The molecular mechanisms regulating expression of the CIITA gene have been the subject of intense research by immunologists for more than a decade because it encodes the ‘class II transactivator’, a transcriptional coactivator that functions as the master regulator of major histocompatibility complex (MHC) class II gene expression1-3. The results reported in this issue by Ni et al.4 bring to light a novel and unanticipated level of complexity in the regulation of interferon gamma (IFN-γ)-induced CIITA expression by demonstrating that it involves a series of five distant enhancers that promote the formation of a complex and dynamic three dimensional chromatin structure.
It is now well established that transcription of the CIITA gene is regulated by multiple promoters2,3,5. Four promoters – referred to as pI, pII, pIII and pIV - have been defined in the human CIITA gene (Fig. 1). Each of these promoters is responsible for distinct tissue specific expression modes of CIITA2,3. Promoters pI and pIII are responsible for myeloid and lymphoid specific expression patterns, respectively6. A specific role for pII is not currently known. Promoter pIV is the major IFN-γ responsive promoter functioning in non-hematopoietic cells7. The finding that triggered the studies of Ni et al.4 was that reporter gene constructs driven by the known regulatory sequences of pIV did not exhibit a dependency on BRG1, an ATP-dependent chromatin remodeling factor previously shown to be required for IFN-γ induced expression of the endogenous CIITA gene8,9. This prompted a search for distant BRG1-dependent enhancers required for IFN-γ-induced activation of pIV.
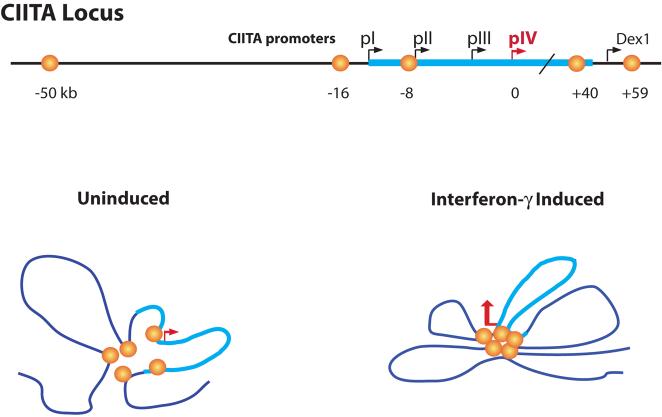
Figure 1.
The CIITA locus is depicted with its four promoters termed pI, pII, pIII, and pIV. The thicker cyan colored line represents the entire CIITA gene from pI through to its last exon. Orange globes represent the relative locations of the newly discovered BRG1-dependent distal enhancers that collectively regulate IFN-γ-induced expression through pIV. In the uninduced state, weak or transient interactions between several of the enhancers and pIV occur. Following IFN-γ induction, the interactions between the enhancers, and between the enhancers and pIV, are stabilized to form a discrete structure that is required for CIITA expression. What is dex1? Please identify.
To pinpoint candidate enhancers the authors first performed ChIP (chromatin immunoprecipitation) and ChIP-chip (ChIP coupled to microarray) experiments4. These approaches were used to identify regions in the CIITA locus that exhibit IFN-γ-induced histone modifications associated with gene activation (acetylation of histones H3 and H4, methylation of lysine 4 of H3), are associated with a histone acetyltransferase (p300) implicated in IFN-γ-induced CIITA expression, and/or are bound by transcription factors (STAT1 and IRF1) known to mediate IFNγ-induced CIITA expression10. Five candidate enhancers spread out over a 110 kb region encompassing the entire CIITA gene were identified (Fig. 1). To show that these five regions indeed function as transcriptional enhancers required for IFN-γ-induced CIITA expression, an impressive series of functional experiments was performed using an elegant and sophisticated reporter gene system that relied on the engineering and transfection of BAC (bacterial artificial chromosome) constructs containing 194 kb of genomic DNA from the CIITA locus. Finally, the 3C (chromatin conformation capture) approach was used to demonstrate that the newly identified distal enhancers engage in multiple long-distance interactions with each other and with pIV to generate a complex and dynamic three-dimensional chromatin structure. The results suggest that in non-induced cells, or in the absence of BRG1, the long-distance chromatin interactions are weak, transient or present in only a fraction of the cells (Fig. 1). However, the presence of BRG1 and/or exposure of the cells to IFN-γ promotes stabilization and remodeling of the three dimensional structure (Fig. 1). Remodeling of the chromatin structure appears to be a requirement for CIITA expression.
Until now, the individual promoters of the CIITA gene were believed to function as independent units and have therefore generally been depicted as separate boxes arrayed along a linear DNA molecule (as shown in the top line of Fig. 1). However, the results reported by Ni et al.4 demonstrate that this one dimensional and promoter-centric view of the regulatory region of the CIITA gene needs to be revised. It is evident from their work that a full understanding of the complexity of CIITA gene regulation must integrate the establishment and function of long-distance and dynamic chromatin interactions between regulatory elements that can be situated more than 100 kb apart. This view is becoming more common with genes that, like CIITA, were once thought to be regulated exclusively by their promoter proximal regulatory regions11,12. Recent examples include the MHC class II genes that CIITA regulates, where CIITA itself participates in long-range interactions with insulator binding factors13.
We can anticipate that future research will concentrate on the establishment and function of chromatin structures at the CIITA locus. In the case of IFN-γ-induced CIITA expression, a number of important questions remain. At the top of the list are questions involving the precise role of BRG1; does it move the nucleosomes to allow STAT1 and IRF1 binding, or are other proteins involved? We currently do not know the identity of the proteins that mediate the establishment of the long distance chromatin loops between the distal enhancers and pIV. While these long-distant loops are clearly formed and required, it is not clear why the proximal promoter regulatory elements at pIV are not sufficient for activation. One possibility suggested by the data is that repressive mechanisms are at work to ensure that the region cannot be activated without the aid of the other enhancer elements. Finally, it will be imperative to determine whether the lessons learned from IFN-γ-induced cells are also relevant to the regulation of CIITA in key professional antigen presenting cells - including dendritic cells, macrophages and B cells - and in cortical and medulary thymic epithelial cells. Do three-dimensional chromatin structures also play a critical role in regulating CIITA expression in antigen presenting cells and medulary thymic epithelial cells? Are the structures formed in these cells similar to or different from those implicated in IFN-γ-induced cells? Are the same or different distal enhancers involved in different cell types? The answers to these questions are bound to provide fascinating new insights into the molecular mechanisms that regulate CIITA expression and thereby control MHC class II-mediated antigen presentation. Progress in these directions will contribute to propelling the regulation of CIITA expression to the forefront among the mammalian gene regulatory systems that have been dissected genetically and biochemically in the greatest detail.
References
- 1.Steimle V, Otten LA, Zufferey M, Mach B. Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 2.Wright KL, Ting JP. Trends Immunol. 2006;27:405–412. doi: 10.1016/j.it.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Leibundgut-Landmann S, et al. Eur J Immunol. 2004;34:1513–1525. doi: 10.1002/eji.200424964. [DOI] [PubMed] [Google Scholar]
- 4.Ni Z, Abou El Hassan M, Xu Z, Yu T, Bremner R. Nat. Immunol. 9:XXX–XXX. doi: 10.1038/ni.1619. [DOI] [PubMed] [Google Scholar]
- 5.Muhlethaler-Mottet A, Otten LA, Steimle V, Mach B. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LeibundGut-Landmann S, Waldburger JM, Reis e Sousa C, Acha-Orbea H, Reith W. Nat Immunol. 2004;5:899–908. doi: 10.1038/ni1109. [DOI] [PubMed] [Google Scholar]
- 7.Waldburger JM, Suter T, Fontana A, Acha-Orbea H, Reith W. J.Exp.Med. 2001;19:393–406. doi: 10.1084/jem.194.4.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mudhasani R, Fontes JD. Mol Cell Biol. 2002;22:5019–5026. doi: 10.1128/MCB.22.14.5019-5026.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pattenden SG, Klose R, Karaskov E, Bremner R. EMBO J. 2002;21:1978–1986. doi: 10.1093/emboj/21.8.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris AC, Beresford GW, Mooney MR, Boss JM. Mol.Cell.Biol. 2002;22:4781–4791. doi: 10.1128/MCB.22.13.4781-4791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Laat W, Grosveld F. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 12.Schoenborn JR, et al. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majumder P, Gomez JA, Chadwick BP, Boss JM. J Exp Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]