Abstract
Interleukin-31, produced mainly by activated CD4+ T cells, is a newly discovered member of the gp130/IL-6 cytokine family. Unlike all the other family members, IL-31 does not engage gp130. Its receptor heterodimer consists of a unique gp130-like receptor chain IL-31RA, and the receptor subunit OSMRβ that is shared with another family member oncostatin M (OSM). Binding of IL-31 to its receptor activates JAK/STAT, PI3K/AKT and MAPK pathways. IL-31 acts on a broad range of immune- and non-immune cells and therefore possesses potential pleiotropic physiological functions, including regulating hematopoiesis and immune response, causing inflammatory bowel disease, airway hypersensitivity and dermatitis. This review summarizes the recent findings on the biological characterization and physiological roles of IL-31 and its receptors.
Keywords: IL-31, receptors, biological function, immunological regulation
1. Introduction
IL-31 is a recently discovered helical cytokine [1], belonging to the gp130/IL-6 cytokine family that includes IL-6, viral IL-6, IL-11, IL-27 [2], leukemia inhibitory factor (LIF), oncostatin M (OSM), ciliary neurotrophic factor (CNTF), cardiotrophin-1 (CT-1), cardiotrophin-like cytokine (CLC) [3,4] (also reported as neurotrophin-1 (NNT-1)/B cell-stimulating factor-3 (BSF-3) [5]), and neuropoietin (NP) [6]. All the members of this family share the common chain of glycoprotein 130 (gp130) in their multi-unit receptor complexes (except for IL-31 that uses gp130-like receptor [7]), and are involved in a variety of fundamental physiological processes, such as neuronal growth, bone metabolism, cardiac development, and immune regulation [8]. Members of this family share very low sequence homology, and consequently the identification of novel family members has proved challenging. The receptors for IL-6 family are type I receptors, and they share a number of common structural motifs, such as the cytokine-binding domain with two pairs of conserved cysteine residues and a WSXWS sequence motif in the extracellular domain. Novel receptors are therefore more readily identified and they have been utilized as a means to uncover new members of the gp130/IL-6 cytokine family. IL-31 was identified following the discovery of its receptor, IL-31RA [1] (also named as GLM-R [9] or GPL [7]). IL-31 is expressed preferentially by activated Th2 CD4+ T cells, signaling through a heterodimeric receptor complex composed of IL-31RA and OSMRβ [1]. Its pleiotropic effects on the immune system are just beginning to be examined.
2. Discovery of IL-31 and IL-31 receptors
The gene encoding human IL-31 is located on chromosome 12q24.31 and the mouse ortholog is situated in a syntenic region of chromosome 5. The IL-31 cDNA is composed of an open reading frame encoding a 164 amino acid (aa) precursor and a predicted 141 aa mature polypeptide containing the four α-helix structure [1]. Based on overall length and secondary structure, IL-31 is suggested to belong to the short-chain cytokine group, although it has no apparent sequence homology to other known four helical-bundle cytokines [1,10]. At amino acid level, mature mouse IL-31 protein shares 31% identity with its human counterpart [1]. However, there is no cross-species activity, i.e., mouse IL-31 fails to interact with human IL-31 receptor and vice versa [11]. Dillon et al. cloned IL-31 gene using a functional cloning approach, based on the proliferation of cells bearing the novel IL-31RA and other known receptors of the gp130 family [1]. IL-31 mRNA is preferentially but not exclusively expressed by Th2 CD4+ T cells after activation [1], and by skin-homing CD45RO+ (memory) cutaneous lymphocyte-associated antigen (CLA)-positive T cells in patients with atopic dermatitis [12]. IL-31 mRNA was also found in testis, bone marrow, skeletal muscle, kidney, colon, thymus, small intestine, trachea [1] and dorsal root ganglia [13].
IL-31RA is a novel type I cytokine receptor that mediates IL-31 signaling when coupled with OSMR [1]. The IL-31R was discovered independently by three research groups using bioinformatic tools to search gene databases for novel cytokine receptors that share the conserved structural motifs of a type I cytokine receptor. Ghilardi et al. scanned the public database for putative cytokine receptors and a cDNA encoding full-length human gp130-like monocyte receptor (GLM-R) was subsequently cloned from a pooled tissue cDNA library. Murine GLM-R was obtained by a combination of cross-species library screening and PCR cloning from a murine spleen cDNA library [9].
Diveu et al. used gp130 cDNA sequence to screen the human genomic database at NCBI. cDNAs encoding a novel cytokine receptor were isolated from the U937 myelomonocytic and the GO-G-UVM glioblastoma cell lines. According to its homology to gp130, this novel cytokine receptor was named GP130-like receptor (GPL) [7].
When searching the translated human genomic sequence database with known cytokine receptor sequence, Dillon et al. identified an exon encoding part of a leukemia inhibitory factor receptor-like (LIFR-like) cytokine receptor. Subsequent cDNA cloning from activated peripheral blood mononuclear cells (PBMC) produced four splice variants of a type 1 cytokine receptor, which were later named as IL-31RAv1-v4. They also isolated two splice variants of mouse IL-31RA from a mouse testis cDNA library [1]. Next, Dillon et al. constructed a series of BaF3 cell lines expressing human IL-31RA alone or in combination with gp130, IL-12Rβ1, IL-12Rβ2, IL-27RA (WSX-1), IL-23R or OSMR. They assayed each cell line for proliferation in response to conditioned media from activated human peripheral T cells or from an activated T cell line CCRFCEM, and found that only the cells expressing GPL together with OSMR were able to proliferate in response to a soluble factor in the conditioned media, which was identified to be IL-31. Furthermore, the addition of an anti-OSMR antibody to the cultures abrogated the proliferation of BaF3 cells in response to IL-31 [1]. Hence, the functional receptor complex for IL-31 is composed of IL-31RA and OSMR.
3. Characterization of IL-31 receptors
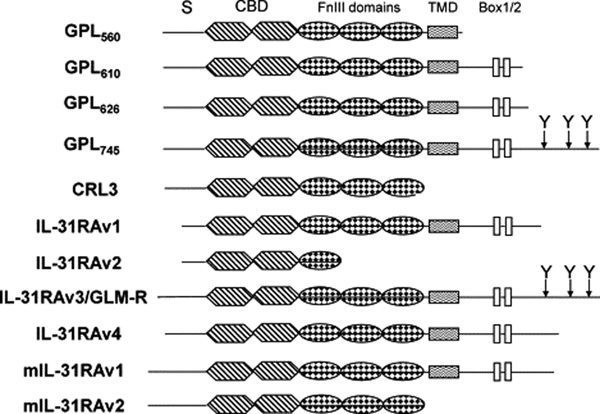
IL-31RA belongs to the gp130-subfamily of type I cytokine receptors. It displays 28% amino acid identity with the common receptor component glycoprotein 130 (gp130) shared by the IL-6 family of cytokines, but lacks the Ig-like domain present at the N-terminus of gp130 [1,7,9]. Human IL-31RA gene is located on 5q11.2, only 24 kb downstream of gp130, with opposite transcriptional orientations to each other [7,9]. To date, IL-31 receptor exists in several different isoforms as follows: GPL560, GPL610, GPL626, GPL745, CRL3, GLM-R and IL-31RAv1-v4 [1,7,9] (Fig. 1). GPL560, GPL610, GPL626 and GPL745 consist of 560 aa, 610 aa, 626 aa and 745 aa, respectively [7]. CRL3 is the soluble form of GPL that only encodes its truncated 509 aa extracellular domain [7,14]. IL-31RAv1, v2, v3, and v4 contain 649 aa, 324 aa, 764 aa and 662 aa, respectively [1]. GLM-R was initially reported to have 732 aa [9], but now has been revised to include the additional 32 aa upstream, making it 100% identical with IL-31RAv3. IL-31RAv1 and IL-31RAv2 all have a potential hydrophobic signal peptide of 19 aa [1]. GPL560, GPL610, GPL626 and GPL745 all include a signal peptide of 33 aa [7]. The signal peptides of GLM-P/IL-31RAv3 and IL-31RAv4 consist of 51 aa and 32 aa respectively [1]. After the signal peptides, GLM-R/IL-31RAv3, GPL610, GPL626, GPL745, IL-31RAv1 and IL-31RAv4 all have the characteristic features of type I cytokine receptors: a cytokine receptor homology domain with two pairs of conserved cysteine residues and a WSDWS signature motif, followed by three fibronectin type III-like domains and a single transmembrane region connected to an intracellular tail. Within the cytoplasmic tail, there is a box-1 motif typically involved in the association with cytoplasmic tyrosine kinases of the Jak family. GLM-R/IL-31RAv3 and GPL745 are different from each other only in signal peptides. They contain three additional tyrosine residues in the cytoplasmic tail, which serve as docking sites for downstream signaling molecules with SH2 domains [1,7,9]. GPL560 only has extracellular and transmembrane domains, whereas CRL3 and IL-31RAv2 are two soluble receptors without transmembrane region [1,7]. Additional studies determined that only GLM-R/IL-31RAv3, GPL745 and IL-31RAv4 can transduce an intracellular signal [1,7,9]. In mouse, IL-31RA is separated by only 19 kb from gp130 in a syntenic region on mouse chromosome 13 [7]. It has two splice variants. One is homologous to human IL-31RAv4, and the other is a soluble form consisting of the cytokine-binding domain (CBD) and fibronectin type III domains. Full-length mouse IL-31RA shares 61% identity with the human IL-31RA in amino acid sequence [1].
Fig. 1.
Isoforms of human and mouse IL-31RA. S, signal peptide; CBD, cytokine-binding domain; FnIII, fibronectin type III; TMD, transmembrane domain; Box1/2, cytoplasmic box 1 and box 2 signaling motifs; Y, location of tyrosine in the cytoplasmic domain. GLM-R and IL-31RAV3 are exactly the same. GPL745 differs from GLM-R only at the signal peptide region with GPL745 shorter for 19 aa. IL-31RAv4 starts at the same aa with CRL3 and GP745. GenBank accession numbers: IL-31RAv1, AY499339; IL-31RAv2, AY499340; IL-31RAv3, AY499341; IL-31RAv4, AY499342; GLM-R, NM_139017; CRL3, AF106913; mIL-31RAv1, AY509150; mIL-31RAv2, AY509151.
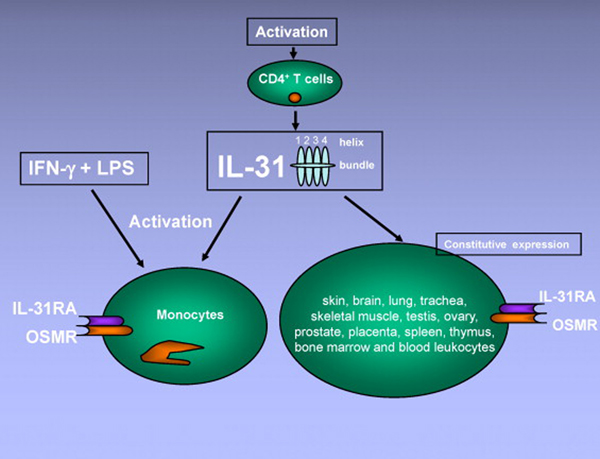
Northern blot experiments showed that constitutive expression of human and mouse IL-31RA mRNA can be detected in skin, brain, lung, trachea, skeletal muscle, testis, ovary, prostate, placenta, spleen, thymus, bone marrow and blood leukocytes [1,7]. In addition, some human cell lines also express IL-31RA, for instance glia-derived cell lines GO-G-UVM and U87MG, melanoma cell line A375, myelomonocytic cell lines U937 and THP1 [7]. Human IL-31RA mRNA, although undetectable in fresh peripheral blood monocytes, is upregulated substantially in monocytes cultured with Interferon-γ (IFN-γ) [1]. OSMR mRNA is more ubiquitously expressed, with the highest levels in trachea, thymus and skin. OSMR mRNA expression can be induced in monocytes treated with lipopolysaccharide (LPS). Thus, IFN-γ and LPS together induce the expression of both receptor chains of IL-31R in human monocytes [1] (Fig. 2). The expression profile of functional IL-31R on mouse immune cells is less clear. One study showed that, compared to wild type counterparts, CD4+ T cells from IL-31RA−/− mice proliferate stronger and secrete more Th2 cytokines when stimulated under neutral or Th2 conditions, suggesting the presence of IL-31R on mouse CD4+ T cells [15]. Nevertheless, the kinetic expression of functional IL-31R on different T cell subsets and antigen-presenting cells, under resting and activation conditions, warrants further detailed study.
Fig. 2.
Interleukin-31 is produced mainly by activated CD4+ T cells, and targets a broad range of cells expressing IL-31RA and OSMR. Monocytes express IL-31RA and OSMR when activated by IFN-γ and LPS. Constitutive expression of both receptor units is found in many tissues.
The receptor chains of the IL-6 family have a modular organization containing in their extracellular part at least one Ig-like domain, at least one CBD with conserved cysteine positions and a WSXWS motif, and FnIII domains [16–18] (Fig. 1). Structural analyses of this receptor family revealed that high affinity binding of the cytokine to its receptor complex involves on one side the CBD of a first receptor subunit and on the other side the Ig-like domain of the second receptor component [17–21]. The IL-31RA is devoid of Ig-like module and binds IL-31 through its CBD. Although OSMR may also recognize IL-31 through its Ig-like domain, immunoprecipitation experiments suggest that IL-31 binds predominantly to IL-31RA but not OSMR in the IL-31R complex [22]. OSMR nevertheless increases IL-31 binding when coupled with IL-31RA [22]. In this regard, IL-31 is very similar to OSM. In human, OSM (as well as LIF) binds to the common LIF/OSM type I receptor complex composed of the gp130 and LIF receptor β (LIFRβ) subunits. In addition, human OSM also specifically recognizes a type II receptor in which the gp130 receptor chain is associated with the OSMR β-chain [23–25]. In mouse, OSM and LIF do not share functional receptors, and OSM is only recognized by the type II receptor gp130-OSMRβ complex [26–28]. Regardless of the species difference and unlike the other family members, OSM and IL-31 are similar in that both show predominant binding to gp130 or its high homology relative GPL (IL-31RA) over individual LIFR or OSMR, which are only converted into high-affinity receptors after forming heterodimer complexes [22–25]. Although IL-31 and OSM do not appear to cross-activate the receptor of each other, as indicated by receptor transfection and immunoprecipitation assays [22], it will be of interest to further study to what extent IL-31 and OSM share similar intracellular signaling pathways.
4. Signal transduction pathways of IL-31
Initial Northern blot and PCR experiments showed that IL-31RA is abundantly expressed in tissues involved in reproduction, in spleen, thymus, lung, skin and trachea, and in cells of myelomonocytic lineage [1,7,9]. These tissues are also normally positive for OSMR, and thus potentially responsive to IL-31. However, Diveu et al. reported that in human only the longest form (GPL745) contains full signaling capacity whilst the truncated form (GPL560) that lacks Jak binding box 1 motif serves as a dominant negative [22]. Given the fact that a mere 2-fold expression in favor of GPL560 over GPL745 strongly neutralizes the signals of the latter, it would be prudent to re-evaluate the tissue expression profiles of signaling vs. antagonizing IL-31RA isoforms in determining IL-31 responsiveness.
4.1 MAPK and PI3K/AKT pathways
The extracellular signal regulated kinase (ERK)1/ERK2 MAP kinases play important roles in mediating the mitogenic effects of the IL-6 family members [29]. The study in glioblastoma cell line expressing both IL-31RA and OSMR showed that stimulation of the cells with IL-31 quickly increased MAPK phosphorylation [22]. Nevertheless, IL-31RA or OSMR alone is insufficient to activate ERK1/2. In this signaling pathway, it seems that IL-31RA contributes only indirectly by cytokine binding and recruitment of OSMR, whereas the important MAPK-activating activity exclusively relies on the signal-transducing partner of receptor OSMR [14].
This theme is reiterated in the activation of PI3K/AKT pathway. When the IL-31RA/OSMR complex is activated by IL-31, a slight but significant tyrosine phosphorylation of PI3K (phosphoinositide-3-kinase) and a marked increase of AKT phosphorylation could be observed [22]. Yet, tyrosine-phosphorylated IL-31RA itself is incompetent in recruiting tyrosine phosphatase SHP-2 and the adaptor protein Shc, both of which are recruited by OSMR [14].
4.2 Jak/STAT pathway
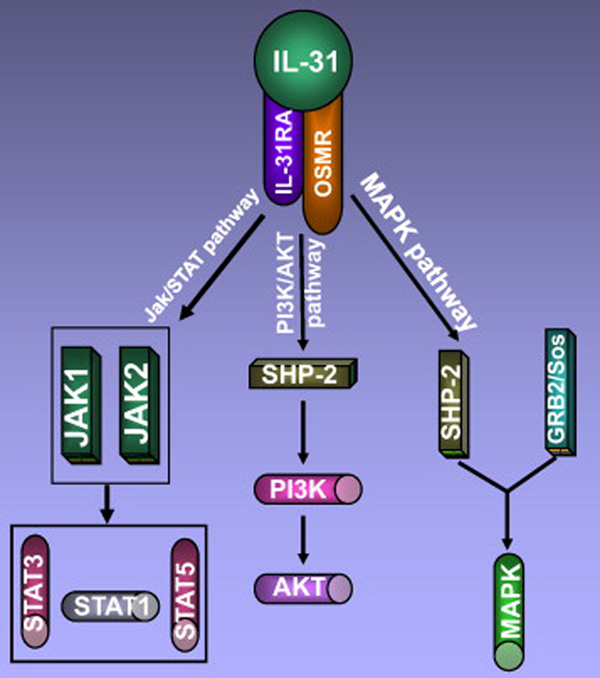
IL-31 can activate Janus kinase (Jak) 1 and Jak2 signaling molecules after binding to its receptor complex. Once activated, Jaks are known to stimulate the phosphorylation of downstream signaling molecules signal transducer and activator of transcription (STAT)-3, STAT-5 and to a lesser degree STAT-1 [1,7,14] (Fig. 3). Dillon et al. showed that STAT-1, STAT-3 and STAT-5 were activated in transfectants expressing IL-31RA and OSMR, but cell lines and primary cells expressing native receptors signaled mainly through STAT-3 [1].
Fig. 3.
Interleukin-31 acts through three signaling pathways: Jak/STAT pathway, PI3K/AKT pathway and MAPK pathway.
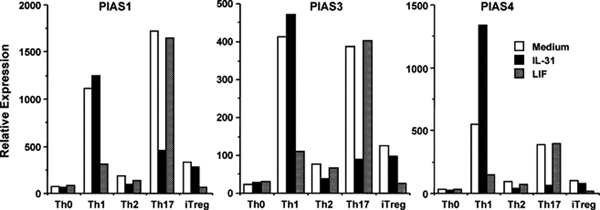
Except for the expression of dominant negative isoforms of IL-31RA, intracellular molecules responsible for fine-tuning IL-31 signaling are not well-studied. It is known that STAT signaling can be negatively regulated through the following three main mechanisms: 1) dephosphorylation of Jaks or STATs by various protein tyrosine phosphatases; 2) inactivation of Jaks by the suppressor of cytokine signaling (SOCS) proteins; and 3) inhibition of the transcriptional activity of STATs by Protein Inhibitor of Activated STAT (PIAS) proteins (for review, see ref. [30]). PIAS1 and PIAS3 function to block the DNA-binding activity of STAT-1 and STAT-3, respectively [31,32], whereas PIAS4 suppresses STAT-1 transcriptional activity without interfering its DNA-binding capacity [33]. Regulation by PIAS proteins may fine-tune the signals from diverse cytokines that converge at STAT activation. For example, we found that IL-31 and LIF each regulated distinctive sets of PIAS proteins in naïve mouse CD4+ T cells differentiated into Th1 and Th17 cells (Fig. 4). Compared to medium control, LIF inhibited PIAS1, PIAS3, and PIAS4 induction under Th1 condition, whereas IL-31 inhibited PIAS1, PIAS3, and PIAS4 induction under Th17 condition. IL-31 even induced more than 2-fold PIAS4 in Th1 cells. By real-time PCR, we also found that LIF inhibited the transcription of IFN-γ in Th1 cells, but not IL-17A/F in Th17 cells; whereas IL-31 increased IFN-γ message in Th1 cells, and dramatically inhibited IL-17A/F mRNA synthesis in Th17 cells (manuscript in preparation). These polar opposite effects by IL-31 and its related family member LIF offer a platform to further study in greater detail the signals from STAT, PIAS to downstream molecules that shape the differentiation of T helper subsets.
Fig. 4.
IL-31 and LIF regulate distinctive sets of PIAS proteins in mouse CD4+ T cells differentiated into Th1 and Th17 cells. Purified mouse CD4+ T cells were stimulated with plate-bound anti-CD3 (10 µg/mL) and soluble anti-CD28 (1 µg/mL) in the presence of various cytokines and neutralizing antibodies. Th0: anti-CD3/28 only; Th1: +IL-12+anti-IL-4; Th2: +IL-4+anti-IFN-γ; Th17: +TGF-β+IL-6+anti-IL-4+anti-IFN-γ; induced regulatory T cells (iTreg): +TGF-β. TGF-β was added at 1 ng/mL. All the other cytokines were added at 50 ng/mL. Neutralizing antibodies were added at 10 µg/mL. PIAS messages are compared by real-time PCR after 3 days.
5. Biological activities of IL-31
5.1 Regulation of cell proliferation and hematopoiesis
In vitro, the effect of IL-31 on cell proliferation is cell type-, cell density- and cytokine concentration-dependent. Although IL-31 was originally identified to stimulate the proliferation of BaF3 cells (a mouse pro-B cell line), a recent study demonstrated that IL-31 is highly effective in suppressing the proliferation of lung epithelial cells by up-regulating p27Kip1 and down-regulating cyclin B1, cdc2, cdk6, mcm4 and Rb [34]. In the colorectal cancer cell line HCT116, IL-31 at high doses significantly suppresses cell proliferation when the cell density is low. However, it loses antiproliferative activity and even stimulates cell proliferation when the cell density is high [35]. It is likely that additional signals from different cell types modulate IL-31 activity in cell proliferation.
Helical cytokines and their receptors play important roles in regulating hematopoiesis in vivo. Oncostatin M, a cytokine sharing a receptor subunit with IL-31, is implicated in the homeostasis of myeloid progenitor cells (MPC) [36]. Compared to wild type animals, OSMR−/− mice have reduced frequencies of erythroid and megakaryocyte progenitors in bone marrow [37]. IL-31 signals through STAT-3 and STAT-5, the activation of which by a number of cytokines has already been shown to be required for hematopoiesis. Direct evidence on IL-31 regulation of hematopoiesis came from the studies comparing IL-31R−/− mice and their littermate wild type controls, which showed that IL-31R deficiency significantly decreased absolute numbers and cycling status of immature subsets of hematopoietic progenitor cells (HPC) in bone marrow and spleen [11]. Thus, IL-31 and its receptors are positively involved in the regulation of hematopoietic progenitor cell homeostasis.
5.2 Induction of cytokines and chemokines
Study in normal human epidermal keratinocytes (NHEKs) showed that several chemokine genes were induced (3- to 9.5-fold) in the cells stimulated with human IL-31, including those encoding GRO1α (CXCL1), TARC (CCL17), MIP-3β (CCL19), MDC (CCL22), MIP-3 (CCL23), MIP-1β (CCL4) and I-309 [1]. Another studies in human colonic subepithelial myofibroblasts (SEMFs) and bronchial epithelial cells also showed that IL-31 stimulated secretion of proinflammatory cytokines, chemokines and matrix metalloproteinases (MMPs) [38,39]. These data indicate that IL-31 may function as a proinflammatory cytokine involved in recruitment of polymorphonuclear cells, monocytes and T cells to an inflammatory site in vivo.
5.3 Regulation of inflammation and immune response
Both subunits of IL-31 receptor are expressed in human monocytes activated with IFN-γ and LPS [1]. LPS activates cells through Toll-like receptor 4 and is required for adaptive Th1 responses [40], but can also induce a Th2 response in a mouse model of allergic sensitization [41,42]. Our own in vitro observation indicates that IL-31 can positively and negatively affect Th1 and Th17 differentiation in an APC-free system (Fig. 4). These data suggest a potential function for IL-31 in regulating immune responses through modulating antigen-presenting cells or more directly T cell themselves.
In a recent study, investigators found that after intravenous injection of Schistosoma mansoni eggs, IL-31RA−/− mice developed more severe pulmonary inflammation, characterized by enlarged area of granulomatous inflammation, increased number of cells positive for resistin-like molecule α, and enhanced collagen deposition, than wild type animals [15]. Under neutral or Th2-polarizing conditions, IL-31RA−/− T cells produced significantly more IL-4, IL-5 and IL-13 than their wild type counterparts. When co-stimulating T cells under Th2-polarizing condition, IL-31RA−/− macrophages exhibited enhanced accessory function compared to wild type cells. In contrast, the generation of CD4+ T cell-mediated Th1 responses was normal in IL-31RA−/− mice. These data implicate IL-31/IL-31R signaling as a negative regulatory pathway that specifically limits type 2 inflammation [15]. However, one potential caveat of the study is that the effects on Th subset differentiation were not examined under the influence of exogenously added IL-31 cytokine. As IL-31 is mainly produced by activated Th2 cells, lack of in vitro effect by IL-31RA deficiency in non-Th2 subsets does not necessarily exclude the possibility that these cells can be affected by Th2-derived IL-31 in vivo. In addition, these findings were based on an acute model of type 2 inflammation in the lung, whether this pathway influences pathologic consequences of chronic type 2 inflammation such as fibrosis and tissue remodeling will require further investigation.
6. Role of IL-31 in diseases
6.1 IL-31 in dermatitis
It is well established that T cell-mediated inflammation plays a major role in the development of most skin diseases, including atopic dermatitis (AD), allergic contact dermatitis (ACD), and psoriasis. Atopic dermatitis is a chronic relapsing inflammatory skin disease characterized by skin lesions with ‘lichenification’, pruritic excoriations and a susceptibility to cutaneous infections [43]. Atopic dermatitis is mediated by T cells, characteristic with a predominant Th2 type response [44], and is often associated with other Th2-mediated allergic diseases such as asthma [43]. Transgenic mice over-expressing IL-31 or wild type mice administrated with recombinant IL-31 protein all developed a notable skin phenotype that closely mimicked that of patients with atopic dermatitis [1]. Higher levels of IL-31 expression were found in biopsy specimens taken from patients with atopic dermatitis than those from healthy individuals [12,45]. These data imply an important role for IL-31 in the pathogenesis of atopic dermatitis.
An examination of IL-31 mRNA expression in NC/Nga mice as an animal modal of atopic dermatitis showed that the expression of IL-31 mRNA in the skin of NC/Nga mice with scratching behavior was significantly higher than that in NC/Nga mice without scratching behavior [46,47]. Comparison of IL-31 mRNA expression in healthy individuals and in patients with pruritic or nonpruritic inflammatory skin diseases showed that human IL-31 was over-expressed in the pruritic atopic dermatitis but not in the nonpruritic psoriatic lesions [13]. Notably, lesional skin of patients with prurigo nodularis, one of the most pruritic forms among inflammatory skin diseases, showed the highest levels of IL-31 mRNA expression [13]. These results suggest that IL-31 mainly plays a role in inducing pruritus but not in directly causing lesions per se. The development of lesions may arise over time from the excessive scratching behavior of the patients [13].
In unstimulated PBMCs from patients with atopic dermatitis, IL-31 levels did not differ from those in PBMCs from healthy, nonatopic controls. However, after staphylococcal enterotoxin B (SEB) or anti-CD3 stimulation, PBMCs from patients with atopic dermatitis expressed higher levels of IL-31 mRNA compared with nonatopic subjects [13]. Recently, an IL-31 gene haplotype has been identified to be linked to the genetic susceptibility to nonatopic eczema. Upon activation, PBMCs from individuals homozygous for this haplotype produced 3.8-fold more IL-31 than those from heterozygous carriers [48]. These observations suggest that PBMCs from susceptible individuals have an increased tendency to produce higher levels of IL-31 in response to superantigen stimulation or polyclonal activation, which may contribute to the development of pruritus in these patients.
In human keratinocytes, IL-31 induces several chemokine genes that have been associated with atopic skin inflammation such as CCL1, CCL17, and CCL22 [1]. Hence, elevated levels of IL-31 in atopic dermatitis lesions may enhance skin inflammation through the induction of chemokines, which subsequently lead to the recruitment of T cells. In turn, activated skin-infiltrating T cells may become new sources of IL-31, thereby amplifying atopic skin inflammation and pruritus.
Analysis of the tissue distribution of IL-31 receptor heterodimer revealed that IL-31RA transcripts are most abundantly expressed in dorsal root ganglia of 63 different human tissues, where the cell bodies of primary sensory neurons reside [13]. This finding is particularly interesting because the sensation of itch, similar to pain, is directly mediated by unmyelinated C fibers of primary sensory neurons whose cell bodies reside within dorsal root ganglia [49,50]. Similar to IL-31RA, several cytokine and chemokine receptors have been detected in dorsal root ganglia, and signaling through these receptors is thought to contribute to hyperalgesia during inflammation [51–53]. Moreover, functional OSMR, the other essential receptor subunit for IL-31 signaling, has been detected on a specific subset of nociceptive neurons in dorsal root ganglia [54,55], which project to the dermis of the skin [55]. These data suggest that IL-31 might induce pruritus by directly modulating the function of sensory neurons. Alternatively, IL-31 may exert its pruritic effects via indirect activation of IL-31 R-expressing keratinocytes, which may induce a yet unknown keratinocyte-derived mediator that subsequently activates unmyelinated C fibers in the skin [56]. Regardless the mechanism, IL-31 and the IL-31R are promising targets for the treatment of inflammatory and itchy dermatoses such as atopic dermatitis.
6.2 IL-31 in inflammatory bowel disease
Colorectal cancer derived intestinal epithelial cell (IEC) lines express both subunits of the IL-31 receptor, while their expression in unstimulated primary murine IEC is low. LPS and the proinflammatory cytokines TNF-α, IL-1β and IFN-γ can increase IL-31, IL-31RA and OSMR mRNA expression. IL-31 can mediate ERK-1/2, Akt, STAT-1 and STAT-3 activation and enhance IL-8 expression in IEC [35]. There are reports showing that activation of Akt and STAT proteins mediates IEC proliferation and migration [57–60]. Likewise, IL-31 shows chemotactic properties and increases IEC migration. In addition, IL-31 can effectively induce chemokines [IL-8, GRO-α (growth-related oncogene-α), MCP-3 (monocyte chemoattractant protein-3), CXCL3, CCL13 and CCL15], proinflammatory cytokines (IL-6, IL-16 and IL-32) and matrix metalloproteinases (MMP-1, MMP-3, MMP-25 and MMP-7) in human colonic subepithelial myofibroblasts. The effects of IL-31 are comparable to IL-17A, and these two cytokines show additive effects in stimulating the secretion of cytokines and chemokines [38]. These data suggest a role for this cytokine in the pathogenesis of IBD by promoting proinflammatory gene expression and modulating IEC barrier function [35].
6.3 IL-31 in airway hypersensitivity
Bronchial and alveolar epithelial cells, pulmonary fibroblasts and macrophages are the primary targets of IL-31 in the lung [61]. IL-31 can significantly elevate the gene and protein expressions of epidermal growth factor (EGF), vascular endothelial growth factor (VEGF) and monocyte chemoattractant protein-1 (MCP-1/CCL2) in human bronchial epithelial cells in a time- and dose-dependent manner [39]. The activation of MAPKs is crucial for IL-31 induced bronchial inflammation. Serum concentrations of IL-31 in allergic asthmatic patients, but not control patients, were significantly higher than those in normal control subjects (50.15 vs 10.01 pg/ml, P < 0.001) [62]. These observations suggest IL-31 may play a pathogenic role in airway hypersensitivity.
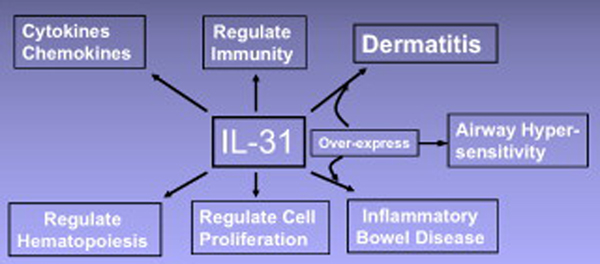
7. Perspectives
The production of IL-31 mainly by CD4+ T helper cells, and the broad spectrum of its receptor expression on immune- and non-immune cells suggest that this novel cytokine may have potential multiple, pleiotropic physiological functions (Fig. 5). Several studies now support a role for IL-31 in dermatitis and epithelial pathologies [1,13,15,45–47]. However, how IL-31 acts in concert with known mediators of pruritus/dermatitis, such as IL-4, IL-7, histamines and neuropeptides, remains to be determined. A precise understanding of the involvement of this novel cytokine in the development of these diseases is critical for more effective strategies for disease intervention.
Fig. 5.
Interleukin-31 produces a number of biological effects, and causes several diseases when over-expressed.
The gene cluster for the cytokine receptors of IL-6 family is very conserved during evolution. IL-31RA displays homology with Dome, a cytokine receptor in Drosophila. The CBDs and FnIII domains of IL-31RA and Dome are 29.7% similar, suggesting that they may evolve from a common ancestor [7]. It is noteworthy that Dome and its ligand, UPD, play important functions in Drosophila embryonic segmentation and maintenance of the niche for germ-line stem cells in the follicles and testis [63,64]. Interestingly, IL-31RA transcripts are also present in ovary, prostate, and placenta. Furthermore, very high level of human IL-31RA mRNA expression is detected in testis [1,7]. The tissue distribution and protein homology between human IL-31RA and Drosophila receptor Dome suggest that the IL-31/IL-31R pathway might have important functions in mammalian reproduction as well. Given the lack of a gross phenotype in naive IL-31RA−/− mice [1,15], it is likely that some other immune mediators may have redundant functions with IL-31 and could compensate the loss of IL-31RA signaling.
Elucidation of the biological activities of IL-31 has only just begun. Further investigation of its signal transduction pathways and in vivo functions will undoubtedly result in a better understanding of the intricate regulation of the immune system, and the role of this cytokine in health and disease.
Acknowledgements
This work was supported by grants from National Science Foundation of China (grant no. 30371307, Q. L.) and Juvenile Diabetes Research Foundation (1-2007-551 and 5-2007-690, W.G.)
Biographies
Wenda Gao, Ph.D., is currently an Assistant Professor in Medicine at Beth Israel Deaconess Medical Center, Harvard Medical School. He got his Ph.D. from Tufts University School of Medicine, and received postdoc training in Dr. Terry B. Strom’s lab at Harvard Medical School on negative co-stimulation and transplantation tolerance. He is now studying the role of graft-protecting regulatory T cells (Treg) and graft-destroying Th1/Th17 cells in transplantation. In recent years, he and collaborators have published a series of papers in Nature, Nature Medicine, Journal of Experimental Medicine, Journal of Clinical Investigation, and American Journal of Transplantation, describing the effects of pro-inflammatory cytokines on the development of Treg and Th17 cells. Particularly, he is interested in the role of IL-6 family cytokines in CD4+ T helper subset differentiation.

Quansheng Liu, Ph.D., is currently an Associate Professor in Sichuan University in China. He received postdoc training in Stanford University on the role of 24p3 (NGAL) in inflammation. Through the years, Dr. Liu and Dr. Gao have collaborated in multiple areas in translational medicine, particularly in the cytokine effects on inflammation and diseases.

Qing Zhang is a graduate student in Dr. Liu’s lab at Sichuan University in China, studying the role of IL-31 in animal models.

Prabhakar Putheti, M.Sc. Ph.D., got his M.Sc. in biochemistry in India (1998–2000) and subsequently moved to the Karolinska Institute, Stockholm, Sweden for Ph.D. study (2001–04). His thesis was on the role of human CD4+CD25+ regulatory T cells in multiple sclerosis (MS). He is currently working as a postdoctoral fellow in Dr. Terry B. Strom’s lab, where he is developing a molecular signature of transplant rejection.

Qiang Zhou studied immunology earning his Ph.D. in 2003 at the Peking University, China. He did postdoctoral research with Dr. Bryon Johnson at the Medical college of Wisconsin, WI from 2004–2006, and then joined the group of Dr. Wenda Gao in Transplant Research Center at Beth Israel Deaconess Medical Center, Harvard Medical School in 2007. Mechanisms and modulation of Treg differentiation are his major research interest.

Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interest statement: The authors declare that they have no competing financial interests.
References
- 1.Dillon SR, Sprecher C, Hammond A, et al. Interleukin 31, a cytokine produced by activated T cells, induces dermatitis in mice. Nat Immunol. 2004;5:752–760. doi: 10.1038/ni1084. [DOI] [PubMed] [Google Scholar]
- 2.Pflanz S, Hibbert L, Mattson J, et al. WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27. J Immunol. 2004;172:2225–2231. doi: 10.4049/jimmunol.172.4.2225. [DOI] [PubMed] [Google Scholar]
- 3.Shi Y, Wang W, Yourey PA, et al. Computational EST database analysis identifies a novel member of the neuropoietic cytokine family. Biochem Biophys Res Commun. 1999;262:132–138. doi: 10.1006/bbrc.1999.1181. [DOI] [PubMed] [Google Scholar]
- 4.Elson GC, Lelievre E, Guillet C, et al. CLF associates with CLC to form a functional heteromeric ligand for the CNTF receptor complex. Nat Neurosci. 2000;3:867–872. doi: 10.1038/78765. [DOI] [PubMed] [Google Scholar]
- 5.Senaldi G, Varnum BC, Sarmiento U, et al. Novel neurotrophin-1/B cell-stimulating factor-3: a cytokine of the IL-6 family. Proc Natl Acad Sci U S A. 1999;96:11458–11463. doi: 10.1073/pnas.96.20.11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derouet D, Rousseau F, Alfonsi F, et al. Neuropoietin, a new IL-6-related cytokine signaling through the ciliary neurotrophic factor receptor. Proc Natl Acad Sci U S A. 2004;101:4827–4832. doi: 10.1073/pnas.0306178101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diveu C, Lelievre E, Perret D, et al. GPL, a novel cytokine receptor related to GP130 and leukemia inhibitory factor receptor. J Biol Chem. 2003;278:49850–49859. doi: 10.1074/jbc.M307286200. [DOI] [PubMed] [Google Scholar]
- 8.Taga T, Kishimoto T. Gp130 and the interleukin-6 family of cytokines. Annu Rev Immunol. 1997;15:797–819. doi: 10.1146/annurev.immunol.15.1.797. [DOI] [PubMed] [Google Scholar]
- 9.Ghilardi N, Li J, Hongo JA, et al. A novel type I cytokine receptor is expressed on monocytes, signals proliferation, and activates STAT-3 and STAT-5. J Biol Chem. 2002;277:16831–16836. doi: 10.1074/jbc.M201140200. [DOI] [PubMed] [Google Scholar]
- 10.Boulay JL, O'Shea JJ, Paul WE. Molecular phylogeny within type I cytokines and their cognate receptors. Immunity. 2003;19:159–163. doi: 10.1016/s1074-7613(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 11.Broxmeyer HE, Li J, Hangoc G, et al. Regulation of myeloid progenitor cell proliferation/survival by IL-31 receptor and IL-31. Exp Hematol. 2007;35:78–86. doi: 10.1016/j.exphem.2007.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bilsborough J, Leung DY, Maurer M, et al. IL-31 is associated with cutaneous lymphocyte antigen-positive skin homing T cells in patients with atopic dermatitis. J Allergy Clin Immunol. 2006;117:418–425. doi: 10.1016/j.jaci.2005.10.046. [DOI] [PubMed] [Google Scholar]
- 13.Sonkoly E, Muller A, Lauerma AI, et al. IL-31: a new link between T cells and pruritus in atopic skin inflammation. J Allergy Clin Immunol. 2006;117:411–417. doi: 10.1016/j.jaci.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 14.Dreuw A, Radtke S, Pflanz S, et al. Characterization of the signaling capacities of the novel gp130-like cytokine receptor. J Biol Chem. 2004;279:36112–36120. doi: 10.1074/jbc.M401122200. [DOI] [PubMed] [Google Scholar]
- 15.Perrigoue JG, Li J, Zaph C, et al. IL-31-IL-31R interactions negatively regulate type 2 inflammation in the lung. J Exp Med. 2007;204:481–487. doi: 10.1084/jem.20061791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bravo J, Staunton D, Heath JK, Jones EY. Crystal structure of a cytokine-binding region of gp130. Embo J. 1998;17:1665–1674. doi: 10.1093/emboj/17.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boulanger MJ, Bankovich AJ, Kortemme T, Baker D, Garcia KC. Convergent mechanisms for recognition of divergent cytokines by the shared signaling receptor gp130. Mol Cell. 2003;12:577–589. doi: 10.1016/s1097-2765(03)00365-4. [DOI] [PubMed] [Google Scholar]
- 18.Boulanger MJ, Chow DC, Brevnova EE, Garcia KC. Hexameric structure and assembly of the interleukin-6/IL-6 alpha-receptor/gp130 complex. Science. 2003;300:2101–2104. doi: 10.1126/science.1083901. [DOI] [PubMed] [Google Scholar]
- 19.Plun-Favreau H, Perret D, Diveu C, et al. Leukemia inhibitory factor (LIF), cardiotrophin-1, and oncostatin M share structural binding determinants in the immunoglobulin-like domain of LIF receptor. J Biol Chem. 2003;278:27169–27179. doi: 10.1074/jbc.M303168200. [DOI] [PubMed] [Google Scholar]
- 20.Aasland D, Oppmann B, Grotzinger J, Rose-John S, Kallen KJ. The upper cytokine-binding module and the Ig-like domain of the leukaemia inhibitory factor (LIF) receptor are sufficient for a functional LIF receptor complex. J Mol Biol. 2002;315:637–646. doi: 10.1006/jmbi.2001.5282. [DOI] [PubMed] [Google Scholar]
- 21.Hammacher A, Richardson RT, Layton JE, et al. The immunoglobulin-like module of gp130 is required for signaling by interleukin-6, but not by leukemia inhibitory factor. J Biol Chem. 1998;273:22701–22707. doi: 10.1074/jbc.273.35.22701. [DOI] [PubMed] [Google Scholar]
- 22.Diveu C, Lak-Hal AH, Froger J, et al. Predominant expression of the long isoform of GP130-like (GPL) receptor is required for interleukin-31 signaling. Eur Cytokine Netw. 2004;15:291–302. [PubMed] [Google Scholar]
- 23.Gearing DP, Thut CJ, VandeBos T, et al. Leukemia inhibitory factor receptor is structurally related to the IL-6 signal transducer, gp130. Embo J. 1991;10:2839–2848. doi: 10.1002/j.1460-2075.1991.tb07833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gearing DP, Comeau MR, Friend DJ, et al. The IL-6 signal transducer, gp130: an oncostatin M receptor and affinity converter for the LIF receptor. Science. 1992;255:1434–1437. doi: 10.1126/science.1542794. [DOI] [PubMed] [Google Scholar]
- 25.Mosley B, De Imus C, Friend D, et al. Dual oncostatin M (OSM) receptors. Cloning and characterization of an alternative signaling subunit conferring OSM-specific receptor activation. J Biol Chem. 1996;271:32635–32643. doi: 10.1074/jbc.271.51.32635. [DOI] [PubMed] [Google Scholar]
- 26.Ichihara M, Hara T, Kim H, Murate T, Miyajima A. Oncostatin M and leukemia inhibitory factor do not use the same functional receptor in mice. Blood. 1997;90:165–173. [PubMed] [Google Scholar]
- 27.Lindberg RA, Juan TS, Welcher AA, et al. Cloning and characterization of a specific receptor for mouse oncostatin M. Mol Cell Biol. 1998;18:3357–3367. doi: 10.1128/mcb.18.6.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka M, Hara T, Copeland NG, et al. Reconstitution of the functional mouse oncostatin M (OSM) receptor: molecular cloning of the mouse OSM receptor beta subunit. Blood. 1999;93:804–815. [PubMed] [Google Scholar]
- 29.Fukada T, Hibi M, Yamanaka Y, et al. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 30.Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol. 2005;5:593–605. doi: 10.1038/nri1667. [DOI] [PubMed] [Google Scholar]
- 31.Chung CD, Liao J, Liu B, et al. Specific inhibition of Stat3 signal transduction by PIAS3. Science. 1997;278:1803–1805. doi: 10.1126/science.278.5344.1803. [DOI] [PubMed] [Google Scholar]
- 32.Liu B, Liao J, Rao X, et al. Inhibition of Stat1-mediated gene activation by PIAS1. Proc Natl Acad Sci U S A. 1998;95:10626–10631. doi: 10.1073/pnas.95.18.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu B, Gross M, ten Hoeve J, Shuai K. A transcriptional corepressor of Stat1 with an essential LXXLL signature motif. Proc Natl Acad Sci U S A. 2001;98:3203–3207. doi: 10.1073/pnas.051489598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chattopadhyay S, Tracy E, Liang P, et al. Interleukin-31 and oncostatin-M mediate distinct signaling reactions and response patterns in lung epithelial cells. J Biol Chem. 2007;282:3014–3026. doi: 10.1074/jbc.M609655200. [DOI] [PubMed] [Google Scholar]
- 35.Dambacher J, Beigel F, Seiderer J, et al. Interleukin 31 mediates MAP kinase and STAT1/3 activation in intestinal epithelial cells and its expression is upregulated in inflammatory bowel disease. Gut. 2007;56:1257–1265. doi: 10.1136/gut.2006.118679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broxmeyer HE, Bruns HA, Zhang S, et al. Th1 cells regulate hematopoietic progenitor cell homeostasis by production of oncostatin M. Immunity. 2002;16:815–825. doi: 10.1016/s1074-7613(02)00319-9. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka M, Hirabayashi Y, Sekiguchi T, et al. Targeted disruption of oncostatin M receptor results in altered hematopoiesis. Blood. 2003;102:3154–3162. doi: 10.1182/blood-2003-02-0367. [DOI] [PubMed] [Google Scholar]
- 38.Yagi Y, Andoh A, Nishida A, et al. Interleukin-31 stimulates production of inflammatory mediators from human colonic subepithelial myofibroblasts. Int J Mol Med. 2007;19:941–946. [PubMed] [Google Scholar]
- 39.Ip WK, Wong CK, Li ML, et al. Interleukin-31 induces cytokine and chemokine production from human bronchial epithelial cells through activation of mitogen-activated protein kinase signalling pathways: implications for the allergic response. Immunology. 2007;122:532–541. doi: 10.1111/j.1365-2567.2007.02668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnare M, Barton GM, Holt AC, et al. Toll-like receptors control activation of adaptive immune responses. Nat Immunol. 2001;2:947–950. doi: 10.1038/ni712. [DOI] [PubMed] [Google Scholar]
- 41.Eisenbarth SC, Piggott DA, Huleatt JW, et al. Lipopolysaccharide-enhanced, toll-like receptor 4-dependent T helper cell type 2 responses to inhaled antigen. J Exp Med. 2002;196:1645–1651. doi: 10.1084/jem.20021340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trautmann A, Akdis M, Brocker EB, Blaser K, Akdis CA. New insights into the role of T cells in atopic dermatitis and allergic contact dermatitis. Trends Immunol. 2001;22:530–532. doi: 10.1016/s1471-4906(01)02004-x. [DOI] [PubMed] [Google Scholar]
- 43.Leung DY, Bieber T. Atopic dermatitis. Lancet. 2003;361:151–160. doi: 10.1016/S0140-6736(03)12193-9. [DOI] [PubMed] [Google Scholar]
- 44.Woodward AL, Spergel JM, Alenius H, et al. An obligate role for T-cell receptor alphabeta+ T cells but not T-cell receptor gammadelta+ T cells, B cells, or CD40/CD40L interactions in a mouse model of atopic dermatitis. J Allergy Clin Immunol. 2001;107:359–366. doi: 10.1067/mai.2001.112695. [DOI] [PubMed] [Google Scholar]
- 45.Neis MM, Peters B, Dreuw A, et al. Enhanced expression levels of IL-31 correlate with IL-4 and IL-13 in atopic and allergic contact dermatitis. J Allergy Clin Immunol. 2006;118:930–937. doi: 10.1016/j.jaci.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 46.Takaoka A, Arai I, Sugimoto M, et al. Expression of IL-31 gene transcripts in NC/Nga mice with atopic dermatitis. Eur J Pharmacol. 2005;516:180–181. doi: 10.1016/j.ejphar.2005.04.040. [DOI] [PubMed] [Google Scholar]
- 47.Takaoka A, Arai I, Sugimoto M, et al. Involvement of IL-31 on scratching behavior in NC/Nga mice with atopic-like dermatitis. Exp Dermatol. 2006;15:161–167. doi: 10.1111/j.1600-0625.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- 48.Schulz F, Marenholz I, Folster-Holst R, et al. A common haplotype of the IL-31 gene influencing gene expression is associated with nonatopic eczema. J Allergy Clin Immunol. 2007;120:1097–1102. doi: 10.1016/j.jaci.2007.07.065. [DOI] [PubMed] [Google Scholar]
- 49.Yosipovitch G, Greaves MW, Schmelz M. Itch. Lancet. 2003;361:690–694. doi: 10.1016/S0140-6736(03)12570-6. [DOI] [PubMed] [Google Scholar]
- 50.Greaves MW, Khalifa N. Itch: more than skin deep. Int Arch Allergy Immunol. 2004;135:166–172. doi: 10.1159/000080898. [DOI] [PubMed] [Google Scholar]
- 51.Lee HL, Lee KM, Son SJ, Hwang SH, Cho HJ. Temporal expression of cytokines and their receptors mRNAs in a neuropathic pain model. Neuroreport. 2004;15:2807–2811. [PubMed] [Google Scholar]
- 52.Zhang N, Inan S, Cowan A, et al. A proinflammatory chemokine, CCL3, sensitizes the heat- and capsaicin-gated ion channel TRPV1. Proc Natl Acad Sci U S A. 2005;102:4536–4541. doi: 10.1073/pnas.0406030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hermann GE, Holmes GM, Rogers RC. TNF(alpha) modulation of visceral and spinal sensory processing. Curr Pharm Des. 2005;11:1391–1409. doi: 10.2174/1381612053507828. [DOI] [PubMed] [Google Scholar]
- 54.Morikawa Y, Tamura S, Minehata K, et al. Essential function of oncostatin M in nociceptive neurons of dorsal root ganglia. J Neurosci. 2004;24:1941–1947. doi: 10.1523/JNEUROSCI.4975-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamura S, Morikawa Y, Miyajima A, Senba E. Expression of oncostatin M receptor beta in a specific subset of nociceptive sensory neurons. Eur J Neurosci. 2003;17:2287–2298. doi: 10.1046/j.1460-9568.2003.02681.x. [DOI] [PubMed] [Google Scholar]
- 56.Steinhoff M, Bienenstock J, Schmelz M, et al. Neurophysiological, neuroimmunological, and neuroendocrine basis of pruritus. J Invest Dermatol. 2006;126:1705–1718. doi: 10.1038/sj.jid.5700231. [DOI] [PubMed] [Google Scholar]
- 57.Brand S, Sakaguchi T, Gu X, Colgan SP, Reinecker HC. Fractalkine-mediated signals regulate cell-survival and immune-modulatory responses in intestinal epithelial cells. Gastroenterology. 2002;122:166–177. doi: 10.1053/gast.2002.30329. [DOI] [PubMed] [Google Scholar]
- 58.Brand S, Dambacher J, Beigel F, et al. CXCR4 and CXCL12 are inversely expressed in colorectal cancer cells and modulate cancer cell migration, invasion and MMP-9 activation. Exp Cell Res. 2005;310:117–130. doi: 10.1016/j.yexcr.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 59.Brand S, Olszak T, Beigel F, et al. Cell differentiation dependent expressed CCR6 mediates ERK-1/2, SAPK/JNK, and Akt signaling resulting in proliferation and migration of colorectal cancer cells. J Cell Biochem. 2006;97:709–723. doi: 10.1002/jcb.20672. [DOI] [PubMed] [Google Scholar]
- 60.Brand S, Beigel F, Olszak T, et al. IL-22 is increased in active Crohn's disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am J Physiol Gastrointest Liver Physiol. 2006;290:G827–G838. doi: 10.1152/ajpgi.00513.2005. [DOI] [PubMed] [Google Scholar]
- 61.Jawa RS, Chattopadhyay S, Tracy E, et al. Regulated expression of the IL-31 receptor in bronchial and alveolar epithelial cells, pulmonary fibroblasts, and pulmonary macrophages. J Interferon Cytokine Res. 2008;28:207–219. doi: 10.1089/jir.2007.0057. [DOI] [PubMed] [Google Scholar]
- 62.Lei Z, Liu G, Huang Q, et al. SCF and IL-31 rather than IL-17 and BAFF are potential indicators in patients with allergic asthma. Allergy. 2008;63:327–332. doi: 10.1111/j.1398-9995.2007.01566.x. [DOI] [PubMed] [Google Scholar]
- 63.Hombria JC, Brown S. The fertile field of Drosophila Jak/STAT signalling. Curr Biol. 2002;12:R569–R575. doi: 10.1016/s0960-9822(02)01057-6. [DOI] [PubMed] [Google Scholar]
- 64.Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]