Abstract
ATP-dependent chromatin-remodeling complexes, such as RSC, can reposition, evict or restructure nucleosomes. A structure of a RSC–nucleosome complex with a nucleosome determined by cryo-EM shows the nucleosome bound in a central RSC cavity. Extensive interaction of RSC with histones and DNA seems to destabilize the nucleosome and lead to an overall ATP-independent rearrangement of its structure. Nucleosomal DNA appears disordered and largely free to bulge out into solution as required for remodeling, but the structure of the RSC–nucleosome complex indicates that RSC is unlikely to displace the octamer from the nucleosome to which it is bound. Consideration of the RSC–nucleosome structure and published biochemical information suggests that ATP-dependent DNA translocation by RSC may result in the eviction of histone octamers from adjacent nucleosomes.
The mobilization of nucleosomes is a prerequisite for DNA transactions. Nucleosomes are repeatedly removed and reassembled in promoter regions, resulting in transient exposure of the DNA for interaction with the transcription machinery1,2. Chromatin modification is also essential in the process of double-strand break repair3. The prime candidates for nucleosome removal are the SWI/SNF family of chromatin-remodeling complexes, which relieve repression by nucleosomes in vivo and perturb nucleosome structure in an ATP-dependent manner in vitro.
Whereas SWI and SNF genes are nonessential in yeast, and their protein products are present at low levels, homologs of these genes encode the components of the essential, abundant RSC complex4. Biochemical and structural studies of RSC have illuminated the chromatin-remodeling process. RSC binds a nucleosome core particle with nanomolar affinity5 and reduces digestion of nucleosomal DNA by nucleases6. A three-dimensional reconstruction of RSC calculated from EM images of single particles preserved in stain revealed a cavity able to accommodate a nucleosome and likely to afford the observed nuclease protection6. Addition of ATP to a RSC–nucleosome complex leads to sliding of the histone octamer along the DNA7 or to transfer of the octamer, either to a histone chaperone8 or to another DNA molecule9. The underlying principle of these activities10 is the coupling of ATP hydrolysis to DNA translocation by Sth1, a member of the Swi2/Snf2 family of DNA-dependent ATPases conserved among chromatin-remodeling complexes. Sth1 contacts the DNA approximately two turns from the dyad of the nucleosome and propels it across the histone octamer surface10. Sth1 alone, or even a catalytic fragment, is capable of this effect. Movement of the DNA occurs by bending, not twisting, of the double helix11,12, and recent studies have indicated that upon ATP hydrolysis RSC can form large (>100 base pairs (bp)) loops on both free DNA13 and nucleosomal14 substrates.
How can RSC both protect nucleosomal DNA and facilitate its movement and accessibility for transcription? Why are additional subunits of RSC required, when the Sth1 subunit alone is sufficient for remodeling activity? How is RSC action restricted to promoter nucleosomes and other limited regions of chromosomes? Here we report the structure of a Saccharomyces cerevisiae RSC–nucleosome complex to gain structural insight into such questions.
RESULTS
Calculation of RSC cryo-EM reconstruction
A previous EM reconstruction of RSC preserved in stain revealed its surface topography at 25–30-Å resolution6. Protein densities were seen surrounding a central cavity, large enough to accommodate a nucleosome. Reconstruction of a RSC–nucleosome complex in stain revealed additional density in the cavity, but less than expected for a nucleosome (Supplementary Fig. 1 online). To improve the quality of the EM reconstruction and avoid artifacts related to staining, we turned to unstained specimens of RSC and RSC–nucleosome complexes.
We applied RSC particles to carbon-coated EM grids, which were then frozen in amorphous ice and imaged under low-dose conditions (Fig. 1a). Using a previously published three-dimensional reconstruction of RSC6 as initial reference, we refined the RSC cryo-images by conventional projection matching15 and obtained a structure that seemed reasonable (Supplementary Fig. 2 online) but that was noisy and failed to increase in resolution as the refinement progressed. Detailed testing revealed that, whereas images corresponding to front views of the RSC complex (prevalent in stained samples) were correctly aligned, alignment of other views was mostly incorrect. This was almost certainly due to a lack of structural details in the front and back of the initial reference volume, resulting from a combination of particle flattening and incomplete sampling in stained RSC specimens.
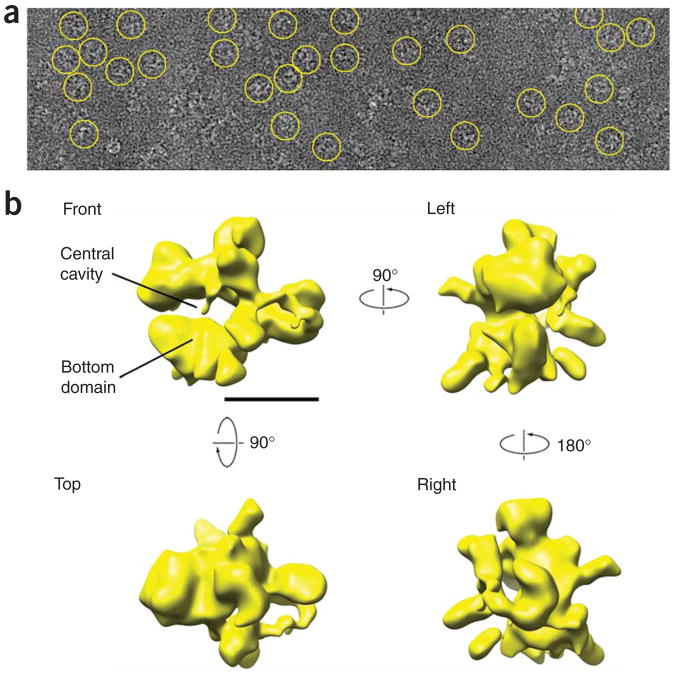
Figure 1.
Cryo-EM RSC data and reconstruction of the RSC complex. (a) Raw images of individual RSC particles preserved in a thin layer of amorphous ice. Individual RSC particles are highlighted by yellow circles. Several different views of the complex are apparent. (b) Three-dimensional structure of RSC calculated from EM images of particles preserved in amorphous ice. Four views of the RSC reconstruction are shown at a threshold value corresponding to the total mass of the complex (~1.3 MDa). The scale bar represents 100 Å.
To address this problem, we subjected about half of the images in the data set (5,200 images with the highest defocus values and therefore highest contrast) to alternating cycles of supervised (guided by the three-dimensional reconstruction of RSC in stain) and unsupervised alignment and classification, in which new projections were merged with previous ones using a modified common-lines algorithm (Supplementary Discussion, Supplementary Methods and Supplementary Fig. 3 online). This procedure yielded an initial low-resolution cryo-reconstruction of RSC (Supplementary Fig. 4 online) that was then used as an initial reference for refinement by projection matching15 of ~26,000 images of frozen, hydrated RSC particles. This resulted in a final RSC reconstruction (Fig. 1b and Supplementary Movie 1 online) that was contoured at a threshold level chosen to give a volume appropriate for the mass of a RSC particle (~1.3 MDa). The resolution of the RSC cryo-EM reconstruction was estimated at ~25 Å by the 0.5 Fourier shell cutoff criterion16 (Supplementary Fig. 5a online, left).
The accuracy of the final cryo-EM RSC reconstruction is supported by two observations. First, comparison of the cryo-EM RSC reconstruction to three independent RSC reconstructions calculated from specimens preserved in stain revealed an appreciable level of similarity to two of them6,17 and an overall resemblance to a third one18, establishing that the overall shape of the cryo-EM reconstruction is correct. Second, reprojections of the cryo-EM reconstruction of RSC closely resembled two-dimensional averages obtained directly from the EM images by reference-free alignment (Supplementary Fig. 5b), demonstrating that the different views and finer details of the cryo-EM reconstruction are also correct.
Conformational changes in the RSC complex
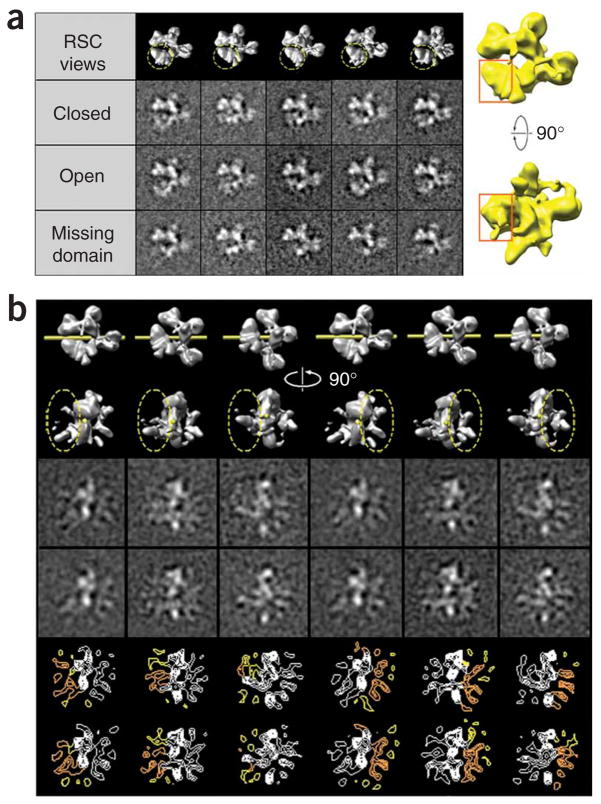
As observed in previous EM studies, RSC particles show different conformations, most notably, changes in the position of the domain that forms the bottom portion of the structure6,17. These changes are most reliably characterized in projections related to the front view of the RSC structure, where the cavity is easily distinguished. In agreement with previous reports6,17, statistical analysis and classification of RSC images related to the front view of the complex revealed open and closed conformations (Fig. 2a), and a small proportion (~16%) of particles in which part of the bottom domain seems to be missing. We observed the most appreciable changes in the distal portion of the bottom domain, which varies in position and/or conformation (Fig. 2a, right, and Supplementary Movie 2 online).
Figure 2.
Statistical analysis of domain mobility in the RSC complex. (a) Multivariate statistical analysis was used to characterize the mobility of the bottom portion of the RSC structure (as seen in front views of the complex). Two-dimensional image classification was carried out focusing on pixels inside a mask surrounding the bottom domain (‘RSC views’, broken yellow contour). Class averages corresponding to ‘closed’ and ‘open’ conformations of the complex were obtained, as well as a third type of class average in which part of the bottom domain was undetectable (‘Missing’). Appreciable changes in the position and/or conformation of the domain are restricted to its left half (right, highlighted by the red rectangle). (b) Focused classification using a mask on either the front or back of side projections of the RSC complex revealed mobility of domains protruding forward and backward from the central RSC density. Contour plots help to show that features apparent in the three-dimensional reconstruction appear in slightly different positions in class averages generated by classification of images corresponding to a single projection of the RSC structure. Varying positions for flexible features evident in the three-dimensional RSC reconstruction are depicted in orange. Yellow contours highlight density that is consistently detected after classification of two-dimensional images but that is too mobile to appear in the three-dimensional structure at the threshold used for rendering.
No substantial changes in the position of other major RSC domains were observed, but it seemed likely that mobility would also affect smaller portions of the RSC structure. In fact, statistical analysis of RSC images corresponding to various side views of the complex indicated that smaller densities extending from the front and back of the RSC structure (and that were not resolved in any of the previous RSC reconstructions from stained specimens) show appreciable mobility (Fig. 2b). Class averages obtained through focused classification show densities that correspond to features in the cryo-EM reconstruction, but whose position changes from one class average to the next. Reproducibility of the features at varying positions indicates that they are real, but correspond to mobile RSC domains. Indeed, overall mobility of the RSC structure is the most likely explanation for the relatively limited resolution attained by the cryo-EM analysis of the complex.
Statistical analysis of RSC–nucleosome cryo-EM images
For EM analysis of the RSC–nucleosome complex, we mixed RSC complex with a four-fold molar excess of nucleosomes. Quantitative analysis of the RSC–nucleosome interaction, performed under the conditions used for EM sample preparation, gave a Kd value of ~7 nM (Supplementary Fig. 6a online). Previous analysis in stain had detected density in the central RSC cavity after incubation of the complex with nucleosomes (Supplementary Fig. 1). However, the detection of partially disordered structural elements near the cavity made it crucial to establish whether any changes observed after incubation with the nucleosomes were directly related to the presence of a nucleosome.
First, it was important to determine whether incubation of RSC with nucleosomes led to an overall rearrangement of the RSC structure. To this end, we separated images in the RSC–nucleosome data set (~37,000) roughly according to their orientation by aligning them to reprojections of the RSC reconstruction. Comparison of the outcomes from repeated rounds of reference-free alignment within the resulting RSC–nucleosome classes indicated that RSC–nucleosome images had been successfully segregated into homogeneous groups by alignment to projections of the RSC structure and established an overall similarity between images of the RSC–nucleosome complex and RSC alone.
Having established that incubation with nucleosomes did not cause large-scale changes in RSC structure, it was important to determine whether smaller differences between images of RSC alone and the RSC–nucleosome complex were directly related to the presence of a nucleosome. To this end, we combined the RSC and RSC–nucleosome data sets to use the RSC images as an internal control during image analysis. Statistical analysis was focused on projections related to the front view of the RSC structure, in which the central cavity where the nucleosome was likely to bind was most apparent. We used correspondence analysis and classification19 to separate images according to density variations in and around the central cavity. The particle images were clearly separated between groups with and without density in the cavity, with the first eigenvector (corresponding to the most statistically significant source of variability in the images) accounting for most of the observed differences and apparently relating directly to the presence of additional density (Supplementary Fig. 6b). Plotting the distribution of images classified along the first eigenvector indicated a single class of particles with an empty cavity for the RSC data set, whereas images from the RSC–nucleosome data set were divided into two classes, with ~65% of the particles classified as having an occupied cavity and ~35% assigned to a group with an empty cavity (Supplementary Fig. 6c). Final classification of the mixed RSC and RSC–nucleosome data set resulted in a clear distinction between particles with and without density in the central cavity (Supplementary Fig. 6d, first and second rows, respectively). Differences between occupied and empty averages for a given orientation revealed the shape of the density in the RSC cavity (Supplementary Fig. 6d, below).
Refinement of RSC–nucleosome cryo-EM data
Having obtained direct evidence that the RSC–nucleosome images contained additional density in the cavity and that incubation of RSC with nucleosomes did not cause an appreciable change in RSC structure, we subjected the RSC–nucleosome data to three-dimensional reconstruction by projection matching, using a low-pass filtered RSC volume as initial reference. Density in the cavity was immediately apparent. Because the concentration of RSC used to prepare EM samples (~20 nM) was close to the estimated Kd for formation of the RSC–nucleosome complex (Supplementary Fig. 6a), and because two-dimensional image analysis indicated that a fraction (about one-third) of images in the RSC–nucleosome data set corresponded to RSC alone (Supplementary Fig. 6c), we analyzed images in the RSC–nucleosome data set using a competitive projection-matching protocol20,21 in which they were compared to projections of the initial RSC–nucleosome complex and RSC alone reconstructions. In agreement with the results obtained from two-dimensional classification, 65 ± 10% of the RSC–nucleosome images were matched to projections of the RSC–nucleosome structure, with the rest showing higher cross-correlation to projections of RSC alone.
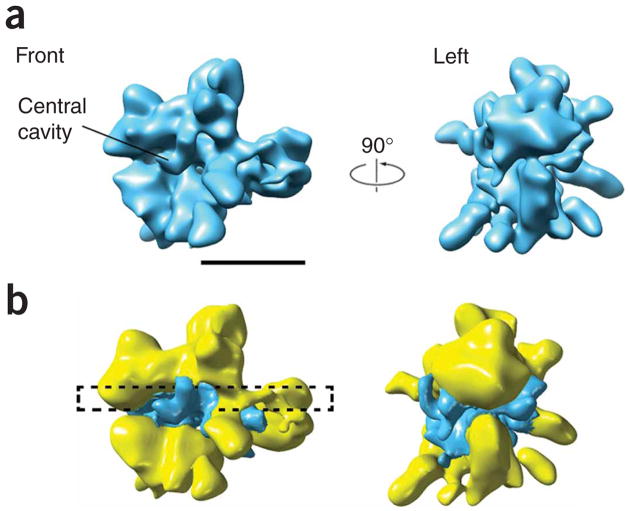
Finally, we subjected the images corresponding to actual RSC–nucleosome complexes (~20,000) to refinement by projection matching using the RSC reconstruction as an initial reference. The resulting RSC–nucleosome reconstruction was close in overall appearance to the RSC reconstruction. However, when contoured at a threshold level at which the RSC portion of the reconstruction matched the reconstruction of RSC alone, the RSC–nucleosome structure showed an occupied central cavity (Fig. 3a). Difference mapping revealed negligible local changes throughout the structure (Supplementary Movie 3 online) and, in agreement with the results from two-dimensional image analysis, the presence of additional density in the central cavity and its immediate vicinity (Fig. 3b).
Figure 3.
Cryo-EM analysis of the RSC–nucleosome complex. (a) Three-dimensional reconstruction of the RSC–nucleosome complex. Two views of the complex (corresponding to views of the RSC structure shown in Figure 1) show a close correspondence to the structure of RSC alone. The threshold for the RSC–nucleosome reconstruction was set to match the size of the RSC density in the three-dimensional reconstruction of RSC. The only substantial difference between the RSC and RSC–nucleosome structures is the presence of additional density occupying the central cavity in the RSC–nucleosome reconstruction. (b) A difference map (surface represented in blue) obtained by subtracting the RSC reconstruction from that of the RSC–nucleosome complex shows the shape and location of extra density related to the presence of a nucleosome. A black, broken rectangle highlights the portion (depicted in Figure 4a) of the RSC–nucleosome structure that contains most of the nucleosome-related density.
Analysis of the RSC–nucleosome cryo-EM reconstruction
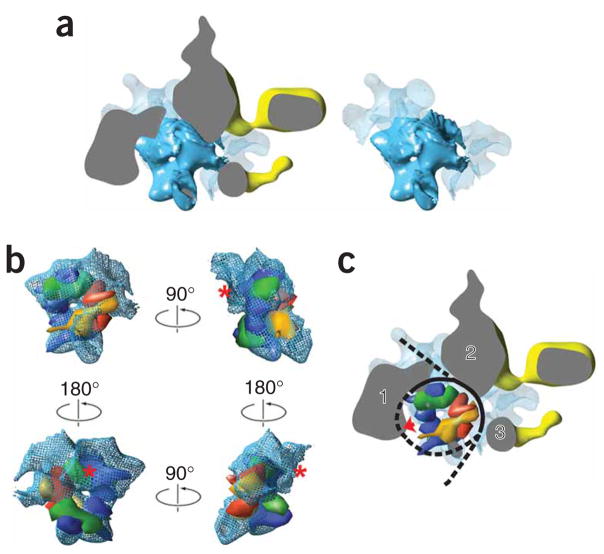
Image analysis conclusively established that density in the center of the RSC–nucleosome reconstruction resulted from incubation of RSC with nucleosomes. However, unexpectedly, the shape of the density in the cavity of the RSC–nucleosome reconstruction did not match the shape of a nucleosome. The central portion of this density seemed to correspond in size and shape to a histone octamer. This was particularly apparent in a top view, where the outline of the histones and a characteristic dimple at the center of the octamer could be easily identified, immediately surrounded by either RSC, or what seemed to be poorly ordered DNA density (Fig. 4a). Docking of a resolution-matched model of the histone octamer derived from its X-ray structure confirmed the match and indicated the orientation of the octamer in the cavity (Fig. 4b and Supplementary Movie 4 online). We were only partially able to resolve the density for one of the H2A-H2B dimers (marked by a red asterisk in Fig. 4b). This could be due to partial dissociation or to inadequate segmentation of the dimer and RSC density resulting from the limited resolution of the cryo-EM reconstruction compounded with close association of the dimer with the mobile bottom domain of RSC. Finally, the fit of the histones in the central density is not perfect, and the possibility that interaction with RSC might induce changes in the structure of the octamer cannot be excluded.
Figure 4.
Analysis of the density in the central cavity of the RSC–nucleosome reconstruction and comparison with the X-ray structure of the nucleosome. (a) A top view of the RSC–nucleosome complex (left) showing only a central slab (denoted by the broken rectangle in Figure 3b) that includes density resulting from nucleosome binding. Density presumably corresponding to the histone core (shown in solid blue) comes into close contact with RSC density (shown in yellow and gray) in several places. Non-RSC density adjacent to the central histone density is rendered as a semitransparent light blue surface. The distribution of density resulting from nucleosome binding is best appreciated after the removal (by difference mapping) of RSC density (right). (b) Comparison of presumed histone density with a low-resolution (25 Å) model of the histone core derived from its X-ray structure39 shows a close correspondence in shape and size. In these views, the histones are shown in space-filling representation: H2A, yellow; H2B, red; H3, blue; H4, green. See text for details. (c) The same slabbed top view of the RSC–nucleosome structure shown in a but with a model of the histones docked in place. RSC densities in close contact with the nucleosome are designated 1–3. The proximity of density 1 to the dyad suggests that it probably corresponds to the Sth1 ATPase subunit. Absence of DNA density around the dyad suggests that binding of Sth1 pulls DNA away from the histones (as indicated by the red arrow), which would explain the origin of a reported DNase I hypersensitivity site near the dyad generated by RSC binding in the absence of ATP10. Possible changes in the arrangement of nucleosomal DNA are schematically represented by a black line, with regions where no DNA density is apparent in the RSC–nucleosome reconstruction shown with a broken line.
A determination of the distribution of DNA density surrounding the octamer was complicated by limited resolution and the close proximity of the RSC domains. Density adjacent to the histones and probably corresponding to nucleosomal DNA was readily apparent in two areas that have no RSC density nearby, but not in areas where RSC comes into close contact with the nucleosome or near the dyad (Fig. 4a). In contrast, reconstructions of nucleosomes on their own, preserved in stain (Y.C. and F.J.A., unpublished data) or in the frozen, hydrated state22, show clear DNA density surrounding the histone octamer. Because the RSC–nucleosome images were recorded under conditions where no DNA translocation was possible (no ATP was present), we surmise that our detection of DNA density was limited by a combination of mobility and limited segmentation from the closely associated RSC density. Interaction of the nucleosome with RSC might alter the balance of histone-DNA contacts that underlie nucleosome stability and result in changes in nucleosome structure.
The histone octamer fits tightly into the RSC cavity, leading to an extensive interface between RSC and the free (top and bottom) surfaces of the octamer (Supplementary Movie 4). RSC–nucleosome interactions along the periphery of the nucleosome are more localized, with RSC density coming into contact with the sides of the nucleosome at a site near the dyad (Fig. 4c, protein density 1) and also interacting with it three to four helical turns to the right and left of the dyad (Fig. 4c, protein densities 2 and 3, respectively). These positions must correspond to some of the multiple sites of RSC-DNA interaction previously revealed by cross-linking23, and one of them should correspond to the location of Sth1. No high-resolution structure of Sth1 is available, but the X-ray structure of the core domain of the Sulfolobus solfataricus Swi2/Snf2 ATPase complexed with DNA24 (30% identity and 32% similarity to the core domain of Sth1) could be docked into protein densities 1 or 2 in our model, resulting in a reasonable alignment of the DNA of the X-ray structure with the expected position of nucleosomal DNA. However, only docking into density 1 is consistent with the position for interaction of Sth1 with the nucleosome suggested by functional studies. Interaction of RSC with the nucleosome results in marked sensitivity to DNase I digestion at a site approximately two turns from the nucleosomal dyad (that is, the center of the 146-bp DNA fragment), which seems to result directly from binding of the ATPase portion of Sth1 (ref. 10) at that position. Features in the RSC–nucleosome reconstruction are consistent with this observation. The outline of the H3-H4 tetramer is apparent in the RSC–nucleosome reconstruction, consistent with the idea that RSC binding leads to separation of DNA from the histones and formation of a bulge around the dyad. Such disruption of DNA-histone contacts near the dyad may also contribute to the apparent disordering of nucleosomal DNA near the ends of the nucleosome (Fig. 4c).
DISCUSSION
Effect of RSC binding on the nucleosome
Previous reports attest to the specific effects of RSC binding on the nucleosome. Besides the aforementioned generation of a hypersensitivity site10, reexamination of our original DNase I footprinting results6 and recent analysis by others10 indicate that RSC binding alone (in the absence of ATP hydrolysis) results in increased sensitivity (that is, diminished interaction with the histone octamer) of DNA near the entry and exit points. These changes might be relevant to a general remodeling mechanism, as binding of the ATPase subunits of the SWI/SNF and ISW2 remodelers also seems to occur near the dyad12,25, and nucleosome binding by the ATP-dependent chromatin assembly factor (ACF) remodeler also results in ATP-independent detachment of nucleosomal DNA26. Such results support our interpretation of the RSC–nucleosome cryo-EM reconstruction: although the overall structure of the nucleosome is maintained after RSC binding in the absence of ATP, interaction with RSC results in extensive ATP-independent rearrangement of the nucleosome. The relevance of these observations to the mechanism of RSC actions is supported by the ability of RSC to remodel nucleosomes such as those used in this analysis8. A recent study reported on interaction of the SWI/SNF complex with linker DNA. Although our model is not inconsistent with the possibility of such interactions for RSC, it is important to note that nucleosome binding, DNA translocation and histone eviction by RSC are not dependent on the presence or length of linker DNA10.
Implications for chromatin remodeling
Our model of the RSC–nucleosome complex has several implications for the regulation and mechanism of chromatin remodeling. First, the embrace of the nucleosome by RSC would preclude complex formation in condensed chromosomal material and, on the basis of our structure, render the great majority of nucleosomes inaccessible to RSC. This could conceivably help to direct RSC activity toward promoter nucleosomes. Indeed, it has been reported that the linker histone H1 renders chromatin fibers resistant to remodeling27 and that reduced H1 binding following its phosphorylation promotes remodeling28. Additional targeting specificity may arise from extensive RSC-histone interactions apparent in the RSC–nucleosome reconstruction. Second, partial disordering and exposure to the solution of DNA in the RSC–nucleosome complex would allow for formation and propagation of a DNA bulge that could mediate translocation13,14. Formation of a DNA bulge might result directly from Sth1 binding near the dyad, where the RSC–nucleosome cryo-EM reconstruction shows diminished DNA-histone interactions (Fig. 4c). Finally, the tight fit of the wedge-shaped histone octamer in the RSC cavity (Supplementary Movie 4) would prevent rotation of the octamer and unspooling of the DNA. Therefore, RSC would be unable to dislodge the octamer from the nucleosome with which it is associated. A recent study of chromatin remodeling at the yeast PHO5 promoter revealed the removal of all but one nucleosome upon activation29. This finding could be explained by the sliding of a nucleosome across the promoter region by a remodeler such as RSC, unraveling DNA and ejecting histone octamers from nucleosomes in its path25,30 and leaving only the nucleosome bound to the remodeling complex at the end of the process.
METHODS
Electron microscopy sample preparation and data collection
We purified RSC as described4. Nucleosomes were prepared from rat liver histone octamers and a 160-bp DNA fragment containing the nucleosome-positioning sequence of the Xenopus laevis 5S ribosomal RNA (rRNA gene) as described8, and they were stored in buffer containing 10mM Tris, 10mM NaHSO3, 0.01 mM EDTA, pH 7.5. To prepare RSC EM samples, we diluted RSC aliquots (~420 μg ml−1 in 100 mM potassium acetate, 20mM HEPES, 10% (v/v) glycerol, 1 mM DTT, 1 mM EDTA, 0.01% (v/v) Nonidet P-40 and 1× protease inhibitors, pH 7.5) to a final concentration of ~25 μg ml−1 with a buffer containing 15 mM HEPES, 3 mM MgCl2 and 100 mM potassium acetate, pH 7.5. About 3 μl of protein was applied to a freshly glow-discharged (in the presence of amyl amine) carbon-coated Maxtaform, 300-mesh Cu/Rh grids (Ted Pella, Inc.) and preserved by flash freezing in amorphous ice31.
We assessed nucleosome binding by RSC by gel electrophoresis as described5. The Kd value (~7 nM) was determined from the slope of a double reciprocal plot of the intensity of the free-nucleosome band in the gel as a function of RSC concentration. To prepare the RSC–nucleosome complex, nucleosomes were mixed with RSC at a molar ratio of ~4:1 and diluted (15 mM HEPES, 3 mM MgCl2, 50 mM potassium acetate, pH 7.5) to obtain a final RSC concentration of ~25 μg ml−1. The mixture was incubated at ~25 °C for 15 min. We prepared EM samples as described for RSC alone.
We collected EM images under low-dose conditions using either a CM200 FEG or a Tecnai F20 microscope (Philips/FEI) equipped with field emission gun, operating at an accelerating voltage of 120 kV. Images were recorded on Kodak SO-163 film, at a magnification of ×66,000 or ×62,000 and with underfocus values between 1.3 and 3.2 μm. Micrographs were digitized on a Zeiss/SCAI flat bed densitometer (ZI/Zeiss) using a step size of 7 μm. Digitized images were two-fold pixel-averaged, resulting in a final pixel size corresponding to 2.06 Å.
A total of 159 micrographs of the RSC complex and 198 micrographs of the RSC–nucleosome complex were digitized, yielding ~26,000 and ~37,000 particle images, respectively. The data sets were divided into defocus groups according to defocus values calculated independently for 12 distinct sections of every micrograph. All image analysis was carried out using the SPIDER software package32.
Calculation of the RSC cryo–electron microscopy reconstruction
We used a published reconstruction of RSC obtained by using the Random Conical Tilt (RCT) method33 and images of RSC particles preserved in stain6 as a starting point for determination of an initial cryo-EM RSC reconstruction using a hybrid strategy for particle-orientation determination. In this hybrid strategy, supervised and unsupervised alignment and classification were used in combination with a modified version of the common-lines algorithm implemented in the SPIDER image analysis package32, to progressively compensate for distortion of the initial RCT RSC volume caused by particle dehydration and flattening and missing cone artifacts (Supplementary Fig. 3). We further refined the resulting RSC cryo-EM reconstruction using an iterative projection-matching algorithm15 with angular separation between reference projections progressively decreasing from 15° to 2°. Consistency of the reconstructed volume with the cyro-EM data was monitored by comparing reprojections of the volume to averages obtained directly from the original images by multiple rounds of the reference-free alignment34 performed in corresponding particle classes defined by supervised classification.
RSC structure-flexibility analysis
For domain-flexibility analysis, the effect of the contrast transfer function (CTF) was minimized by phase flipping, and RSC images were low-pass filtered to a spatial frequency corresponding to one-fourteenth of an angstrom. Images were divided into 195 reference groups according to alignment parameters determined by matching to reference projections calculated from the three-dimensional RSC reconstruction using an angular separation of 10°. We generated three-dimensional masks covering regions of interest and, from them, obtained two-dimensional masks for each specific projection. Correspondence analysis35 of the densities under the two-dimensional masks, followed by Hierarchical Ascendant Classification using the Ward’s criterion36, were used to generate class averages.
Two-dimensional occupancy analysis in RSC–nucleosome images
Images of RSC–nucleosome particles were CTF corrected, low-pass filtered and divided into groups by alignment to projections of the RSC cryo-EM reconstruction, as described above for the RSC data. RSC and RSC–nucleosome images corresponding to the same projection directions were combined, keeping track of the data set from which each image originated. We created a cylindrical three-dimensional mask including the central RSC cavity and surrounding regions and used it to generate twp-dimensional masks for each projection direction. Multivariate statistical analysis and classification were performed as described above. Analysis of eigenimages, as well as the distribution of particle images along eigenvectors, immediately indicated sharp differences between RSC and RSC–nucleosome data sets. Final image classification using 12 factors yielded class averages showing well-defined differences in the central cavity, as highlighted by difference mapping.
Calculation of the RSC–nucleosome cryo–electron microscopy reconstruction
We subjected RSC–nucleosome data to ten rounds of refinement by projection matching using as the initial reference a low-pass filtered RSC volume. Additional density in the RSC central cavity became immediately apparent. However, because the two-dimensional image analysis suggested heterogeneity of the RSC–nucleosome data set, we used a competitive-refinement strategy20,21 to identify images in the RSC–nucleosome data corresponding to full complexes. Images in the RSC–nucleosome data set were iteratively cross-correlated to projections of the RSC and RSC–nucleosome volumes and classified as occupied (nucleosome bound) or empty (without a nucleosome). The portion of the RSC–nucleosome data classified as occupied was then refined using the structure of RSC alone as the initial reference. As part of the nucleosome density in the RSC–nucleosome reconstruction is disordered, the appropriate threshold for rendering the RSC–nucleosome reconstruction could not be established simply from the anticipated mass of the complex and was instead determined by matching the RSC portion of the two volumes.
Difference mapping and comparison with the nucleosome X-ray structure
We carried out volume inspection and comparison with the X-ray structure of the nucleosome using Chimera37, which was also used to generate figures. Before difference mapping, all volume densities below the threshold value chosen for surface representation were set to zero. Volume densities were then normalized, and the RSC volume was subtracted from the RSC–nucleosome volume. The only substantial feature in the difference map was a large central density (several small, disconnected peripheral densities representing minor changes in the position or surface appearance of poorly ordered domains were removed from subsequent analysis by flood filling the main density of the difference map using the Situs software package38). The structure of the nucleosome39, rendered at a resolution comparable to the resolution of the RSC–nucleosome model was docked into the difference volume using the program COAN40.
Supplementary Material
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Footnotes
AUTHOR CONTRIBUTIONS
B.M.-D. purified RSC and nucleosomes; Y.L. established optimal conditions and determined affinity for RSC–nucleosome interaction; C.E. and W.-H.C. worked on the initial RSC and RSC–nucleosome cryo–reconstructions; F.Z. collected a portion of the RSC cryo-EM data; Y.C. collected additional cryo-EM data and was responsible for refinement and analysis of the final cryo-EM reconstructions; R.D.K. and F.J.A. interpreted the results and wrote the manuscript.
References
- 1.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 2.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Karagiannis TC, El-Osta A. Chromatin modifications and DNA double-strand breaks: the current state of play. Leukemia. 2007;21:195–200. doi: 10.1038/sj.leu.2404478. [DOI] [PubMed] [Google Scholar]
- 4.Cairns BR, et al. RSC, an abundant and essential chromatin remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 5.Lorch Y, Cairns BR, Zhang M, Kornberg RD. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 6.Asturias FJ, Chung WH, Kornberg RD, Lorch Y. Structural analysis of the RSC chromatin-remodeling complex. Proc Natl Acad Sci USA. 2002;99:13477–13480. doi: 10.1073/pnas.162504299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lorch Y, Zhang M, Kornberg RD. RSC unravels the nucleosome. Mol Cell. 2001;7:89–95. doi: 10.1016/s1097-2765(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 8.Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc Natl Acad Sci USA. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorch Y, Zhang M, Kornberg RD. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- 10.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 11.Lorch Y, Davis B, Kornberg RD. Chromatin remodeling by DNA bending, not twisting. Proc Natl Acad Sci USA. 2005;102:1329–1332. doi: 10.1073/pnas.0409413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 13.Lia G, et al. Direct observation of DNA distortion by the RSC complex. Mol Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, et al. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Penczek PA, Grassucci RA, Frank J. The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 16.Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J Microsc. 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 17.Skiniotis G, Moazed D, Walz T. Acetylated histone tail peptides induce structural rearrangements in the RSC chromatin remodeling complex. J Biol Chem. 2007;282:20804–20808. doi: 10.1074/jbc.C700081200. [DOI] [PubMed] [Google Scholar]
- 18.Leschziner AE, et al. Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc Natl Acad Sci USA. 2007;104:4913–4918. doi: 10.1073/pnas.0700706104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. Academic Press; San Diego: 1996. p. 342. [Google Scholar]
- 20.Craighead JL, Chang WH, Asturias FJ. Structure of yeast RNA polymerase II in solution. Implications for enzyme regulation and interaction with promoter DNA. Structure. 2002;10:1117–1125. doi: 10.1016/s0969-2126(02)00813-4. [DOI] [PubMed] [Google Scholar]
- 21.Gao H, Valle M, Ehrenberg M, Frank J. Dynamics of EF-G interaction with the ribosome explored by classification of a heterogeneous cryo-EM dataset. J Struct Biol. 2004;147:283–290. doi: 10.1016/j.jsb.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Leforestier A, Dubochet J, Livolant F. Bilayers of nucleosome core particles. Biophys J. 2001;81:2414–2421. doi: 10.1016/S0006-3495(01)75888-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sengupta SM, et al. The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J Biol Chem. 2001;276:12636–12644. doi: 10.1074/jbc.m010470200. [DOI] [PubMed] [Google Scholar]
- 24.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 25.Dechassa ML, et al. Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol. 2008;28:6010–6021. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strohner R, et al. A ‘loop recapture’ mechanism for ACF-dependent nucleosome remodeling. Nat Struct Mol Biol. 2005;12:683–690. doi: 10.1038/nsmb966. [DOI] [PubMed] [Google Scholar]
- 27.Saeki H, et al. Linker histone variants control chromatin dynamics during early embryogenesis. Proc Natl Acad Sci USA. 2005;102:5697–5702. doi: 10.1073/pnas.0409824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horn PJ, et al. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat Struct Biol. 2002;9:263–267. doi: 10.1038/nsb776. [DOI] [PubMed] [Google Scholar]
- 29.Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133:716–726. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dubochet J, et al. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 32.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 33.Radermacher M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J Electron Microsc Tech. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- 34.Penczek P, Radermacher M, Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- 35.Borland L, Vanheel M. Classification of image data in conjugate representation spaces. J Opt Soc Am A Opt Image Sci Vis. 1990;7:601–610. [Google Scholar]
- 36.Ward JH. Hierarchical grouping to optimize an objective function. Am Stat Assoc J. 1963;58:236–244. [Google Scholar]
- 37.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Wriggers W, Birmanns S. Using situs for flexible and rigid-body fitting of multi-resolution single-molecule data. J Struct Biol. 2001;133:193–202. doi: 10.1006/jsbi.2000.4350. [DOI] [PubMed] [Google Scholar]
- 39.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 40.Volkmann N, Hanein D. Docking of atomic models into reconstructions from electron microscopy. Methods Enzymol. 2003;374:204–225. doi: 10.1016/S0076-6879(03)74010-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.