Summary
We investigated recruitment of the yeast mRNA export factor Yra1 to the transcription elongation complex (TEC). Previously, the Sub2 helicase subunit of TREX was proposed to recruit Yra1. We report that Sub2 is dispensable for Yra1 recruitment, but the cleavage/polyadenylation factor, CF1A, is required. Yra1 binds directly to the Zn-finger/Clp1 region of Pcf11, the pol II CTD binding subunit of CF1A, and this interaction is conserved between their human homologues. Tethering of Pcf11 to nascent mRNA is sufficient to enhance Yra1 recruitment. Interaction with Pcf11 can therefore explain Yra1 binding to the TEC independently of Sub2. We propose that after initially binding to Pcf11, Yra1 is transferred to Sub2. Consistent with this idea, Pcf11 binds the same regions of Yra1 that also contact Sub2, indicating a mutually exclusive interaction. These results suggest a new mechanism for co-transcriptional assembly of the export-competent mRNP and for coordinating export with 3′-end processing.
Keywords: mRNA export, Pcf11, co-transcriptional recruitment, coupled 3′-end processing, Yra1, REF/Aly
Introduction
Transcription by RNA polymerase II (pol II) is coupled to capping, splicing and cleavage/polyadenylation of the nascent transcript, as well as packaging of the transcript with mRNA export factors. How these various co-transcriptional events are coordinated with one another is still largely unknown (for review see (Bentley, 2005; Jensen et al., 2003; Luna et al., 2008; Maniatis and Reed, 2002)). In yeast, the essential mRNA export factor Yra1 is recruited to genes co-transcriptionally, (Lei et al., 2001) consistent with the hypothesis that packaging of the mRNP begins during synthesis of the mRNA precursor (Daneholt, 2001). Yra1 is a member of the highly conserved REF (RNA and Export Factor) family of RNA binding proteins, including mammalian Aly, that convey mRNA to the export receptors, Mex67/TAP/Nxf (Strasser and Hurt, 2000; Stutz, 2000; (Iglesias and Stutz, 2008; Kohler and Hurt, 2007). While Yra1/Aly is required for export of many yeast mRNAs (Hieronymus and Silver, 2003), its role in mRNA export in metazoan systems is probably redundant with other factors (Gatfield and Izaurralde, 2002; MacMorris et al., 2003).
Several studies suggest a major role for the TREX (TRanscription/EXport) complex in co-transcriptional recruitment of Yra1 (Abruzzi et al., 2004; Kohler and Hurt, 2007; Strasser et al., 2002; Zenklusen et al., 2002). TREX comprises the THO complex in association with the DEAD-box protein Sub2, and Yra1. Sub2 and its mammalian homologue UAP56 are ATPase/RNA helicases necessary for splicing and mRNA export (Fleckner et al., 1997; Gatfield et al., 2001; Kistler and Guthrie, 2001; Libri et al., 2001; MacMorris et al., 2003). TREX is thought to be assembled by binding of the THO complex to the pol II transcription elongation complex, followed by Sub2 recruitment via interaction with the Hpr1 subunit (Abruzzi et al., 2004; Strasser et al., 2002; Zenklusen et al., 2002). Because Sub2 binds Yra1 and is necessary for mRNA export, it was proposed that Sub2 is primarily responsible for recruiting Yra1 to pol II transcription complexes (Strasser and Hurt, 2001). A similar mechanism appears to operate in metazoans in that Aly interacts with the UAP56-THO complex (Luo et al., 2001; Strasser et al., 2002), and UAP56 and Aly are recruited to actively transcribed genes (Kiesler et al., 2002). Following recruitment to the transcription elongation complex, Sub2 and Yra1 are transferred to the nascent RNA, marking the mRNP as export competent (Abruzzi et al., 2004). The mRNA is then handed-off from Yra1 to Mex67, which escorts the mRNP to the nuclear pore. Importantly, Sub2/UAp56 and Mex67/TAP bind to the same regions of Yra1/Aly, and they are therefore likely to bind in a mutually exclusive way (Hautbergue et al., 2008; Kohler and Hurt, 2007; Rodrigues et al., 2001; Strasser and Hurt, 2000, 2001; Stutz et al., 2000). Whether or not Sub2 is actually necessary for Yra1 localization on transcribed genes in vivo is a critical unresolved question. Sub2 is not absolutely essential in yeast in all cases (Kistler and Guthrie, 2001) and some mRNA export can be supported in its absence (Strasser and Hurt, 2001) suggesting the possibility that another protein might participate in Yra1 recruitment (Keys and Green, 2001).
While the TREX model can explain co-transcriptional export factor recruitment and the function of Sub2/UAP56 in coupling splicing to export, it does not account for the important role of 3′-end processing in mRNA export (Eckner et al., 1991; Huang and Carmichael, 1996; Long et al., 1995; Lu and Cullen, 2003). Mutation of yeast cleavage factor 1A (CF1A) subunits, RNA14, RNA15 and Pcf11, as well as poly (A) polymerase, inhibit mRNA export (Brodsky and Silver, 2000; Hammell et al., 2002), and cause transcript retention in foci at sites of transcription (Hilleren et al., 2001; Libri et al., 2002). Reciprocally, mutants of some export proteins result in transcripts with improperly processed 3′-ends (Hammell et al., 2002; Jensen et al., 2001). Furthermore in yeast and mammalian cells, export can be inhibited if 3′-end formation at a poly(A) site is substituted by a self-cleaving ribozyme (Dower et al., 2004; Huang and Carmichael, 1996; Libri et al., 2002). Consistent with a role for cleavage/polyadenylation in export, Lei and Silver showed that co-transcriptional Yra1 recruitment to yeast genes was inhibited in a mutant of the CF1A subunit RNA15 (Lei and Silver, 2002); however, it was not clear if this defect could have been due to reduced transcription at the non-permissive temperature, RNA degradation at the site of transcription (Andrulis et al., 2002), or destabilization of the Yra1 protein. A link between 3′-end processing and export is also suggested by genetic interactions between cleavage/polyadenylation factors and TREX subunits, and a severe 3′-end processing defect in extracts from Sub2 and other THO complex mutants (Saguez et al., 2008). Moreover when the TREX complex is inactivated, the 3′ ends of a subset of genes are specifically sequestered within stalled RNP intermediates containing nuclear pore components and chromatin (Rougemaille et al., 2008). Finally, evidence for a link between export and 3′-end processing is provided by the observation that localization of transcribed genes to the nuclear periphery depends on cleavage/polyadenylation signals (Abruzzi et al., 2006; Taddei et al., 2006). In summary, although many studies support the idea that 3′-end processing and export are linked in yeast and metazoans, the mechanisms by which this coupling occurs are poorly understood.
Yeast cleavage-polyadenylation complexes, like export factors, localize to genes co-transcriptionally and this recruitment is facilitated by direct binding of the Pcf11 subunit of CF1A to pol II (Ahn et al., 2004; Barilla et al., 2001; Licatalosi et al., 2002; Sadowski et al., 2003). CF1A is recruited to transcribed genes progressively with low levels at the 5′ end and high levels at the poly (A) site (Calvo and Manley, 2005; Kim et al., 2004; Licatalosi et al., 2002). Pcf11 is a conserved 3′-end processing factor that plays a central role in coupling 3′-end processing with transcription via its N-terminal CTD interaction domain (CID) (Sadowski et al., 2003; Steinmetz and Brow, 1996), which binds to Ser2 phosphorylated CTD heptad repeats and to RNA (Hollingworth et al., 2006; Licatalosi et al., 2002). Pcf11 also interacts directly with the Clp1, RNA14 and RNA15 subunits of CF1A (Amrani et al., 1997; Gross and Moore, 2001; Noble et al., 2007). Human homologue of Pcf11 is a subunit of the CFII(m) complex (de Vries et al., 2000), and is required for transcription termination (West and Proudfoot, 2008) consistent with an in vivo 3′-end processing function.
We have investigated co-transcriptional recruitment of the yeast mRNA export factor Yra1. Unexpectedly, recruitment of Yra1 to actively transcribed genes is independent of the TREX subunit Sub2, but dependent on the 3′-end processing factor CF1A. Furthermore, we demonstrate a conserved protein-protein interaction between yeast and human Pcf11 and Yra1/Aly. Tethering of Pcf11 to RNA in vivo was sufficient to re-distribute Yra1 along a transcription unit. We propose a new model for co-transcriptional recruitment of Yra1 based on this new connection between export and 3′-end processing factors.
Results
Yra1 Recruitment to Actively Transcribed Genes is Independent of Sub2
A central prediction of the TREX model is that disruption of Sub2 recruitment to actively transcribed genes will prevent Yra1 recruitment. To test this prediction, pol II, Sub2, and Yra1 occupancy was monitored by ChIP in WT and sub2 mutant cells. The sub2-201 and sub2-206 mutants we examined are defective in both splicing and export (Libri et al., 2001). Multiplex ChIP PCR analysis at three positions along the TEF1 gene (Fig. 1A) revealed that in both mutants, Sub2 recruitment to the TEF1 gene was greatly reduced at the restrictive temperature, 37°C (Fig. 1A, lanes 10-12), while pol II occupancy was largely unaffected (Fig. 1A, lanes 7-9). Sub2 protein levels measured by immunoblotting were unchanged at 37°C in the mutants compared to the wild type (Fig. 1B), demonstrating that these mutants specifically diminish recruitment to the gene rather than protein stability. In these experiments we monitored co-transcriptional recruitment by ChIP of untagged endogenous Yra1 using antibody raised against the C-terminus (Fig. S4) because C-terminal tagging of Yra1 with GFP caused a ts growth phenotype in WT W303 cells (data not shown). Consistent with previous reports, (Lei et al., 2001; Zenklusen et al., 2002) Yra1 ChIP signals were greater at the 3′-end than at the 5′-end of the gene (Fig. 1A, lanes 13-15) suggesting that recruitment is to some extent progressive as the transcript elongates. Unexpectedly, loss of Sub2 from the TEF1 gene in both sub2 mutants at 37°C did not cause a detectable change in recruitment of Yra1 (Fig. 1A, lanes 13-15).
Figure 1. Yra1 recruitment to actively transcribed genes is independent of Sub2.

(A) ChIP analysis of WT, sub2-201 and sub2-206 cells at 37°C (90 minutes) with anti-pol II, anti-Sub2, anti-Yra1 and normal rabbit serum (NRS). 32P-labeled PCR products correspond to the indicated amplicons of TEF1. (B) Anti-Sub2 immunoblot (top panel) of WT and sub2 ts cells (10ug of whole cell extract (WCE)) at 25 and 37°C (90 minutes). Anti-Pgk1 immunoblot (bottom panel) serves as a loading control. Note that both Sub2 and Pgk1 protein levels increase slightly at 37°C in ALL cell types, including the WT. (C) ChIP analysis of PMA1 as in (A) analyzed by real-time PCR with amplicons shown in the map. Red lines on the map indicate putative poly A sites. Relative ChIP signals are plotted for Sub2 (top right), pol II (bottom left), and Yra1 (bottom right). Mean values, normalized to the maximal signal in each data set, +/- SEM are shown. (D) ChIP signals plotted as ratios of Yra1:pol II for PMA1and ACT1, +/- SEM.
We next examined recruitment of Yra1 in the absence of functional Sub2 on the PMA1 gene by ChIP using high-resolution quantitative real-time PCR. Protein occupancy in WT and sub2 mutant cells was assayed at nine positions on PMA1 and its flanking sequences (Fig. 1C). As we observed at TEF1, there was a near complete loss of Sub2 within the PMA1 ORF in both mutant strains at 37°C (Fig. 1C top right). Pol II and Yra1 were also diminished on PMA1 at 37°C in the mutants relative to WT, but they remained well above the background in the flanking sequences (Fig. 1C, bottom left and right). To control for the reduction in transcription that often occurs in ts mutants under restrictive conditions, Yra1 ChIP signals were normalized to pol II. Normalization revealed that Yra1 occupancy on PMA1 was unaffected in sub2-201 relative to WT, and it actually increased relative to WT in sub2-206 (Fig. 1D left). More Yra1 is recruited to intronless genes like PMA1 than to most intron-containing genes like ACT1 (Abruzzi et al., 2004; Lei and Silver, 2002). We therefore examined how recruitment of Yra1 on ACT1 was affected by sub2 mutants compared to PMA1. As expected, at 37°C Sub2 occupancy was greatly reduced on ACT1 in sub2-201 and sub2-206 (data not shown). In contrast, Yra1 occupancy normalized to pol II was increased on ACT1 and PGK1 in the mutants relative to WT at 37°C (Figs. 1D right, S1). We conclude that recruitment of Yra1 to actively transcribed genes occurs independently of Sub2 on genes with and without introns, and that in some cases inactivation of Sub2 can stabilize Yra1 on a gene. This result, therefore, contrasts with the predictions of the conventional TREX model for co-transcriptional Yra1 recruitment.
Recruitment of Yra1 Requires 3′-end Processing Factors
If Sub2 is not required for Yra1 recruitment to actively transcribed genes, then what is responsible for co-transcriptional recruitment of this mRNA export factor? To investigate further the role of cleavage/polyadenylation factors in Yra1 recruitment, we assessed pol II and Yra1 occupancy in ts mutants of CF1A subunits (Figs. 2 and 3). In agreement with previous results (Lei and Silver, 2002), we observed that Yra1 recruitment on the TEF1 gene was reduced in rna15-2 relative to WT at 37°C (Fig 2A top, lanes 14 and 16). Importantly, this effect is not accounted for by reduced transcription in rna15-2 at the non-permissive temperature. Normalization to pol II revealed that Yra1 occupancy was reduced approximately two fold by inactivation of RNA15 (Fig. 2A, bottom). Note that inactivation of the 3′-end processing factor RNA15 reduced Yra1 recruitment approximately equally at the 5′ and 3′ ends of the gene (Fig. 2A). Total Yra1 protein levels were virtually unaffected by shifting the CF1A mutants to the non-permissive temperature as determined by immunoblotting, thereby confirming that the reduced Yra1 occupancy on TEF1 at 37°C was indeed due to its impaired recruitment (Fig. 2B).
Figure 2. Yra1 Recruitment is inhibited by inactivation of RNA15 and RNA14, and is not restored by deletion of the nuclear exosome.

(A) ChIP analysis of WT rna15-2 and rna15-2Δrrp6 (DBY569) cells with anti-pol II, anti-Yra1 and NRS control. 32P-labeled PCR products of TEF1 amplicons as in Fig. 1A (see map) are shown. Asterisk (*) indicates non-specific product. Graph shows the ratio of Yra1:pol II ChIP signals determined by multiplex PCR, +/- SEM. (B) Anti-Yra1 immunoblot (top panel) of WT and mutant cells (10ug of WCE) at 25° and 37°C (60 minutes). The anti-Pgk1 immunoblot (bottom panel) serves as a loading control. (C) ChIP analysis of WT and rna14-1 and rna14-1Δrrp6 (DBY568) cells as in (A).
Figure 3. Co-transcriptional Recruitment of Yra1 Requires Functional Pcf11.

(A) ChIP analysis of WT and pcf11-2 cells at 37° (60 minutes) with anti-pol II, anti-Sub2, anti-Yra1 and NRS control. 32P-labeled PCR products of TEF1 amplicons as in Fig. 1A (see map) are shown. In this experiment, after normalization to pol II, Yra1 occupancy at TEF1-5′ showed no change, TEF1-mid decreased 1.5 fold, and TEF1-3′ decreased 2.2 fold. (B) ChIP analysis of PMA1 as in Fig. 1C analyzed by real-time PCR with amplicons shown in the map. Red lines on the map indicate putative poly A sites. Relative ChIP signals are plotted for pol II (top left) and Yra1 (top right). Mean values, normalized to the maximal signal in each data set, +/- SEM are shown. (C) ChIP signals plotted as a ratios of Yra1:pol II and Sub2:pol II for PMA1, +/- SEM.
The nuclear exosome degrades transcripts with aberrant 3′-ends (Hilleren et al., 2001), and interacts genetically with Yra1 (Zenklusen et al., 2002). It is possible, therefore, that inactivation of cleavage/polyadenylation factors results in loss of the nascent RNA chain and associated proteins such as Yra1. We asked whether deletion of the nuclear exosome subunit Rrp6 in rna15-2 could stabilize recruitment of Yra1. As shown in Fig. 2A (top), deletion of RRP6 did not restore Yra1 occupancy on TEF1 at 37°C (lane 18). Hence, the loss of Yra1 from the gene when RNA15 is inactivated, is probably not a consequence of nascent chain degradation by the exosome.
Next, we examined mutants in two other CF1A subunits, RNA14 and Pcf11 to determine if they also affected Yra1 recruitment. Normalization to pol II revealed that Yra1 occupancy on TEF1 was reduced more than two fold by inactivation of RNA14 (Fig. 2C, bottom). As we observed in rna15-2, deletion of RRP6 in the rna14-1 mutant did not rescueYra1 recruitment to TEF1 at 37°C (Fig. 2C, lanes 16, 18). In the pcf11-2 ts mutant, Yra1 occupancy on TEF1 was almost eliminated at 37°C compared to WT (Fig. 3A, lanes 7, 8), whereas pol II was only moderately reduced (Fig. 3A, lanes 5, 6). A similar reduction in Yra1 recruitment was observed in the pcf11-9 mutant, which unlike pcf11-2, has a defect in binding to the pol II CTD (Sadowski et al., 2003) (Fig. S2).
To investigate recruitment of Yra1 in the absence of functional Pcf11 in greater detail, we mapped pol II, Yra1, and Sub2 on PMA1 in pcf11-2 cells at 37°C. As expected, the transcription termination defect in this mutant at 37°C resulted in elevated pol II occupancy in the 3′ flanking region relative to WT (Fig. 3B, left). We also noted that Yra1 occupancy fell off near the second poly (A) site, before pol II occupancy started to decline (Fig. 3B, right), in agreement with previous results (Kim et al., 2004; Zenklusen et al., 2002). As we observed at TEF1, Yra1 occupancy within the PMA1 ORF normalized to pol II was reduced about two fold in pcf11-2 relative to WT (Fig. 3C, left).
We investigated the possibility that reduced Yra1 recruitment upon Pcf11 inactivation is linked to loss of Sub2 from the gene. Sub2 occupancy was measured by ChIP and normalized to pol II on PMA1 at 37°C in WT and pcf11-2 strains. Figure 3C shows that whereas Yra1 recruitment relative to pol II was reduced by inactivation of Pcf11 (left), Sub2 occupancy was almost unaffected (right). Similar results were observed at TEF1 (Fig. 3A, lanes 9, 10). In summary, consistent with our observations in sub2 ts mutants (Fig. 1), Yra1 occupancy changed independently of Sub2 when Pcf11 was inactivated. Furthermore, the experiments in Figures 2 and 3 show that Yra1 recruitment, when normalized to pol II, was specifically reduced by inactivation of the CF1A subunits RNA15, RNA14 or Pcf11, and was not restored by deletion of a nuclear exosome subunit.
Yra1 protein interacts with CF1A
To examine whether poor recruitment of Yra1 to actively transcribed genes in the absence of functional CF1A could be caused by disruption of a physical interaction between these proteins, we asked whether Yra1 and CF1A co-immunoprecipitate. As shown in Figure 4A, anti-Yra1 effectively co-immunoprecipitated HA-tagged Pcf11 and RNA15 subunits of CF1A, but not the Pgk negative control. Notably, the interaction between Yra1 and Pcf11 withstood treatment with RNAse and DNAse, and washing with 300mM NaCl (Fig. 4A, red box panel 1); RNA15 co-immunoprecipitation with Yra1 was less robust than Pcf11, but still detectable (panel 2). Conversely, immunoprecipitation of Pcf11 with anti-HA co-precipitated Yra1 and RNA15, but not Pgk. As expected, co-immunoprecipitation between the CF1A subunits Pcf11 and RNA15 was very robust and was unaffected by treatment with nucleases or high salt. The fraction of Yra1 co-immunoprecipitating with Pcf11-HA (Fig. 4A, red box panel 4) relative to the input was considerably less than RNA15. This difference probably reflects, in part, the four fold higher abundance of Yra1 (Oeffinger et al., 2007) relative to RNA15 (yeastgfp.ucsd.edu), and a weaker interaction. In summary, co-immunoprecipitation demonstrates a novel protein:protein interaction between Yra1 and CF1A, consistent with the fact that recruitment of Yra1 to actively transcribed genes requires this cleavage/polyadenylation factor.
Figure 4. Yra1 Physically Interacts with CF1A via Pcf11.

(A) Co-immunoprecipitation between Yra1 and CF1A from whole cell extracts (WCE). Antibodies used for IP are listed at the top, and those used for blotting are listed below each panel. IP's treated with RNase and DNase are indicated (+). IPs were washed with increasing concentrations of [Na+] (50mM, 100mM, 300mM, and 1000mM). Each sample represents the precipitation of 1mg WCE using 5ug antibody. (B) Yra1 binds specifically to the Pcf11 subunit of CF1A. GST pull-down assays using 35S-labeled in vitro translated Pcf11, RNA14, RNA15 and luciferase (Luc) negative control. Autoradiogram (top panel) and corresponding coomassie blue stained loading control (bottom panel) are shown. (C) Diagram of Pcf11 truncations used in (D) (see (Sadowski et al., 2003)). (D) Yra1 interacts with a C-terminal fragment of Pcf11 independent of the CID. GST pull-down analysis as in (B).
A conserved binding interaction between Pcf11 and Yra1/Aly
To determine whether Yra1 makes direct or indirect contact with CF1A, we tested whether radiolabeled in vitro translated Pcf11, RNA14 or RNA15 bound to GST-Yra1 in a pull-down assay. Figure 4B shows that GST-Yra1, but not GST alone, effectively captured Pcf11, but not RNA14, RNA15 or the luciferase control (top panel, lanes 3, 6, 9, and 11). This in vitro interaction is therefore consistent with the fact that in vivo inactivation of Pcf11 reduces co-transcriptional Yra1 recruitment (Fig. 3). The rna14-1 and rna15-2 mutations that have similar effects in vivo (Fig. 2) may disrupt Yra1 interaction with Pcf11 by perturbing the stability or conformation of the CF1A complex.
We next wished to determine which domain(s) of Pcf11 interacts with Yra1. Pcf11 contains a N-terminal pol II CTD Interaction Domain (CID), a polyglutamine domain (Q20), and two zinc fingers (C2H2 and C2HC) (Fig. 4C) on either side of the Clp1 binding domain (Noble et al., 2007; Sadowski et al., 2003). In pull-down assays, GST-Yra1, but not GST alone, bound to a C-terminal fragment of Pcf11 (residues 141-626) deleted for the CID (ΔCID-Pcf11), and not to the CID alone (Fig. 4D, top panel lanes 3, 6, 9, and 11). To pinpoint the segment of Pcf11 that binds Yra1, we hypothesized that a functional interaction might be conserved between the human homologues of these proteins. In the region C-terminal of the CID, human and yeast Pcf11 are most closely related in the region of the two zinc fingers and Clp1 interacting domain comprising residues 420-608 and 1342-1487 of the respective proteins (Fig. 5A). We purified recombinant His-tagged proteins corresponding to these conserved segments of yeast and human Pcf11 (Fig. 5B), and tested them for binding to GST fusions of yeast Yra1 and human Aly. To ensure that binding was not influenced by the presence of nucleic acids, immobilized GST proteins were pre-treated with RNAse, and EtBr (400μg/ml) was added to the binding reaction. As shown in Figure 5C, GST-Yra1, but not GST alone, effectively captured the Zn-finger/Clp1 segment (420-608) of yeast Pcf11 in a concentration dependent manner (top panel), consistent with a direct protein:protein interaction. Importantly, this interaction is conserved between the human homologues of these proteins as shown by the concentration-dependent binding of huPcf11 (1342-1487) to GST-Aly, but not GST alone (Fig. 5D, top). Cross-species interactions also occur such that GST-Yra1 binds huPcf11 (1342-1487) and GST-Aly binds yeast Pcf11 (420-608) (data not shown), consistent with complementation of a YRA1 deletion by Aly (Strasser and Hurt, 2000). We conclude therefore that a conserved region of yeast and human cleavage/polyadenylation factor Pcf11 is sufficient for direct binding to the cognate mRNA export adaptor proteins Yra1 and Aly.
Figure 5. Conserved Binding of yeast and human Pcf11 to REF Family Proteins.

(A) Alignment of yeast and human Pcf11 zinc finger/Clp1 regions. (B) Coomassie blue stained gel of His-tagged yeast and human Pcf11 fragments in (A). (C) Direct binding of yeast Pcf11 zinc finger/Clp1 region to Yra1. GST or GST-Yra1 bound to beads (see stained gel, bottom panel) was incubated with increasing amounts (1.25, 2.5, 5, 10ug) of yeast Pcf11(420-608), and bound protein was detected by anti-His immunoblot (top panel). Note the Pcf11(420-608) eluted from the beads migrates more slowly than the input (lane 1) because these samples contain 0.75M NaCl. (D). Direct binding of human Pcf11 zinc finger/Clp1 region to Aly. GST or GST-Aly bound to beads (see stained gel, bottom panel) was incubated with increasing amounts (1.25, 2.5, 5, 10ug) of human Pcf11(1342-1487), and bound protein was detected by anti-His immunoblot (top panel). (E) Domain diagram of full length Yra1 and truncations used in (F). The N/C REF protein fragment contains a linker (residues 92-132) from λ repressor. (F) Pull-down assays as in (C, D) with GST alone and fusions with full-length -Yra1, -N + RRM, -RRM, -RRM + C, -N, -C, and -N/C REF (see bottom panel). Yeast Pcf11(420-608) bound to the beads was eluted in high salt and visualized by anti-His immunoblot (top panel).
Pcf11 Binds the Sub2/Mex67 Interacting Domains in Yra1
Yra1 contains a RRM (RNA Recognition Motif), two highly conserved N- and C-terminal REF domains, and two moderately conserved regions rich in glycine, serine and positively charged residues flanking the RRM termed N-variable and C-variable regions, respectively (Stutz et al., 2000). Both variable regions, amino acids 14-77 and 167-210, can mediate binding of Yra1 to Sub2 and Mex67 (Strasser and Hurt, 2001; Zenklusen et al., 2001). To determine which domain(s) of Yra1 binds to Pcf11, we made GST fusions with fragments of Yra1 (Fig. 5E) and performed pull-down assays with the Pcf11 420-608 fragment that binds full-length Yra1. This analysis showed that amino acids 1-66, comprising most of the first variable region and the N-REF domain, and amino acids 167-226, comprising the second variable region and the C-REF domain, of Yra1 are each sufficient for direct binding to Pcf11 (Fig. 5F, top panel). Conversely, neither the RRM, nor the N/C-terminal REF domains in isolation are able to bind Pcf11 (Fig. 5F, top panel lanes 5 and 9). Thus Pcf11, like Sub2 and Mex67, binds the N- and C-terminal variable regions of Yra1. These results suggest the possibility that Sub2 and Mex67 compete with Pcf11 for binding to Yra1, and that Yra1 binding to these three partner proteins is mutually exclusive.
Pcf11 is sufficient for co-transcriptional Yra1 recruitment
The results presented above suggest a model in which Yra1 is initially recruited to the transcription elongation complex through a direct interaction with the Pcf11 subunit of CF1A that associates with the pol II CTD. We tested this model by asking whether Pcf11 is sufficient to recruit Yra1 to an actively transcribed gene in vivo using an RNA tethering strategy (Fig. 6A). Endogenous Pcf11 was tethered to nascent RNA from the chromosomal GAL1-YLR454W gene containing six tandem BoxB hairpins, via a C-terminal tag comprising a 6XHA epitope and the BoxB binding peptide from the N-protein of bacteriophage P22 (Wiegand et al., 2003). As a non-tethered control, endogenous Pcf11 was tagged with 6XHA only. Normally, Pcf11 is recruited progressively in a 5′ to 3′ direction, with its peak density near the poly(A) site (Kim et al., 2004; Licatalosi et al., 2002). Because the BoxB sequences were inserted into the YLR454W ORF at position 1242, about 6600bp upstream of the poly(A) site, Pcf11 prematurely accumulated near the 5′ end in the tethered strain, (Pcf11:HA-N peptide, DBY986) compared to the non-tethered strain (Pcf11:HA, DBY987) (Fig. 6B, lanes 3 and 4, top band). The TEF1 3′ amplicon served as a control, demonstrating that Pcf11:HA-N peptide is recruited normally to another endogenous gene (Fig. 6B, lanes 3 and 4, bottom band).
Figure 6. RNA tethering demonstrates that Yra1 and Pcf11 physically interact in vivo.

(A) Diagram of Pcf11-HA-Npeptide tethering to nascent RNA via 6X BoxB sequences inserted in to the chromosomal GAL-YLR454W. (B) Tethering of Pcf11 to BoxB elements in GAL-YLR454W. ChIP analysis of non-tethered (YLR-6XBoxB/Pcf11:HA) and tethered (YLR-6XBoxB/Pcf11:HA-Npep) cells with anti-HA and NRS control. 32P-labeled PCR products corresponding to a region of GAL-YLR454W immediately adjacent to the tether (amplicon 1423) and to the 3′-end of the TEF1 gene are shown. (C) Pcf11 tethering alters Yra1 recruitment to the chromosomal GAL-YLR454W gene. Anti-Yra1 ChIP signals analyzed by real-time PCR for strains with tethered (purple bars) and non-tethered (blue bars) Pcf11. Diagram of GAL-YLR454W-6XBoxB and the corresponding amplicons are included below the plot. Data are represented as mean values, normalized to the maximal signal in the data set, +/- SEM. (D) Yra1 recruitment on a control gene PMA1 is unaffected by the Pcf11-HANpeptide tag. Plot of Yra1 occupancy quantified by real-time PCR and normalized to maximum values as in (C).
Having established that RNA tethering can prematurely recruit Pcf11 to YLR454W, we asked if tethered Pcf11 affects co-transcriptional recruitment of Yra1. Yra1 was monitored by ChIP at ten positions along the GAL-YLR454W gene in the non-tethered control (DBY987) and the Pcf11 tethered strain (DBY986). Yra1 occupancy was equivalent in both strains at position +753 upstream of the BoxB tethering elements (Fig. 6C). However, in the regions adjacent to the tether and at all downstream amplicons, except +7475, Yra1 recruitment was enhanced by tethered Pcf11 (Fig. 6C). No change in Yra1 occupancy was seen on PMA1, confirming that the effect was specific to the gene containing the BoxB sequences (Fig. 6D). No changes were detected in either pol II (Fig. S3, left) or Sub2 (Fig. S3, right) occupancy on the YLR454W gene when Pcf11 was tethered, arguing that the small enhancement of Yra1 occupancy seen under the same conditions is specific. The lack of effect of Pcf11 tethering on Sub2 recruitment is consistent with the idea that Yra1 binding to Pcf11 is mutually exclusive with Sub2 binding. Together these data demonstrate that enhancement of Pcf11 occupancy by RNA tethering caused a specific increase in Yra1 occupancy. This experiment therefore argues that the protein:protein interaction between Yra1 and Pcf11 observed in vitro functions in localizing Yra1 on genes in vivo.
Discussion
Maturation of the nascent mRNP is coordinated with co-transcriptional pre-mRNA processing so that only properly processed transcripts become competent for export. In particular mRNA export in yeast is dependent on cleavage/polyadenylation, but the molecular basis for the connection between export and 3′ end processing has not been identified. Prior to their export from the nucleus, nascent mRNPs must pass quality control tests to ensure that aberrant mRNAs are not transported into the cytoplasm where they could be translated into deleterious abnormal proteins (Iglesias and Stutz, 2008; Jensen et al., 2003; Schmid and Jensen, 2008). An important step in maturation of mRNPs is the co-transcriptional loading of the conserved export adaptor Yra1/Aly. We report that the mRNA 3′-end processing machinery is closely linked to recruitment of Yra1 in yeast through a direct interaction with the Pcf11 subunit of the cleavage/polyadenylation factor CF1A. Yra1 occupancy, relative to pol II, is reduced in ts mutants of Pcf11 and other subunits of CF1A (Fig. 2, 3) consistent with previous results (Lei and Silver, 2002). Furthermore, we identified a direct protein:protein interaction between Yra1 and the zinc finger/Clp1 interaction region of Pcf11 (Fig. 5C), thereby establishing a possible molecular basis for the connection between export and 3′-end processing. The pcf11-2 mutant contains four substitutions at C424, S538, F562, and S579 while pcf11-9 has one substitution at K435, in this region (Sadowski et al., 2003). The only mutation at a residue conserved in human Pcf11 is C424 in the first Zn finger (see Fig. 5A). pcf11-9, which is defective for CTD binding (Sadowski et al., 2003), appears to have a somewhat greater effect on Yra1 occupancy than pcf11-2 (Fig. S2). We think that the pcf11-2 mutations in the Zn finger/Clp1 interaction domain probably do not play a direct role in binding of Yra1 and that defective recruitment of Yra1p in both PCF11 mutants is due to disruption of CF1A integrity and/or binding to pol II.
This interaction between a yeast export factor and a cleavage/polyadenylation factor is conserved between their human counterparts, Aly and huPcf11 (Fig. 5D). Residues 420-608 of yeast Pcf11 and the homologous residues 1342-1487 of huPcf11 are sufficient for interaction with Yra1/Aly. The conservation of this protein:protein interaction between humans and yeast suggests that it is of general significance for coordination of 3′-end processing with export. How Aly is recruited to mammalian genes is not fully understood, but its interactions with both the UAP56-THO complex (Luo et al., 2001; Strasser et al., 2002) and cap binding complex are likely to be important (Cheng et al., 2006). Our biochemical results suggest that in addition, huPcf11 may play a role in recruitment of Aly. Finally, amino acids 1-66 and 124-226 of Yra1 are each sufficient to bind Pcf11 (Fig. 5E, F). Closely overlapping regions of the protein (residues 1-77 and 167-210) bind to the export receptor Mex67 and RNA (Rodrigues et al., 2001; Zenklusen et al., 2001), and Sub2 (Strasser and Hurt, 2001).
The current model for co-transcriptional recruitment of the export adaptor Yra1 through interaction with the Sub2 subunit of TREX (reviewed in (Iglesias and Stutz, 2008; Kohler and Hurt, 2007; Luna et al., 2008) is based on the fact that Sub2 is required for mRNA export, and interacts with Yra1 genetically and physically (Strasser and Hurt, 2001; Strasser et al., 2002; Zenklusen et al., 2002). We report that Sub2 is not required for Yra1 recruitment to the transcription elongation complex, but the 3′-end processing complex CF1A is required. Mutants of Sub2 that eliminate its recruitment to the gene did not prevent Yra1 recruitment and actually enhanced it in some cases (Figs. 1D, S1). Furthermore the pcf11-2 mutant reduced Yra1 recruitment without affecting Sub2 recruitment (Fig. 3C). To account for these new observations, we suggest a modified model for co-transcriptional recruitment of Yra1 that helps reconcile the TREX model with the importance of 3′-end processing for mRNA export (Fig. 7). This model proposes that an important early step is Yra1 binding to the Pcf11 subunit of CF1A, independently of Sub2 (step 1 in Fig. 7). Pcf11 is associated with the pol II transcription complex through Ser2 phosphorylated CTD heptads (Licatalosi et al., 2002) therefore this interaction can explain how Yra1 binds the TEC independent of Sub2. Following recruitment by Pcf11, we suggest that Yra1 is handed off to Sub2 (step 2 in Fig. 7). In support of this proposal we show that Pcf11 and Sub2 have overlapping binding sites at the N- and C-terminal variable regions of Yra1 (Fig. 5F), suggesting that they bind in a mutually exclusive way. The enhancement of Yra1 ChIP signals after inactivation of Sub2 (Fig. 1D) may be due to inhibition of hand-off to Sub2, and stabilization of the Yra1 interaction with CF1A. Sub2/UAP56 is thought to facilitate the subsequent transfer of Yra1/Aly to RNA (Abruzzi et al., 2004) in an ATP dependent reaction (Taniguchi and Ohno, 2008). Therefore, transcription-independent RNA binding by Yra1/Aly is distinct from transcription-coupled Yra1 recruitment to the pol II elongation complex; the former requires Sub2 and the latter requires CF1A. In a final hand-off reaction (step 3 in Fig. 7), Yra1/Aly binds the export receptor Mex67/TAP via the same conserved N- and C-terminal regions of Yra1/Aly that bind Pcf11 and Sub2/UAP56 (Hautbergue et al., 2008; Rodrigues et al., 2001; Strasser et al., 2002). The requirement for the 3′-end processing factor CF1A for Yra1 loading may provide an opportunity to exert quality control. In the event that a fully functional transcription elongation complex with associated CF1A is not assembled, then Yra1 loading would be limited thereby preventing formation of an export competent mRNP. Similarly, Yra1 loading onto the nascent mRNA would be limited if TREX assembly was defective. While our experiments establish a Pcf11-dependent mechanism of Yra1 recruitment, they do not exclude the possibility of additional mechanisms that could also contribute to Yra1 localization at transcribed genes.
Figure 7. A revised model of co-transcriptional Yra1 recruitment by interaction with Pcf11 followed by hand-off to Sub2 and Mex67.
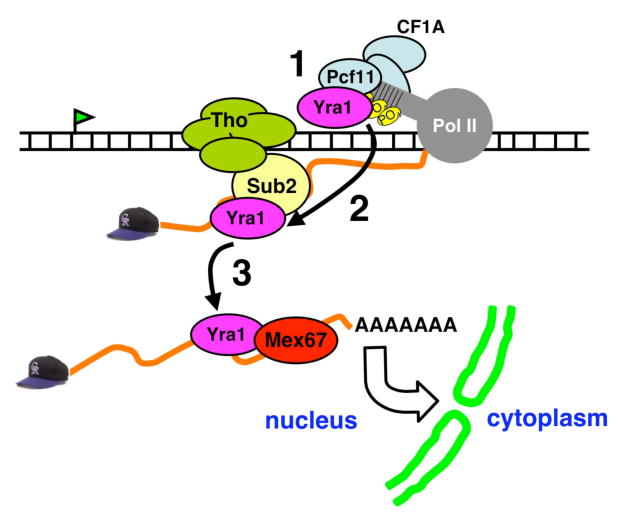
1 = Recruitment of Yra1 via the Pcf11 subunit of CF1A. 2 = Transfer of Yra1 to the nascent RNA facilitated by binding of Sub2. 3 = Loading of the export receptor Mex67 onto the mRNA via interaction with Yra1.
The fact that CF1A mutations affect Yra1 occupancy at 5′ as well as 3′ ends suggests one possible function for the presence of this 3′-end processing factor at 5′ ends (Calvo and Manley, 2005; Licatalosi et al., 2002); namely to help recruit the export factor Yra1. Whether Yra1 association with CF1A affects the 3′-end processing function of this complex is unknown. Yra1 co-localizes with CF1A in the body of the gene and it appears to dissociate near the poly (A) site (Figs. 1C and 3B) (Kim et al., 2004; Zenklusen et al.). These observations suggest the possibility that association with Yra1 negatively regulates the processing function of CF1A. Whether or not enhanced Yra1 retention at the 3′ ends of genes following Sub2 inactivation (Figs. 1D, S1) contributes to cleavage/polyadenylation defects (Saguez et al., 2008) or to accumulation of stalled export intermediates (Rougemaille et al., 2008) remains to be determined.
In summary, we demonstrate a new connection between mRNA export and 3′-end processing mediated by a direct contact between the export adaptor Yra1/Aly and cleavage/polyadenylation factor Pcf11. By physically linking recruitment of Yra1 with Pcf11, this mechanism coordinates the development of mRNA export competence with cleavage/polyadenylation, a processing event that is universal among yeast mRNAs. In contrast only a small fraction of yeast genes produce transcripts that are spliced. Although our results demonstrate a direct link between essential export and cleavage/polyadenylation factors, the details of how export is coordinated with maturation of the mRNA 3′-end per se are still unclear. It is also unknown how interaction of Yra1 with a pol II CTD binding protein, Pcf11, might influence transcription. In future it will be of interest to elucidate how the interaction between Pcf11 and Yra1 affects transcription, assembly of an export competent mRNP, 3′-end processing, and mRNP quality control.
Experimental Procedures
Plasmids and Yeast Strains
Plasmids and strains are described in the supplementary methods and Tables 1, 2.
Antibodies
Rabbit anti-panCTD was described (Schroeder et al., 2000) and anti-RNA15 was a gift of F. Wyers. Anti-Yra1 and -Sub2 were made by immunizing rabbits with GST-Yra1(124-226) (Fig. S4) or full-length Sub2 (a gift of Dr. R. Zhao, UCHSC). Monoclonal antibodies included anti-HA (12CA5), anti-PGK (Molecular Probes), and anti-His (Sigma).
ChIP
WT and ts strains were grown at 25°C in YPD to a density of OD600= 0.8, then grown for an additional 60-90 minutes at 37°C prior to lysis or formaldehyde crosslinking. For galactose induction (Fig. 6) cells were grown in YP raffinose (2%) to OD600=0.8 and induced with 2% galactose for 90 minutes. ChIP, multiplex 32-P labeled PCR, and real-time PCR with SybrGreen (Roche LC-480) were as described (Schroeder et al. 2000)(Glover-Cutter, 2008). Ratios of Yra1/Pol II and Sub2/Pol II were determined on multiple experiments then normalized to the highest value in each data set and means for normalized values. A minimum of two ChIPs and two PCRs from each were used to generate means +/- SEM. Primer sequences are available on request.
Immunoprecipitation
Whole cell extract was made by glass bead disruption in low salt buffer (50mM HEPES pH 7.6, 50mM NaF, 1mM MgCl2, 1mM EGTA, 5% glycerol, 0.25% Tween-20, and protease inhibitors (modified from D. Kellogg) and immunoprecipitated with antibodies coupled to CNBr-sepharose (GE) at 1mg/ml. IP's were washed twice in low-salt buffer and, as necessary, digested (37°, 30 min.) in the same buffer plus 5mM MgCl2, 2mM CaCl2, 10μg/ml RNAse A, and 20U/ml DNase I at (37°, 30 min). Samples were then washed three times in low-salt buffer supplemented with NaCl to 100mM, 300mM, or 1000mM, and eluted with SDS sample buffer.
GST Pull-Down Assays
GST fusion proteins (10 μg) were bound to glutathione sepharose at 1mg/ml and blocked in TIF (150mM NaCl, 20mM Tris pH 8.0, 1mM MgCl2, 0.1% NP40, 10% glycerol, and protease inhibitors) containing 5% non-fat dry milk. [35S]methionine-labeled proteins were made by TNT, (Promega). Binding reactions were for 16 hours at 4°C in 0.5 ml of TIF plus 0.5% non-fat dry milk and 400μg/ml EtBr. Beads were washed five times, in TIF at room temperature and eluted with SDS sample buffer or TIF + 1M NaCl.
Supplementary Material
Acknowledgments
Supported by NIH grant GM58163 to D.B. We thank T. Blumenthal, R. Davis, T. Evans, J. Tyler, S. Chavez, S. Kim, and R. Sclafani for helpful suggestions. B. Cullen (Duke University), F. Stutz (U. Lausanne), D. Libri, F. Lacroute, F. Wyers and M. Minet (CNRS, Gif-sur-Yvette), C. Moore (Tufts U.), and R. Zhao (UCHSC) for generous gifts of plasmids and yeast strains, antibody and proteins, and the UCHSC Cancer Center sequencing facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abruzzi KC, Belostotsky DA, Chekanova JA, Dower K, Rosbash M. 3′-end formation signals modulate the association of genes with the nuclear periphery as well as mRNP dot formation. EMBO J. 2006;25:4253–4262. doi: 10.1038/sj.emboj.7601305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23:2620–2631. doi: 10.1038/sj.emboj.7600261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SH, Kim M, Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell. 2004;13:67–76. doi: 10.1016/s1097-2765(03)00492-1. [DOI] [PubMed] [Google Scholar]
- Amrani N, Minet M, Wyers F, Dufour ME, Aggerbeck LP, Lacroute F. PCF11 encodes a third protein component of yeast cleavage and polyadenylation factor I. Mol Cell Biol. 1997;17:1102–1109. doi: 10.1128/mcb.17.3.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Barilla D, Lee BA, Proudfoot NJ. Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2001;98:445–450. doi: 10.1073/pnas.98.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. doi: 10.1016/j.ceb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Brodsky AS, Silver PA. Pre-mRNA processing factors are required for nuclear export. RNA. 2000;6:1737–1749. doi: 10.1017/s1355838200001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo O, Manley JL. The transcriptional coactivator PC4/Sub1 has multiple functions in RNA polymerase II transcription. EMBO J. 2005;24:1009–1020. doi: 10.1038/sj.emboj.7600575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- Daneholt B. Assembly and transport of a premessenger RNP particle. Proc Natl Acad Sci U S A. 2001;98:7012–7017. doi: 10.1073/pnas.111145498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries H, Ruegsegger U, Hubner W, Friedlein A, Langen H, Keller W. Human pre-mRNA cleavage factor II(m) contains homologs of yeast proteins and bridges two other cleavage factors. EMBO J. 2000;19:5895–5904. doi: 10.1093/emboj/19.21.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower K, Kuperwasser N, Merrikh H, Rosbash M. A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA. 2004;10:1888–1899. doi: 10.1261/rna.7166704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckner R, Ellmeier W, Birnstiel ML. Mature mRNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 1991;10:3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckner J, Zhang M, Valcarcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- Gatfield D, Izaurralde E. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J Cell Biol. 2002;159:579–588. doi: 10.1083/jcb.200207128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Le Hir H, Schmitt C, Braun IC, Kocher T, Wilm M, Izaurralde E. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol. 2001;11:1716–1721. doi: 10.1016/s0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- Gross S, Moore C. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc Natl Acad Sci U S A. 2001;98:6080–6085. doi: 10.1073/pnas.101046598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Gross S, Zenklusen D, Heath CV, Stutz F, Moore C, Cole CN. Coupling of termination, 3′ processing, and mRNA export. Mol Cell Biol. 2002;22:6441–6457. doi: 10.1128/MCB.22.18.6441-6457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue GM, Hung ML, Golovanov AP, Lian LY, Wilson SA. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc Natl Acad Sci U S A. 2008;105:5154–5159. doi: 10.1073/pnas.0709167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Silver PA. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat Genet. 2003;33:155–161. doi: 10.1038/ng1080. [DOI] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- Hollingworth D, Noble CG, Taylor IA, Ramos A. RNA polymerase II CTD phosphopeptides compete with RNA for the interaction with Pcf11. RNA. 2006;12:555–560. doi: 10.1261/rna.2304506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Carmichael GG. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Stutz F. Regulation of mRNP dynamics along the export pathway. FEBS letters. 2008;582:1987–1996. doi: 10.1016/j.febslet.2008.03.038. [DOI] [PubMed] [Google Scholar]
- Jensen TH, Dower K, Libri D, Rosbash M. Early formation of mRNP: license for export or quality control? Mol Cell. 2003;11:1129–1138. doi: 10.1016/s1097-2765(03)00191-6. [DOI] [PubMed] [Google Scholar]
- Jensen TH, Patricio K, McCarthy T, Rosbash M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol Cell. 2001;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- Keys RA, Green MR. Gene expression. The odd coupling. Nature. 2001;413:583–585. doi: 10.1038/35098172. [DOI] [PubMed] [Google Scholar]
- Kiesler E, Miralles F, Visa N. HEL/UAP56 binds cotranscriptionally to the Balbiani ring pre-mRNA in an intron-independent manner and accompanies the BR mRNP to the nuclear pore. Curr Biol. 2002;12:859–862. doi: 10.1016/s0960-9822(02)00840-0. [DOI] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler AL, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 2001;1(5):1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Silver PA. Intron status and 3′-end formation control cotranscriptional export of mRNA. Genes Dev. 2002;16:2761–2766. doi: 10.1101/gad.1032902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Dower K, Boulay J, Thomsen R, Rosbash M, Jensen TH. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol Cell Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Graziani N, Saguez C, Boulay J. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes Dev. 2001;15:36–41. doi: 10.1101/gad.852101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/s1097-2765(02)00518-x. [DOI] [PubMed] [Google Scholar]
- Long RM, Elliott DJ, Stutz F, Rosbash M, Singer RH. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]
- Lu S, Cullen BR. Analysis of the stimulatory effect of splicing on mRNA production and utilization in mammalian cells. RNA. 2003;9:618–630. doi: 10.1261/rna.5260303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna R, Gaillard H, Gonzalez-Aguilera C, Aguilera A. Biogenesis of mRNPs: integrating different processes in the eukaryotic nucleus. Chromosoma. 2008;117:319–331. doi: 10.1007/s00412-008-0158-4. [DOI] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- MacMorris M, Brocker C, Blumenthal T. UAP56 levels affect viability and mRNA export in Caenorhabditis elegans. RNA. 2003;9:847–857. doi: 10.1261/rna.5480803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Noble CG, Beuth B, Taylor IA. Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res. 2007;35:87–99. doi: 10.1093/nar/gkl1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP. Comprehensive analysis of diverse ribonucleoprotein complexes. Nature Methods. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- Rodrigues JP, Rode M, Gatfield D, Blencowe BJ, Carmo-Fonseca M, Izaurralde E. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc Natl Acad Sci U S A. 2001;98:1030–1035. doi: 10.1073/pnas.031586198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, Libri D. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell. 2008;135:308–321. doi: 10.1016/j.cell.2008.08.005. [DOI] [PubMed] [Google Scholar]
- Sadowski M, Dichtl B, Hubner W, Keller W. Independent functions of yeast Pcf11p in pre-mRNA 3′ end processing and in transcription termination. EMBO J. 2003;22:2167–2177. doi: 10.1093/emboj/cdg200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saguez C, Schmid M, Olesen JR, Ghazy MA, Qu X, Poulsen MB, Nasser T, Moore C, Jensen TH. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol Cell. 2008;31:91–103. doi: 10.1016/j.molcel.2008.04.030. [DOI] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. Quality control of mRNP in the nucleus. Chromosoma. 2008 doi: 10.1007/s00412-008-0166-4. [DOI] [PubMed] [Google Scholar]
- Schroeder SC, Schwer B, Shuman S, Bentley D. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol Cell Biol. 1996;16:6993–7003. doi: 10.1128/mcb.16.12.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- Strasser K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, Rondon AG, Aguilera A, Struhl K, Reed R, Hurt E. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun IC, Seraphin B, Wilm M, Bork P, Izaurralde E. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Taniguchi I, Ohno M. ATP-dependent recruitment of export factor Aly/REF onto intronless mRNAs by RNA helicase UAP56. Mol Cell Biol. 2008;28:601–608. doi: 10.1128/MCB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ. Human Pcf11 enhances degradation of RNA polymerase II-associated nascent RNA and transcriptional termination. Nucleic Acids Res. 2008;36:905–914. doi: 10.1093/nar/gkm1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand HL, Lu S, Cullen BR. Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc Natl Acad Sci U S A. 2003;100:11327–11332. doi: 10.1073/pnas.1934877100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Strahm Y, Stutz F. The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol Cell Biol. 2001;21:4219–4232. doi: 10.1128/MCB.21.13.4219-4232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol. 2002;22:8241–8253. doi: 10.1128/MCB.22.23.8241-8253.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


