following the mid-blastula transition and show that they are able to inhibit FGF signalling in Xenopus tissues. DUSP5 is a novel Xenopus MKP which is expressed in the early mesoderm and neural plate in a pattern which is remarkably similar to that of Xenopus FGF3 [44]. Similar correlation with sites of FGF activity has been reported for MKP3, which has also been shown to act as a feedback inhibitor of FGF signaling [34], [56].
The picture that emerges is that activation of FGF signaling induces the production of multiple inhibitors which act to moderate and limit the response to FGF signaling. A number of positive feedback mechanisms also impact on the FGF pathway, including the transcriptional activation the FLRT3 and brachyury genes [6], [57], [58]. FLRT3 is a transmembrane protein that potentiates FGF signal transduction and brachyury is a T-box transcription factor which has been shown to be a component of a positive feedback loop that leads to increased transcription of FGF ligand genes in the early mesoderm [5], [6], [58]. The presence of positive and negative feedback loops which modulate FGF signaling at multiple levels highlights the critical importance for fine tuning the overall levels of FGF signaling during development.
An interesting observation is that expression of the FoxD3 and Lin28 genes are down regulated in response to FGF inhibition. The FoxD3 transcription factor is linked to amphibian organizer function [59] but has also been implicated as a component of the pluripotency circuit of mammalian embryonic stem cells via regulation of the nanog gene [17], [18]. The Lin28 gene codes for an RNA binding protein, which together with the Oct4, Sox2 and Nanog genes, can convert somatic cells to an embryonic stem cell phenotype [14].
Previous studies have indicated that FGF signaling is required as a competence factor necessary for the response of embryonic amphibian cells to activin-like signals during development [39], [40], [60]. We also note a recent study which showed that FGF signaling in murine ES cells is necessary to allow differentiation into multiple lineages, including mesoderm [12]. These observations raise the intriguing possibility that FGF signaling, acting via downstream targets such as Lin28 and FoxD3, might have a general role in regulating pluripotency or the competence of embryonic cells to respond to signals which direct differentiation both during normal development and in culture.
Materials and Methods
Ethics statement
All animal work was undertaken under a licence from the UK Home Office.
Embryological methods
Embryos were staged according to Nieuwkoop and Faber [61]. Normal embryos were cultured in NAM/10. Injection of embryos with synthetic mRNAs was carried out in 33% NAM+5% ficoll (Sigma) at the 2 or 4-cell stage. Animal caps were explanted from embryos in 50% NAM Recombinant FGF4 [43] treatment was in 50% NAM+ 5 mg/ml BSA.
Identification of Xenopus tropicalis full length clones
Clones containing full length coding region of X.tropicalis MKP1 (DUSP1/XCL100) (accession number AL967533) and DUSP5 (accession number AL648624) were identified using the peptide sequence of X.laevis orthologues (accession numbers NM_001088684 and BJ067398 respectively) and BLASTP on the Sanger Institute X.tropicalis EST database (www.sanger.ac.uk/Projects/X_tropicalis/).
mRNA synthesis
Capped mRNA was synthesised using the SP6 Megascript kit (Ambion) and a modified protocol using a 1∶10 ratio of GTP to m7G(5′)Gppp(5′)G cap. All cDNAs used for mRNA were in either pSP64t, Cs2+ or CS107 and were transcribed using SP6 polymerase. The dominant negative X.laevis FGFRI (dnFGFR1) plasmid was a gift from Enrique Amaya [24]. The dominant negative X.laevis FGFR4a (dnFGFR4) plasmid was a gift from Harumasa Okamoto [25]. The X.laevis MKP3 (DUSP6/X17C) plasmid was a gift from Bob Old [62].
In situ hybridisation
Embryos were fixed in MEMFA and in situ hybridizations were carried out as per [63] with the modifications described in [64]. Details for in situ probe plasmids are shown in Table S15. The sources of the plasmids, including Geneservice (www.geneservice.co.uk) and the NIBB/NIG/NBRP X. laevis EST project (xenopus.nibb.ac.jp) are indicated.
RNAase protection analysis
RNA extraction and RNAase protection analysis were carried out according to the methods of Pownall et al. (1996). Data relating to RNAase protection plasmids are shown in Table S16.
Whole-mount immunohistochemistry and Western blotting
dp-ERK immunohistochemistry was carried out according the methods of [22]. Western blot samples were homogenized in PhosphoSafe (Novagen) extraction buffer per embryo. Following centrifugation supernatants were subjected to SDS-PAGE. Gels were blotted onto Immobilon-P (Millipore) transfer membrane. Antibody concentrations were mouse anti-dp-ERK (Sigma), 1∶8000, anti-phosphoSmad1/5/8 (NEB), 1∶500, anti-GAPDH (HyTest), 1∶3000. 1∶8,000, anti-GAPDH (HyTest), 1∶1,000,000, anti-mouse POD (Amersham), 1∶3000 and anti-rabbit POD (Amersham), 1;2000. Peroxidase activity was detected using BM chemiluminescence blotting substrate (Roche) and Hyperfilm (Amersham).
Embryos for microarray experiments
Embryos were injected with 4 ng of dnFGFR1 or dnFGFR4 mRNA and were collected with sibling controls at early gastrula stage 10.5 (11 hours post-fertilization at 23°C). In order to enable statistical analysis three biological replicates were produced by in vitro fertilization from different pairs of male and female frogs. Each replicate set comprised control, dnFGFR1 injected and dnFGFR4 injected embryos. Before processing for microarray analysis each replicate set was assessed for the effectiveness of FGF signaling inhibition by analysing levels of dp-ERK and levels of the known FGF targets Xbra, Cdx4 and myoD expression in sibling embryos.
For the early developmental timecourse embryos from a single mating were cultured at 23°C and collected at 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15 and 16 hours post fertilization (NF stage 8, 8.5, 9, 9.5, 10, 10+, 10.5, 11, 12, 12.5, 13 and 14) for microarray analysis. Sibling embryos were also collected at the same time points for Western blot analysis of dp-ERK.
Preparation of total RNA for microarray analysis
Batches of 10 embryos were extracted in Tri-reagent according manufacturer's protocol (Sigma). RNA was precipitated using isopropanol and cleaned up using the Qiagen RNeasy kit followed by a lithium chloride precipitation. Quality of RNA was assessed using the Agilent 2100 Bioanalyzer.
Preparation of labelled cRNA and chip hybridization
2 µg of total RNA was processed for the microarray by using the Affymetrix GeneChip one-cycle target labelling kit (Affymetrix) according to the manufacturer's recommended protocols. The quality and quantity of the resulting biotinylated cRNA was determined by using NanoDrop ND 1000 (NanoDrop Technologies) and Agilent Bioanalyzer 2100 (Agilent Technologies). Biotin-labelled cRNA samples were fragmented randomly to 35–200 bp at 94°C in fragmentation buffer (Affymetrix) according to the manufacturer's recommended protocols and aliquots of the fragmented cRNA were run on the Agilent 2100 Bioanalyzer to assess the quality of the generated cRNA.
Biotin-labeled fragmented cRNA samples were combined with hybridization buffer containing hybridization Control cRNA and Control Oligo B2 (Affymetrix), before hybridization to GeneChip® Xenopus laevis Genome Array for 16 h at 45°C. The arrays were washed, stained, and scanned using the Affymetrix Model 450 Fluidics Station and Affymetrix Model 3000 scanner using the manufacturer's recommended protocols.
Microarray data analysis
Affymetrix microarray experiments were conducted in accordance with the MIAME standards requirements [65]. Raw data processing was performed by using the Affymetrix GCOS 1.2 software. After hybridization and scanning, probe cell intensities were calculated and summarized for the respective probe sets by means of the MAS5 algorithm. To compare the expression values of the genes from chip to chip, global scaling was performed, which resulted in the normalization of the trimmed mean of each chip to a target intensity (TGT value) of 500 as per manufacturers documentation. Each sample and hybridization underwent a quality control evaluation checking for adequate scaling factors (1–3 for all samples), percentage of probe sets reliably detected (between 40–60% present call), and optimal 3′/5′ hybridization ratios for the housekeeping genes (e.g., GAPDH), poly(A) spike-in controls, and the prokaryotic controls (bioB, bioC, bioD and cre).
Data were imported into BRB ArrayTools software version 3.6.0 (http://linus.nci.nih.gov/BRB-ArrayTools.html). Imported array data were filtered using the following criteria.
Spot filters- Threshold minimum value if spot intensity below 5.
Normalization- Normalize (center) each array using median over entire array.
Exclude a gene under any of the following conditions- Percent of data missing or filtered out exceeds 50% Percent of absent (i.e., Detection Call = A) data exceeds 50%
Scatterplots were generated using the phenotype averages tool of BRB ArrayTools. Gene lists of FGF targets were generated using the BRB between groups of arrays class comparison tool (unpaired, two sample t-test with random variance model and nominal significance level p = 0.01). An additional filter excluded genes with less than 2-fold difference from controls.
Temporal expression profiles for a given gene were generated in Microsoft Excel by plotting relative expression at each time point as a percentage of the maximum expression level within the time course. Cluster analysis was undertaken using the BRB gene cluster analysis tool (complete linkage and centred correlation). The Affymetrix Cel files for all microarray experiments are available at EMBL ArrayExpress, accession numbers E-MEXP-2058 and E-MEXP-2059.
Target gene annotation
Target gene annotation was accomplished using a combination of existing Affymetrix Gene array annotation and BLAST searching of target sequences against Genbank, Swiss-prot (www.ncbi.nlm.nih.gov/BLAST/) and NIBB/NIG/NBRP Xenopus laevis EST project (xenopus.nibb.ac.jp) databases.
Supporting Information
Alignment of amphibian and human DUSP5 peptide sequences. Alignment of the peptide sequences for human and Xenopus tropicalis DUSP5 produced by the Clustal W method. Identical residues are boxed in red
(0.15 MB TIF)
Expression of FGF7, FGF10 and FGF20 during amphibian development. In situ hybridisations for showing the expression of FGF7 (A, B and C), FGF10 (D, E and F) and FGF20 (G) at the indicated stages. In situ hybridizations for FGF7 and FGF10 are on Xenopus tropicalis embryos. In situ hybridization for FGF20 is on a Xenopus laevis embryo. (A, C D, E and F) are lateral views with anterior to the left and dorsal to the top. (B) is a posterior view with dorsal to the top. (G) is a vegetal view with dorsal to the top. (A and B) shows expression in the posterior mesoderm and ectoderm (white arrow) around the closed blastopore (bp). Expression is also detected in the anterior endoderm (end). (C) shows expression in the branchial arch region (bra). (D and E) show expression in a domain juxtaposed to the anterior of the otic vesicle (otv, black arrow) and in the branchial arch (bra) region. (F) shows expression in the tailbud (tlb). (G) shows expression in the circumblastoporal region in a distinct dorsal to ventral gradient. The dorsal blastopore lip (dbl) is indicated.
(1.81 MB TIF)
Genes differentially regulated in dnFGFR1 versus dnFGFR4 injected embryos
(0.03 MB DOC)
Genes positively regulated by FGF signaling involved in transcriptional regulation
(0.07 MB DOC)
Genes positively regulated by FGF signaling involved in cell signalling
(0.08 MB DOC)
Genes positively regulated by FGF signaling involved in metabolism
(0.04 MB DOC)
Genes positively regulated by FGF signaling of other known function
(0.05 MB DOC)
Genes positively regulated by FGF signaling of unknown function
(0.04 MB DOC)
Genes negatively regulated by FGF signaling involved in transcriptional regulation
(0.03 MB DOC)
Genes negatively regulated by FGF signaling involved in cell signalling
(0.03 MB DOC)
Genes negatively regulated by FGF signaling involved in metabolism
(0.03 MB DOC)
Genes negatively regulated by FGF signaling of other known function
(0.03 MB DOC)
Genes negatively regulated by FGF signaling of unknown function
(0.03 MB DOC)
GO terms for genes positively regulated by FGF signalling
(0.10 MB DOC)
GO terms for genes negatively regulated by FGF signalling
(0.04 MB DOC)
Expression of genes positively regulated by FGF signalling
(0.38 MB DOC)
In situ clone data
(0.06 MB DOC)
RNAase protection probe data
(0.04 MB DOC)
Acknowledgments
We would like to thank Naveed Aziz from the York Technology Facility for his help with processing of microarrays and Julie Affleck for technical assistance. This work was funded by a BBSRC project grant to HVI.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Work funded by BBSRC grant BB/D010039/1. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bottcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26:63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- 2.Griffin K, Patient R, Holder N. Analysis of FGF function in normal and no tail zebrafish embryos reveals separate mechanisms for formation of the trunk and the tail. Development. 1995;121:2983–2994. doi: 10.1242/dev.121.9.2983. [DOI] [PubMed] [Google Scholar]
- 3.Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1:37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 4.Mathieu J, Griffin K, Herbomel P, Dickmeis T, Strahle U, et al. Nodal and Fgf pathways interact through a positive regulatory loop and synergize to maintain mesodermal cell populations. Development. 2004;131:629–641. doi: 10.1242/dev.00964. [DOI] [PubMed] [Google Scholar]
- 5.Amaya E, Stein PA, Musci TJ, Kirschner MW. FGF signalling in the early specification of mesoderm in Xenopus. development. 1993;118:477–487. doi: 10.1242/dev.118.2.477. [DOI] [PubMed] [Google Scholar]
- 6.Isaacs HV, Pownall ME, Slack JMW. eFGF regulates Xbra expression during Xenopus gastrulation. EMBO Journal. 1994;13:4469–4481. doi: 10.1002/j.1460-2075.1994.tb06769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Dormann D, Munsterberg AE, Weijer CJ. Cell movement patterns during gastrulation in the chick are controlled by positive and negative chemotaxis mediated by FGF4 and FGF8. Dev Cell. 2002;3:425–437. doi: 10.1016/s1534-5807(02)00256-3. [DOI] [PubMed] [Google Scholar]
- 8.Sheng G, dos Reis M, Stern CD. Churchill, a zinc finger transcriptional activator, regulates the transition between gastrulation and neurulation. Cell. 2003;115:603–613. doi: 10.1016/s0092-8674(03)00927-9. [DOI] [PubMed] [Google Scholar]
- 9.Pera EM, Ikeda A, Eivers E, De Robertis EM. Integration of IGF, FGF, and anti-BMP signals via Smad1 phosphorylation in neural induction. Genes Dev. 2003;17:3023–3028. doi: 10.1101/gad.1153603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fletcher RB, Baker JC, Harland RM. FGF8 spliceforms mediate early mesoderm and posterior neural tissue formation in Xenopus. Development. 2006;133:1703–1714. doi: 10.1242/dev.02342. [DOI] [PubMed] [Google Scholar]
- 11.Ribisi S, Jr, Mariani FV, Aamar E, Lamb TM, Frank D, et al. Ras-mediated FGF signaling is required for the formation of posterior but not anterior neural tissue in Xenopus laevis. Dev Biol. 2000;227:183–196. doi: 10.1006/dbio.2000.9889. [DOI] [PubMed] [Google Scholar]
- 12.Kunath T, Saba-El-Leil MK, Almousailleakh M, Wray J, Meloche S, et al. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. development. 2007;134:2895–2902. doi: 10.1242/dev.02880. [DOI] [PubMed] [Google Scholar]
- 13.Stavridis MP, Lunn JS, Collins BJ, Storey KG. A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development. 2007;134:2889–2894. doi: 10.1242/dev.02858. [DOI] [PubMed] [Google Scholar]
- 14.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 15.Bendall SC, Stewart MH, Menendez P, George D, Vijayaragavan K, et al. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature. 2007;448:1015–1021. doi: 10.1038/nature06027. [DOI] [PubMed] [Google Scholar]
- 16.Levenstein ME, Ludwig TE, Xu RH, Llanas RA, VanDenHeuvel-Kramer K, et al. Basic fibroblast growth factor support of human embryonic stem cell self-renewal. Stem Cells. 2006;24:568–574. doi: 10.1634/stemcells.2005-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Labosky PA. Regulation of embryonic stem cell self-renewal and pluripotency by Foxd3. Stem Cells. 2008;26:2475–2484. doi: 10.1634/stemcells.2008-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 19.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaBonne C, Burke B, Whitman M. Role of MAP kinase in mesoderm induction and axial patterning during Xenopus development. Development. 1995;121:1475–1486. doi: 10.1242/dev.121.5.1475. [DOI] [PubMed] [Google Scholar]
- 21.Umbhauer M, Marshall CJ, Mason CS, Old RW, Smith JC. Mesoderm induction in Xenopus caused by activation of MAP kinase. Nature. 1995;376:58–62. doi: 10.1038/376058a0. [DOI] [PubMed] [Google Scholar]
- 22.Christen B, Slack J. Spatial response to fibroblast growth factor signalling in Xenopus embryos. Development. 1999;126:119–125. doi: 10.1242/dev.126.1.119. [DOI] [PubMed] [Google Scholar]
- 23.Newport J, Kirschner M. A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell. 1982;30:675–686. doi: 10.1016/0092-8674(82)90272-0. [DOI] [PubMed] [Google Scholar]
- 24.Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF receptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- 25.Hongo I, Kengaku M, Okamoto H. FGF signaling and the anterior neural induction in Xenopus. developmental biology. 1999;216:561–581. doi: 10.1006/dbio.1999.9515. [DOI] [PubMed] [Google Scholar]
- 26.Smith JC, Price BM, Green JB, Weigel D, Herrmann BG. Expression of a Xenopus homolog of Brachyury (T) is an immediate-early response to mesoderm induction. Cell. 1991;67:79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]
- 27.Keenan ID, Sharrard RM, Isaacs HV. FGF signal transduction and the regulation of Cdx gene expression. Dev Biol. 2006;299:478–488. doi: 10.1016/j.ydbio.2006.08.040. [DOI] [PubMed] [Google Scholar]
- 28.Fisher ME, Isaacs HV, Pownall ME. eFGF is required for activation of XmyoD expression in the myogenic cell lineage of Xenopus laevis. Development. 2002;129:1307–1315. doi: 10.1242/dev.129.6.1307. [DOI] [PubMed] [Google Scholar]
- 29.Klingbeil P, Frazzetto G, Bouwmeester T. Xwig1, a novel putative endoplasmic reticulum protein expressed during epithelial morphogenesis and in response to embryonic wounding. Int J Dev Biol. 2001;45:379–385. [PubMed] [Google Scholar]
- 30.Levine B, Jean-Francois M, Bernardi F, Gargiulo G, Dobens L. Notch signaling links interactions between the C/EBP homolog slow border cells and the GILZ homolog bunched during cell migration. Dev Biol. 2007;305:217–231. doi: 10.1016/j.ydbio.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Smith WC, Harland RM. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70:829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- 32.Sasai Y, Lu B, Steinbeisser H, Geissert D, Gont LK, et al. Xenopus chordin: a novel dorsalizing factor activated by organizer-specific homeobox genes. Cell. 1994;79:779–790. doi: 10.1016/0092-8674(94)90068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis T, Groom LA, Sneddon AA, Smythe C, Keyse SM. XCL100, an inducible nuclear MAP kinase phosphatase from Xenopus laevis: its role in MAP kinase inactivation in differentiated cells and its expression during early development. J Cell Sci. 1995;108(Pt 8):2885–2896. doi: 10.1242/jcs.108.8.2885. [DOI] [PubMed] [Google Scholar]
- 34.Gomez AR, Lopez-Varea A, Molnar C, de la Calle-Mustienes E, Ruiz-Gomez M, et al. Conserved cross-interactions in Drosophila and Xenopus between Ras/MAPK signaling and the dual-specificity phosphatase MKP3. Dev Dyn. 2005;232:695–708. doi: 10.1002/dvdy.20227. [DOI] [PubMed] [Google Scholar]
- 35.Isaacs HV. New perspectives on the role of the fibroblast growth factor family in amphibian development. Cell Mol Life Sci. 1997;53:350–361. doi: 10.1007/PL00000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sivak JM, Petersen LF, Amaya E. FGF Signal Interpretation Is Directed by Sprouty and Spred Proteins during Mesoderm Formation. Dev Cell. 2005;8:689–701. doi: 10.1016/j.devcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 37.Isaacs HV, Pownall ME, Slack JM. Regulation of Hox gene expression and posterior development by the Xenopus caudal homologue Xcad3. EMBO J. 1998;17:3413–3427. doi: 10.1093/emboj/17.12.3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christian JL, Olson DJ, Moon RT. Xwnt-8 modifies the character of mesoderm induced by bFGF in isolated Xenopus ectoderm. EMBO J. 1992;11:33–41. doi: 10.1002/j.1460-2075.1992.tb05024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornell RA, Kimelman D. Activin-mediated mesoderm induction requires FGF. Development. 1994;120:453–462. doi: 10.1242/dev.120.2.453. [DOI] [PubMed] [Google Scholar]
- 40.LaBonne C, Whitman M. Mesoderm induction by activin requires FGF-mediated intracellular signals. Development. 1994;120:463–472. doi: 10.1242/dev.120.2.463. [DOI] [PubMed] [Google Scholar]
- 41.Burks PJ, Isaacs HV, Pownall ME. FGF signaling modulates transcriptional repression by Xenopus groucho-related-4. Biol Cell. 2008 doi: 10.1042/BC20080136. [DOI] [PubMed] [Google Scholar]
- 42.Christen B, Slack JM. FGF-8 is associated with anteroposterior patterning and limb regeneration in Xenopus. Dev Biol. 1997;192:455–466. doi: 10.1006/dbio.1997.8732. [DOI] [PubMed] [Google Scholar]
- 43.Isaacs HV, Tannahill D, Slack JMW. Expression of a novel FGF in the Xenopus embryo. A new candidate inducing factor for mesoderm formation and anteroposterior specification. Development. 1992;114:711–720. doi: 10.1242/dev.114.3.711. [DOI] [PubMed] [Google Scholar]
- 44.Lombardo A, Isaacs HV, Slack JM. Expression and functions of FGF-3 in Xenopus development. Int J Dev Biol. 1998;42:1101–1107. [PubMed] [Google Scholar]
- 45.Kimelman D, Kirschner M. Synergistic induction of mesoderm by FGF and TGF-beta and the identification of an mRNA coding for FGF in the early Xenopus embryo. Cell. 1987;51:869–877. doi: 10.1016/0092-8674(87)90110-3. [DOI] [PubMed] [Google Scholar]
- 46.Slack JM, Darlington BG, Heath JK, Godsave SF. Mesoderm induction in early Xenopus embryos by heparin-binding growth factors. Nature. 1987;326:197–200. doi: 10.1038/326197a0. [DOI] [PubMed] [Google Scholar]
- 47.Delaune E, Lemaire P, Kodjabachian L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development. 2005;132:299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- 48.Fletcher RB, Harland RM. The role of FGF signaling in the establishment and maintenance of mesodermal gene expression in Xenopus. Dev Dyn. 2008;237:1243–1254. doi: 10.1002/dvdy.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitchell TS, Sheets MD. The FGFR pathway is required for the trunk-inducing functions of Spemann's organizer. Dev Biol. 2001;237:295–305. doi: 10.1006/dbio.2001.0385. [DOI] [PubMed] [Google Scholar]
- 50.Shinga J, Itoh M, Shiokawa K, Taira S, Taira M. Early patterning of the prospective midbrain-hindbrain boundary by the HES-related gene XHR1 in Xenopus embryos. Mech Dev. 2001;109:225–239. doi: 10.1016/s0925-4773(01)00528-7. [DOI] [PubMed] [Google Scholar]
- 51.Zaraisky AG, Ecochard V, Kazanskaya OV, Lukyanov SA, Fesenko IV, et al. The homeobox-containing gene XANF-1 may control development of the Spemann organizer. Development. 1995;121:3839–3847. doi: 10.1242/dev.121.11.3839. [DOI] [PubMed] [Google Scholar]
- 52.Frazzetto G, Klingbeil P, Bouwmeester T. Xenopus marginal coil (Xmc), a novel FGF inducible cytosolic coiled-coil protein regulating gastrulation movements. Mech Dev. 2002;113:3–14. doi: 10.1016/s0925-4773(01)00664-5. [DOI] [PubMed] [Google Scholar]
- 53.Takeuchi M, Nakabayashi J, Sakaguchi T, Yamamoto TS, Takahashi H, et al. The prickle-related gene in vertebrates is essential for gastrulation cell movements. Curr Biol. 2003;13:674–679. doi: 10.1016/s0960-9822(03)00245-8. [DOI] [PubMed] [Google Scholar]
- 54.Winning RS, Scales JB, Sargent TD. Disruption of cell adhesion in Xenopus embryos by Pagliaccio, an Eph-class receptor tyrosine kinase. Dev Biol. 1996;179:309–319. doi: 10.1006/dbio.1996.0262. [DOI] [PubMed] [Google Scholar]
- 55.Nutt SL, Dingwell KS, Holt CE, Amaya E. Xenopus Sprouty2 inhibits FGF-mediated gastrulation movements but does not affect mesoderm induction and patterning. Genes Dev. 2001;15:1152–1166. doi: 10.1101/gad.191301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mason C, Lake M, Nebreda A, Old R. A novel MAP kinase phosphatase is localised in the branchial arch region and tail tip of Xenopus embryos and is inducible by retinoic acid. mechanisms of development. 1996;55:133–144. doi: 10.1016/0925-4773(96)00495-9. [DOI] [PubMed] [Google Scholar]
- 57.Bottcher RT, Pollet N, Delius H, Niehrs C. The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nat Cell Biol. 2004;6:38–44. doi: 10.1038/ncb1082. [DOI] [PubMed] [Google Scholar]
- 58.Schulte-Merker S, Smith JC. Mesoderm formation in response to Brachyury requires FGF signalling. Curr Biol. 1995;5:62–67. doi: 10.1016/s0960-9822(95)00017-0. [DOI] [PubMed] [Google Scholar]
- 59.Steiner AB, Engleka MJ, Lu Q, Piwarzyk EC, Yaklichkin S, et al. FoxD3 regulation of Nodal in the Spemann organizer is essential for Xenopus dorsal mesoderm development. Development. 2006;133:4827–4838. doi: 10.1242/dev.02663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cornell RA, Musci TJ, Kimelman D. FGF is a prospective competence factor for early activin-type signals in Xenopus mesoderm induction. Development. 1995;121:2429–2437. doi: 10.1242/dev.121.8.2429. [DOI] [PubMed] [Google Scholar]
- 61.Nieuwkoop PD, Faber J. Normal Tableof Xenopus laevis (Daudin) Amsterdam: North-Holland; 1967. [Google Scholar]
- 62.Mason C, Lake M, Nebreda A, Old R. A novel MAP kinase phosphatase is localised in the branchial arch region and tail tip of Xenopus embryos and is inducible by retinoic acid. Mech Dev. 1996;55:133–144. doi: 10.1016/0925-4773(96)00495-9. [DOI] [PubMed] [Google Scholar]
- 63.Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- 64.Pownall ME, Tucker AS, Slack JM, Isaacs HV. eFGF, Xcad3 and Hox genes form a molecular pathway that establishes the anteroposterior axis in Xenopus. Development. 1996;122:3881–3892. doi: 10.1242/dev.122.12.3881. [DOI] [PubMed] [Google Scholar]
- 65.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignment of amphibian and human DUSP5 peptide sequences. Alignment of the peptide sequences for human and Xenopus tropicalis DUSP5 produced by the Clustal W method. Identical residues are boxed in red
(0.15 MB TIF)
Expression of FGF7, FGF10 and FGF20 during amphibian development. In situ hybridisations for showing the expression of FGF7 (A, B and C), FGF10 (D, E and F) and FGF20 (G) at the indicated stages. In situ hybridizations for FGF7 and FGF10 are on Xenopus tropicalis embryos. In situ hybridization for FGF20 is on a Xenopus laevis embryo. (A, C D, E and F) are lateral views with anterior to the left and dorsal to the top. (B) is a posterior view with dorsal to the top. (G) is a vegetal view with dorsal to the top. (A and B) shows expression in the posterior mesoderm and ectoderm (white arrow) around the closed blastopore (bp). Expression is also detected in the anterior endoderm (end). (C) shows expression in the branchial arch region (bra). (D and E) show expression in a domain juxtaposed to the anterior of the otic vesicle (otv, black arrow) and in the branchial arch (bra) region. (F) shows expression in the tailbud (tlb). (G) shows expression in the circumblastoporal region in a distinct dorsal to ventral gradient. The dorsal blastopore lip (dbl) is indicated.
(1.81 MB TIF)
Genes differentially regulated in dnFGFR1 versus dnFGFR4 injected embryos
(0.03 MB DOC)
Genes positively regulated by FGF signaling involved in transcriptional regulation
(0.07 MB DOC)
Genes positively regulated by FGF signaling involved in cell signalling
(0.08 MB DOC)
Genes positively regulated by FGF signaling involved in metabolism
(0.04 MB DOC)
Genes positively regulated by FGF signaling of other known function
(0.05 MB DOC)
Genes positively regulated by FGF signaling of unknown function
(0.04 MB DOC)
Genes negatively regulated by FGF signaling involved in transcriptional regulation
(0.03 MB DOC)
Genes negatively regulated by FGF signaling involved in cell signalling
(0.03 MB DOC)
Genes negatively regulated by FGF signaling involved in metabolism
(0.03 MB DOC)
Genes negatively regulated by FGF signaling of other known function
(0.03 MB DOC)
Genes negatively regulated by FGF signaling of unknown function
(0.03 MB DOC)
GO terms for genes positively regulated by FGF signalling
(0.10 MB DOC)
GO terms for genes negatively regulated by FGF signalling
(0.04 MB DOC)
Expression of genes positively regulated by FGF signalling
(0.38 MB DOC)
In situ clone data
(0.06 MB DOC)
RNAase protection probe data
(0.04 MB DOC)