Abstract
Double stranded RNAs (dsRNA) molecules targeted to gene promoter regions can induce transcriptional gene silencing in a DNA cytosine methylation dependent manner in plants (RNA-dependent DNA methylation or RdDM).1-3 Whether a similar mechanism exists in mammalian systems is a vital and currently controversial issue.4-6 DNA methylation is an important component in mammalian gene silencing for normal processes such as gene imprinting and x-chromosome inactivation,7-9 and aberrant CpG island hypermethylation at tumor suppressor promoters is associated with transcriptional silencing and loss of gene function in cancer.10 Hence, we investigated whether RdDM may operate in human cancers to mediate epigenetic silencing using the endogenous CDH1 gene as a potential target. The loss of this cell-cell adhesion factor facilitates the metastatic process, and its promoter is frequently hypermethylated in breast and other cancers.11-14 We find that, although small dsRNAs targete exclusively to the CDH1 promoter can effectively induce transcriptional repression with chromatin changes characteristic of inactive promoters, this is entirely independent of DNA methylation. Moreover, we can accomplish such silencing in a cancer cell line genetically modified such that it lacks virtually any capacity to methylate DNA.
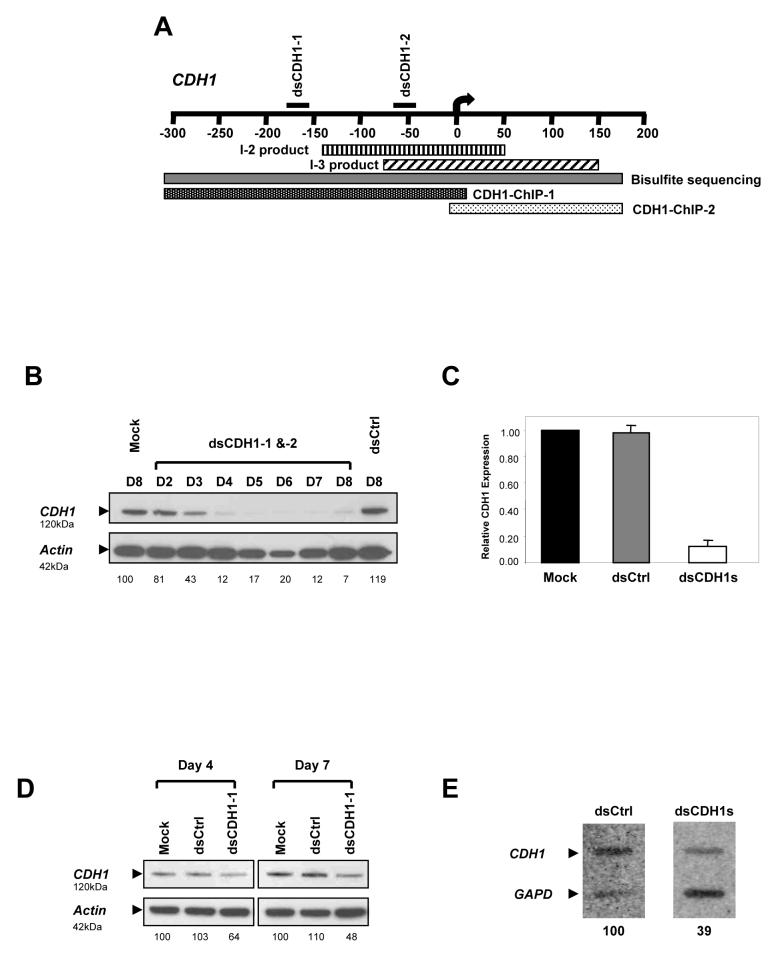
To test whether dsRNAs could induce transcriptional gene silencing at the endogenous CDH1 promoter, we transfected HCT116 human colorectal cancer cells with two 21-nucleotide long dsRNA oligos (dsCDH1-1 and dsCDH1-2). These sequences are homologous to the CpG island of the CDH1 promoter but do not overlap any known transcribed sequences (Fig. 1a and Supplementary Note). When both oligos were administered simultaneously, CDH1 protein was decreased to barely detectable levels by day 7 of treatment as compared to treatments with either the mock or dsCtrl, a scrambled control dsRNA oligo (Fig. 1b). We verified this decrement of CDH1 expression by real time RT-PCR (qRT-PCR) and found that there was a corresponding decline in CDH1 mRNA level in the dsCDH1s treated cells when compared to the untreated cells (Fig. 1c). When used alone, the dsCDH1-1 treatment resulted in a greater down-regulation of CDH1 expression than dsCDH1-2 alone but not greater than the two oligos combined (Fig. 1d). To establish that loss of the CDH1 protein was due to transcriptional silencing, we performed nuclear run-on assays. Analysis on the dsCDH1s treated cells indicated that CDH1 transcription on day 7 was decreased by 61% as detected by a 3′ cDNA probe when compared to the dsCtrl treated cells (Fig. 1e). Thus, promoter-targeting dsRNAs could effectively silence CDH1 transcription resulting in a net decrease in protein production.
Figure 1.
21-nucleotide long dsRNA species can induce transcriptional silencing in human colorectal cancer cells. (a) A schematic diagram showing the target sequence positions of dsCDH1s relative to the transcription start site (0). This region was analyzed for CpG methyaltion by MSP primers that amplify the regions indicated as I-2 (vertical line) and I-3 (diagonal line). The bisulfite sequenced fragment is indicated as a solid gray bar. ChIP analysis for H3 dimethyl-K4 and H3 dimethyl-K9 was performed with 2 sets of primers amplifying the 5′ (dotted black bar) and 3′ (dotted white bar) portions of this promoter. (b) Western blot analysis of HCT116 cells treated with dsCDH1s for 8 consecutive days. CDH1 protein levels were gradually depleted to barely detectable levels by day 7 when compared to the controls. The intensity of the CDH1 bands, stated below the pictures as normalized percentages, was quantified by densitometry and normalized to the β-actin bands. (c) qRT-PCR analysis of CDH1 expression. The dsCDH1s treated cells contained only 13±0.04% of CDH1 mRNA when compared to the mock treated HCT116 cells. (d) Western blot analysis on days 4 and 7 of HCT116 cells treated with dsCDH1-1 for 7 consecutive days. Moderate CDH1 protein depletions were observed when compared to the controls on each day. (e) Nuclear run-on assays of dsCtrl and dsCDH1s treated cells on day 7. CDH1-specific probe binding was reduced to 39% by densitometry quantification in the dsCDH1s treated cells when compared to the dsCtrl treated cells.
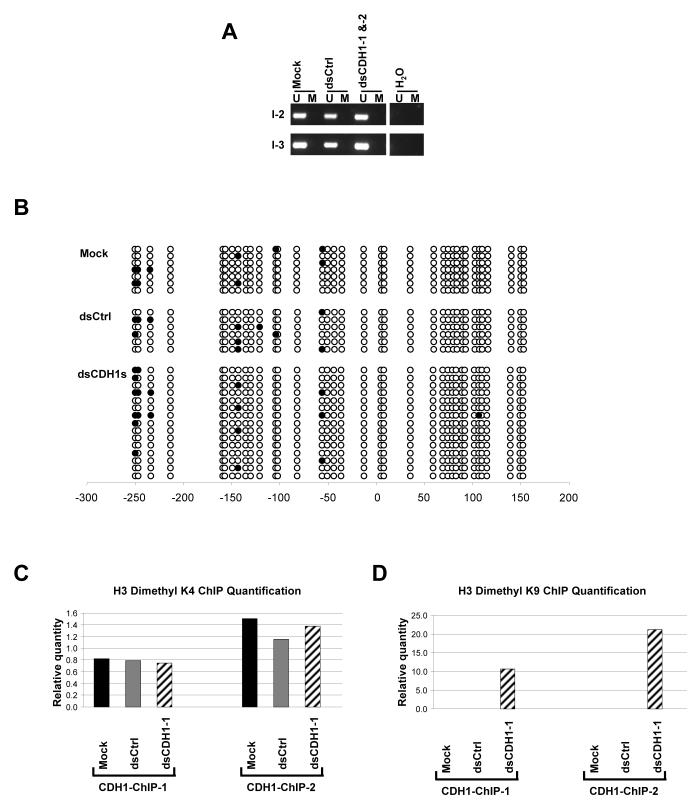
Next, we examined the methylation status of the two regions targeted by the dsCDH1s within the CDH1 CpG island, whose methylation status is closely correlated to the silencing of this promoter.12,13 The two target regions were analysed by methylation specific PCR (MSP) to verify the presence of DNA methylation parallel to the gene silencing.15 MSP distinguishes between unmethylated (U) and methylated (M) alleles by using 2 sets of primers that amplify either the U or M sequences after bisulfite treatment, which specifically converts unmethylated cytosines to uracils. In wildtype as well as the control cells, the CDH1 promoter is fully unmethylated, resulting in a single U band in the MSP analysis (Fig. 2a). To maximize the probability of observing a change in DNA methylation patterns, we examined the cells that were treated with both dsCDH1 oligos for 7 days, which produced the greatest silencing effect, and compared them with the control cells. The MSP pattern of only U alleles was preserved in these dsCDH1s treated cells indicating an absence of DNA methylation at either of the two targeted CpG island sites.
Figure 2.
DNA methylation and ChIP assay analyses on the CDH1 promoter after dsCDH1s treatment in HCT116 cells. (a) MSP analysis of the CpG island within the CDH1 promoter at 2 regions specified in Fig. 1a. The CDH1 promoter in HCT116 is normally unmethylated. DNA methylation patterns showing the presence of only the unmethylated (U) alleles remain unchanged in the mock, dsCtrl, and dsCDH1s treated cells. (b) Bisulfite sequencing analysis of the CpG island in the CDH1 promoter in the mock, dsCtrl, and dsCDH1s treated cells. The CpG sites are shown by their positions relative to the transcription start site (0), the open circles (○) represent unmethylated CpG sites, and closed circles (•) represent methylated CpG sites. The promoter remains unmethylated for each treatment group. (c) Real time PCR analysis of H3 dimethyl-K4 modification at the CDH1 promoter by ChIP assay. Two sets of primers were used in the PCR analysis to span the promoter as indicated in Fig. 1a. H3 dimethyl-K4 residues were observed, as expected, at both regions at the CDH1 promoter in the mock, dsCtrl, and dsCDH1-1 treated cells. (d) Real time PCR analysis of the H3 dimethyl-K9 modification at the CDH1 promoter by ChIP assay. H3 dimethyl-K9 was enriched at the CDH1 promoter in the dsCDH1-1 treated cells when compared to either the mock or the dsCtrl treated cells.
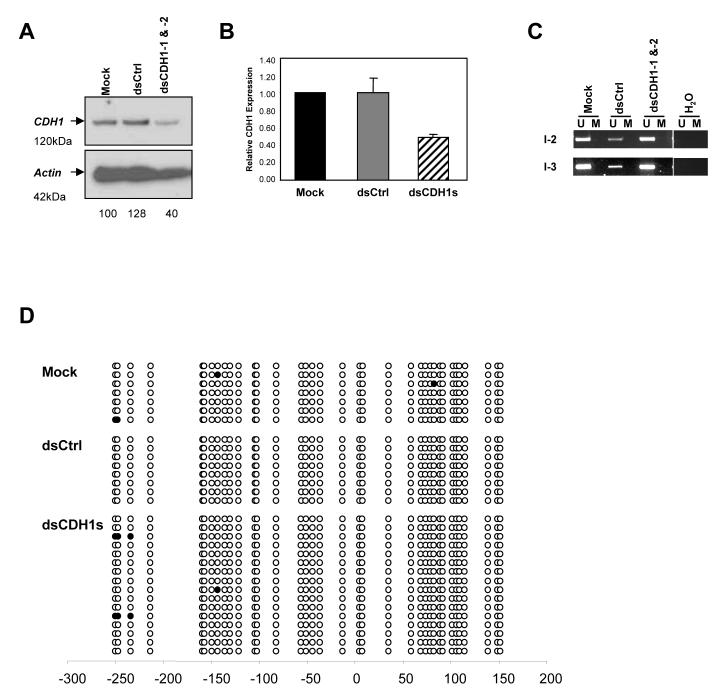
Since MSP analysis only examined the few CpG sites within the sequences recognized by the primers, we performed bisulfite sequencing to verify the methylation status of all 36 CpG sites within the target region. Unbiased primers that do not contain CpG di-nucleotides were used to amplify the bisulfite converted promoter region, and resulting PCR products were cloned and sequenced individually (Fig. 2b). The combined dsCDH1s treated cells showed the same sporadic methylation at only a few CpG sites as in the control cells. These observations verified that although transcriptional silencing was induced by the dsCDH1s treatment, there was no change in DNA methylation. To confirm that our results were not unique to the HCT116 cells, we tested the same breast cancer cells, MCF-7, used in a recent paper by Kawasaki and Taira in which a silencing strategy virtually identical to ours was employed for CDH1 and found to produce transcriptional silencing with simultaneous DNA methylation.4 Western blot analysis showed a 60% reduction in CDH1 protein level with combined treatment using the two oligos for 7 days when compared to either the mock or the dsCtrl treated MCF-7 cells (Fig. 3a). CDH1 expression, as assayed by qRT-PCR, was reduced by 50±0.03% in the dsCDH1s treated MCF-7 cells when compared to the controls (Fig. 3b). However, we saw no DNA methylation by MSP analysis (Fig. 3c), and this was again confirmed by bisulfite sequencing (Fig. 3d). Thus, in both breast and colon cancer cells, we observed CDH1 gene silencing in the absence of DNA methylation, confirming that the dsRNA dependant transcriptional silencing (RdTS) pathway is active in more than one human cancer cell type.
Figure 3.
MCF-7 human breast cancer cells treated with dsCDH1s also exhibit gene silencing in the absence of DNA methylation changes. (a) Western blot analysis of MCF-7 cells on day 7 after either the mock, dsCtrl, or dsCDH1s treatment for 7 days. A marked decrease in CDH1 protein level, 40% remaining by densitometry quantification, is evident in the dsCDH1s treated cells when compared to the controls. (b) qRT-PCR analysis of CDH1 expression in the mock, dsCtrl, and dsCDH1s treated MCF-7 cells. The dsCDH1s treated cells contained 50±0.03% of CDH1 mRNA when compared to the mock treated HCT116 cells. (c) MSP analysis of the CDH1 promoter in MCF-7 cells treated with the mock, dsCtrl, or dsCDH1s. The CDH1 promoter remains unmethylated at either the I-2 or the I-3 region as analyzed by MSP. (d) Bisulfite sequencing analysis of the CpG island in the CDH1 promoter in the mock, dsCtrl, and dsCDH1s treated cells on day 7. The CpG sites are shown by their positions relative to the transcription start site (0), the open circles (○) represent unmethylated CpG sites, and closed circles (•) represent methylated CpG sites. The promoter remains unmethylated for each treatment group.
In Arabidopsis, the RdDM pathway also involves chromatin modifications.16-19 Moreover, emerging evidence shows that chromatin modifications, including histone methylation at key residues, play an important role in modulating gene expression in mammalian systems.20-23 Therefore, we explored the possibility that RdTS may result in histone modification changes in the CDH1 promoter in HCT116 cells. Chromatin Immunoprecipitation (ChIP) assays on the CDH1 promoter, using two sets of PCR primers spanning the CDH1 core promoter, reveal the expected presence of H3 dimethyl-K4, a histone mark generally associated with actively transcribed promoters, in all treatment groups (Fig. 2c). However, H3 dimethyl-K9 residues, a modification present at inactive promoters, were only detected at the CDH1 promoter in dsCDH1-1 treated cells (Fig. 2d). PCR was also performed on the same set of immunoprecipitated DNA fractions for GAPDH and hMLH1 promoters as controls. The relative histone modification enrichments at these non-targeted promoters remained unchanged across the samples (Supplementary Fig. 1). These data confirm that RdTS in human cancer cells also involves histone modifications corresponding to a conversion from an active to silent state as has been found by others.4,5
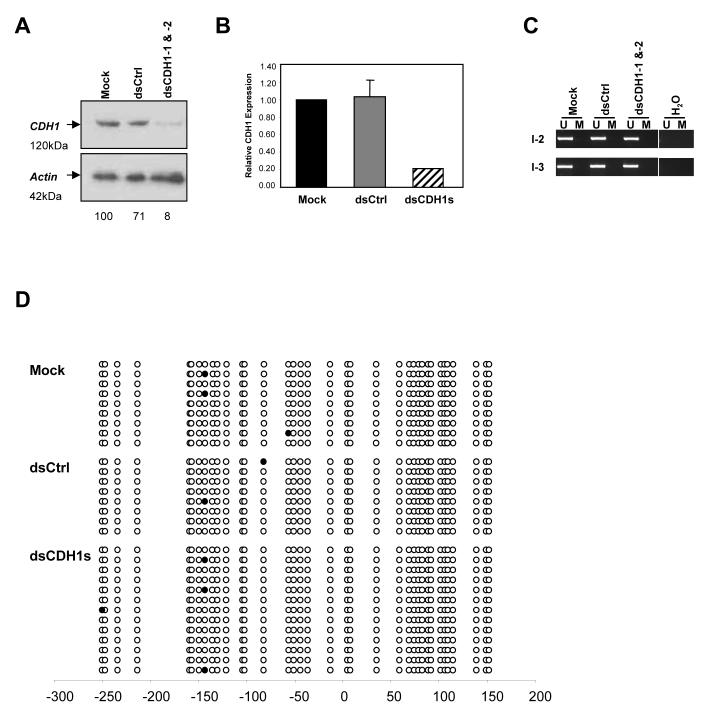
Since we have observed effective RdTS without any DNA methylation changes, we tested whether RdTS can be induced in the absence of the DNA methylation machinery. DNA methylation in humans is catalysed by three known functional enzymes, DNA methyltransferases (DNMT) 1, 3a, and 3b. DNMT1 and 3b cooperate in HCT116 cells to maintain DNA methylation and account for almost all DNMT activities in these cells.24 We transfected DNMT1-/-, DNMT3b-/- double knockout (DKO) HCT116 cells, which have virtually no DNA methylation or capacity to produce it, with the two dsCDH1 oligos for 7 days. Similar to the parental HCT116 cells, CDH1 was almost completely silenced by the dsCDH1s treatment in the DKO cells by day 7 (Fig. 4a). The decrease in protein level was well correlated to a decrease in CDH1 mRNA level, as verified by qRT-PCR (Fig. 4b). MSP analysis of the target promoter indicated a lack of DNA methylation even though a strong silencing effect was apparent (Fig. 4c). Bisulfite sequencing of the CDH1 promoter in these cells also confirmed the absence of DNA methylation (Fig. 4d). This silencing in the absence of the major DNMTs supports the notion that acute RdTS in human colon cancer cells does not require DNA methylation.
Figure 4.
RdTS is achieved in HCT116 cells genetically lacking both DNMT1 and DNMT3b (DKO) cells. (a) Western blot analysis of DKO cells on day 7 after the mock, dsCtrl, or dsCDH1s treatment for 7 days. CDH1 protein level was decreased to only 8% when compared to the mock treated cells, indicating RdTS was effectively achieved in the absence of two major DNA methyltransferases that are responsible of 95% of the total DNA methylation in these cells.24 (b) qRT-PCR analysis of CDH1 expression in the mock, dsCtrl, and dsCDH1s treated DKO cells. The dsCDH1s treated cells contained only 21±0.00% of CDH1 mRNA when compared to the mock treated HCT116 cells. (c) MSP analysis of the CDH1 promoter in DKO cells treated with the mock, dsCtrl, or dsCDH1s. The MSP pattern of only the U band amplification is preserved in all three treatment groups, indicating a lack of DNA methylation changes. (d) Bisulfite sequencing analysis of the CpG island in the CDH1 promoter in the mock, dsCtrl, and dsCDH1s treated cells on day 7. The CpG sites are shown by their positions relative to the transcription start site (0), the open circles (○) represent unmethylated CpG sites, and closed circles (•) represent methylated CpG sites. The promoter remains unmethylated for each treatment group.
In the results above, we have seen promoter silencing by dsRNAs in the absence of DNA methylation while Kawasaki and Taira observed both events for the same gene in some of the same human cancer cell types.4 Morris et al. have also reported RdDM for the EF1α promoter in human fibroblasts.5 What accounts for these opposing results? We believe the answer may lie in the types of DNA methylation assays done and how they were performed. First, Kawasaki and Taira indicated that the DNA methylation they observed included CpNpG methylation as a consequence of dsRNA targeting to the CDH1 promoter in MCF-7 cells. In plants, RdDM induces DNA methylation in the context of CpNpG and CpG in the targeted promoter.2 While CpNpG and CpG methylation both have strong correlations to gene silencing in plants, CpNpG methylation is rare and has no documented functional implications regarding gene silencing in humans. We found no CpNpG methylation within our bisulfite sequenced region, which overlaps the region examined by Kawasaki and Taira.4 Unconverted cytosines in non-CpG contexts often indicate incomplete bisulfite conversions rendering unmethylated cytosine residues in both CpG and non-CpG context to remain as cytosines. Such non-conversions may explain part of the differences in the bisulfite sequencing results between our study and that of Kawasaki and Taira. Furthermore, the PCR primers used by Kawasaki and Taira to obtain the initial bisulfite sequencing template contain CpG sites that may bias the amplification step and produce problematic bisulfite sequencing results that are difficult to interpret.4 A third problem in the sequencing data for CDH1 concerns the methylation status for two Alu repeats upstream of the 5′ CpG island in the CDH1 promoter region. These sequences were found by Kawasaki and Taira to be completely unmethylated while previous characterization of this region in MCF-7 cells revealed, as is true for most Alu repeats, dense methylation even though the adjacent CDH1 proximal promoter region is unmethylated.13 Finally, in the work with the silencing of the EF1α promoter in human fibroblasts, the DNA methylation data remains to be verified since the DNA methylation analyses were not as extensive and the indirect evidence of 5′ aza-deoxycytidine (5′-Aza) and trichostatin A (TSA) co-administration alleviating the silencing was difficult to interpret. TSA is a histone deacetylase inhibitor and 5′-Aza, although a DNMT inhibitor, can reactivate promoters that contain little or no DNA methylation.25
Taken together, our data strongly suggest that dsRNA species can induce effective transcriptional gene silencing in human colon and breast cancer cell lines, and this silencing does not involve DNA methylation changes in the acute time frame our experiments were performed. Clearly, this phenomenon is different from the description of RdDM in plants, namely the silencing effect is not accompanied by DNA methylation changes. Our observations are also consistent with the data in Yeast, where silencing effects and heterochromatin formation induced by dsRNAs were achieved in the absence of DNA methylation.26-28 Also, multiple oligos simultaneously targeting the same promoter have a synergistic silencing effect on the targeted promoter both in our hands and in studies of others. This may be analogous to the plant system, where RdDM triggers are usually long dsRNA molecules that span a few hundred base pairs of the target promoter, and are later processed to smaller 21-30 base pair long species.16
It is important to point out that DNA methylation may still be involved in a prolonged silencing event in human cells since in Arabidopsis, RdDM is always studied in the progenies of plants that received the initial dsRNA trigger and requires intact DNMT’s to propagate the silencing.16,29,30 Therefore, future experiments in mammalian systems should undoubtedly include thorough examination of the long-term outcomes of RdTS. Most importantly, the presence of RdTS signifies the possibility that this may be an existing gene regulatory mechanism in humans even though at the present time, we are unable to verify that it initiates tumor suppressor gene hypermethylation in human cancers.
Methods
Trasfection and cell culture
HCT116 and MCF-7 cells were obtained from American Type Culture Collection (ATCC) and cultured as directed. The DKO cells were derived from HCT116 cells and thus maintained in the same manner.24 The dsRNA oligos were obtained from Dharmacon with the UU overhang option, and the target sequences are 5′-AAC TCC AGG CTA GAG GGT CAC-3′ (dsCDH1-1), 5′-AAT CAG CGG TAC GGG GGG CGG-3′ (dsCDH1-2), and 5′-AAA CCC TAG CGC CAT CGT GCC-3′ (dsCtrl). 60nM dsRNA were transfected into cells with Lipofectamine 2000 (Invitrogen) at 24hr intervals, and the transfection media was replaced with normal complete media after 4 hours.
Western blotting
10μg cell lysates were analysed by western blotting using mouse anti-CDH1 antibody (1/2500 dilution, BD 610181) and mouse anti- β-actin antibody (1/10,000 dilution, Sigma A5441). The resulting blots were quantified by densitometry analysis using Scion Image software.
Real time RT-PCR (qRT-PCR)
Total RNA were extracted from the treated cells with RNeasy Mini Kit (Qiagen 74104) and treated with DNase (Qiagen 79254). qRT-PCR analyses were performed using iScript One-step RT-PCR Kit with SYBR Green (Bio-Rad 170-8892). Primers used for CDH1 qRT-PCR were 5′-CCGCCGGCGTCTGTAGGAA-3′ and 5′-AGGGCTCTTTGACCACCGCTCTC-3′ at 58°C melting temperature. qRT-PCR for GAPDH was also performed on the same samples to facilitate quantification and normalization of the results. GAPDH primers were 5′-GAAGGTCGGAGTCAACGGATTT-3′ and 5′-ATGGGTGGAATCATATTGGAA-3′, and the PCR reactions were carried out at 55°C melting temperature. The data were presented as relative CDH1 expression and were calculated as follows: (Starting quantity of CDH1 /Starting quantity of GAPDH)sample / (Starting quantity of CDH1/Starting quantity of GAPDH)mock.
Nuclear run-on assay
The nuclear run-on assays were performed following the Current Protocol of Molecular Biology. 106 nuclei per samples were used in the reaction with α32p-UTP (NEN BLU507X500UC). The purified transcripts were hybridized to a nylon membrane (PerkinElmer NEF976) containing immobilized nucleic acids corresponding to the 3′ (+3301 to +4517) region of the CDH1 cDNA and to the GAPDH cDNA (+28 to +479).
DNA methylation analysis
Genomic DNA were extracted and treated with sodium bisulfite as previously described.31 Methylation specific PCR (MSP) was performed on the CDH1 promoter with 2 sets of previously described primers spanning the CpG island (I2 and I3) of this region.13 Bisulfite sequencing was performed using a forward primer 5′-AAT AAA AGA ATT TAG TTA AGT GT-3′ and a reverse primer 5′-AAA ACC TAC AAC AAC AAC AAC-3′ at 54°C for 38 cycles. The PCR products were subsequently cloned into the TOPO TA vector (Invitrogen K2000-01SC) and sequenced with the M13R primer.
Chromatin Immunoprecipitation assay and Real time PCR
Cells were cross-linked and processed following the UpState Chromatin Immunoprecipitation (ChIP) Assay Kit protocol (UpState 17-295). 2×106 cells were used for each IP reaction. Rabbit anti-H3 dimethyl-K4 (5μg/IP, Upstate 07-030) and rabbit anti-H3 dimethyl-K9 (10 μg/IP, a gift from Dr. Thomas Jenuwein) antibodies were used for the specific IP of the respective histone residues while rabbit anti-HA antibody (10 μg/IP, Santa Cruz SC805) was used as the control. 50uL of sonicated, pre-IP DNA from each sample were used as input controls. The ChIP results were analyzed by real time PCR using the QuantiTect SYBR Green PCR Kit (QIAGEN 204143) with 2 sets of CDH1 promoter specific primers spanning the CpG island of interest. CDH1-ChIP-1 primers are 5′-AAC AAA AGA ACT CAG CCA AGT G-3′ and 5′-ACG CCA CTG AGA GGG GGT GC-3′ (59°C annealing temperature), and CDH1-ChIP-2 primers are 5′-CCC TCT CAG TGG CGT CGG AAC T-3′ and 5′-AGA CCT GCA GCA GCA GCA GCA-3′ (57°C annealing temperature). A set of serially diluted DNA standards was used to calculate the starting quantities in each PCR reaction. The final results for each sample were normalized to the inputs. GAPDH and hMLH1 PCRs were also performed on the same sets of immunoprecipitated DNA fractions as controls.
Supplementary Material
Acknowledgements
We thank B. Lee for suggestions, support, and critical reading of the manuscript and T. Jenuwein for providing the H3 dimethyl-K9 antibodies used in the ChIP assay. This work was supported by NIH Grant CA43318.
Footnotes
Competing Interest Statement: The authors declare that they have no competing financial interests regarding the particular studies in this manuscript. Drs. Baylin and Herman are paid consultants to OncoMethylome Sciences. Under licensing agreement between the Johns Hopkins University and this company, M.S.P. was licensed to OncoMethylome Sciences and they are entitled to a share of the royalties received by the University from sales of the licensed technology.
References
- 1.Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–76. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 2.Pelissier T, Thalmeir S, Kempe D, Sanger HL, Wassenegger M. Heavy de novo methylation at symmetrical and non-symmetrical sites is a hallmark of RNA-directed DNA methylation. Nucleic Acids Res. 1999;27:1625–34. doi: 10.1093/nar/27.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wassenegger M. RNA-directed DNA methylation. Plant Mol Biol. 2000;43:203–20. doi: 10.1023/a:1006479327881. [DOI] [PubMed] [Google Scholar]
- 4.Kawasaki H, Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2004;431:211–7. doi: 10.1038/nature02889. [DOI] [PubMed] [Google Scholar]
- 5.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–92. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 6.Park CW, Chen Z, Kren BT, Steer CJ. Double-stranded siRNA targeted to the huntingtin gene does not induce DNA methylation. Biochem Biophys Res Commun. 2004;323:275–80. doi: 10.1016/j.bbrc.2004.08.096. [DOI] [PubMed] [Google Scholar]
- 7.Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B. Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet. 2002;36:233–78. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 8.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 9.Reik W, Collick A, Norris ML, Barton SC, Surani MA. Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature. 1987;328:248–251. doi: 10.1038/328248a0. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 11.Graff JR, et al. Distinct patterns of E-cadherin CpG island methylation in papillary, follicular, Hurthle’s cell, and poorly differentiated human thyroid carcinoma. Cancer Res. 1998;58:2063–6. [PubMed] [Google Scholar]
- 12.Graff JR, Herman JG, Myohanen S, Baylin SB, Vertino PM. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J Biol Chem. 1997;272:22322–9. doi: 10.1074/jbc.272.35.22322. [DOI] [PubMed] [Google Scholar]
- 13.Graff JR, et al. E-cadherin Expression is Silenced by DNA Hypermethylation in Human Breast and Prostate Carcinomas. Cancer Research. 1995;55:5195–5199. [PubMed] [Google Scholar]
- 14.Hiraguri S, et al. Mechanisms of inactivation of E-cadherin in breast cancer cell lines. Cancer Res. 1998;58:1972–7. [PubMed] [Google Scholar]
- 15.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matzke M, et al. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim Biophys Acta. 2004;1677:129–41. doi: 10.1016/j.bbaexp.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 17.Kanno T, et al. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–5. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 18.Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJ. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. Embo J. 2002;21:6832–41. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJ. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. Embo J. 2000;19:5194–201. doi: 10.1093/emboj/19.19.5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeddeloh JA, Stokes TL, Richards EJ. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 1999;22:94–7. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 21.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 22.Heard E, et al. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–38. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–3. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 24.Rhee I, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–6. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 25.Schmelz K, et al. Induction of gene expression by 5-Aza-2′-deoxycytidine in acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS) but not epithelial cells by DNA-methylation-dependent and -independent mechanisms. Leukemia. 2005;19:103–11. doi: 10.1038/sj.leu.2403552. [DOI] [PubMed] [Google Scholar]
- 26.Schramke V, Allshire R. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science. 2003;301:1069–74. doi: 10.1126/science.1086870. [DOI] [PubMed] [Google Scholar]
- 27.Volpe T, et al. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–46. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- 28.Volpe TA, et al. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 29.Cao X, et al. Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol. 2003;13:2212–7. doi: 10.1016/j.cub.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 30.Aufsatz W, Mette MF, Matzke AJ, Matzke M. The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant Mol Biol. 2004;54:793–804. doi: 10.1007/s11103-004-0179-1. [DOI] [PubMed] [Google Scholar]
- 31.Frommer M, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc.Natl.Acad.Sci U.S.A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.