Abstract
Terminal continuation (TC) RNA amplification was developed originally to reproducibly and inexpensively amplify RNA. The TC RNA amplification method has been improved further by obviating second strand DNA synthesis, a cost-effective protocol that takes less time to perform with fewer manipulations required for RNA amplification. Results demonstrate that TC RNA amplification without second strand synthesis does not differ from the original protocol using RNA harvested from mouse brain and from hippocampal neurons obtained via laser capture microdissection from postmortem human brains. The modified TC RNA amplification method can discriminate single cell gene expression profiles between normal control and Alzheimer’s disease hippocampal neurons indistinguishable from the original protocol. Thus, TC RNA amplification without second strand synthesis is a reproducible, time- and cost-effective method for RNA amplification from minute amounts of input RNA, and is compatible with microaspiration strategies and subsequent microarray analysis as well as quantitative real-time PCR.
Keywords: Alzheimer’s disease, in vitro transcription, microarray, qPCR, RNA amplification
Introduction
Microarray analysis is an effective approach to evaluate transcript levels in a high-throughput manner, but requires significant amounts of high quality input RNA. Molecular manipulations have been implemented to increase RNA including exponential PCR-based amplification and linear RNA amplification procedures (Ginsberg, 2008; Nygaard and Hovig, 2006; Schneider et al., 2004). PCR-based protocols are not optimal, as exponential amplification can skew the original quantitative relationships between genes from an initial population (Kacharmina et al., 1999). In contrast, linear RNA amplification methods allow for the analysis of relative gene expression levels. A linear RNA amplification procedure typically entails generating quantities of RNA species through in vitro transcription (IVT) (Eberwine et al., 2001; Ginsberg and Mirnics, 2006), although PCR/linear RNA amplification hybrid methods (Wang et al., 2000) as well as isothermal RNA amplification (Dafforn et al., 2004; Kurn et al., 2005) procedures are also available that generate a faithful representation of the original input RNA.
A well known linear amplification method, amplified antisense RNA (aRNA) amplification (Eberwine et al., 1992; VanGelder et al., 1990), enables the quantitation of relative gene expression levels from fairly small amounts of input RNA. There have been modifications of the aRNA procedure to improve efficiency (Iscove et al., 2002; Matz et al., 1999; Wang et al., 2000; Zhumabayeva et al., 2001) and several kits that use aRNA-based technology are available commercially. The principal obstacle of problematic second strand cDNA synthesis remains. This difficulty is not exclusive for the aRNA protocol. Rather, the majority of current RNA amplification methods are afflicted by problems with second strand synthesis efficiency and specificity (Goff et al., 2004; Wang et al., 2000; Zhumabayeva et al., 2001). Key factors to improving RNA amplification include streamlining and/or obviating second strand cDNA synthesis and allowing for flexibility in the placement of bacteriophage transcriptional promoter sequences for sense and antisense amplification. An RNA amplification procedure developed in our laboratory named terminal continuation (TC) RNA amplification satisfies these objectives (Fig. 1A). TC RNA amplification originally consisted of synthesizing first strand cDNA complementary to the mRNA template, subsequent second strand cDNA synthesis complementary to the first strand cDNA, and finally IVT using the double stranded cDNA as template (Che and Ginsberg, 2004). First strand cDNA synthesis complementary to the template mRNA entails the use of two oligonucleotide primers, a first strand poly d(T) primer and a TC primer (Che and Ginsberg, 2004) (Fig. 1A). Transcript orientation with TC RNA amplification procedure can be in the antisense orientation when the bacteriophage promoter sequence is placed on the first strand poly d(T) primer or in a sense orientation when the promoter sequence is attached to the TC primer. One round of amplification is sufficient for downstream genetic analyses (Che and Ginsberg, 2004). TC RNA amplification has been shown to be more sensitive than aRNA amplification using input RNA obtained from neurons from mouse and postmortem human brains (Che and Ginsberg, 2004; Ginsberg and Che, 2002).
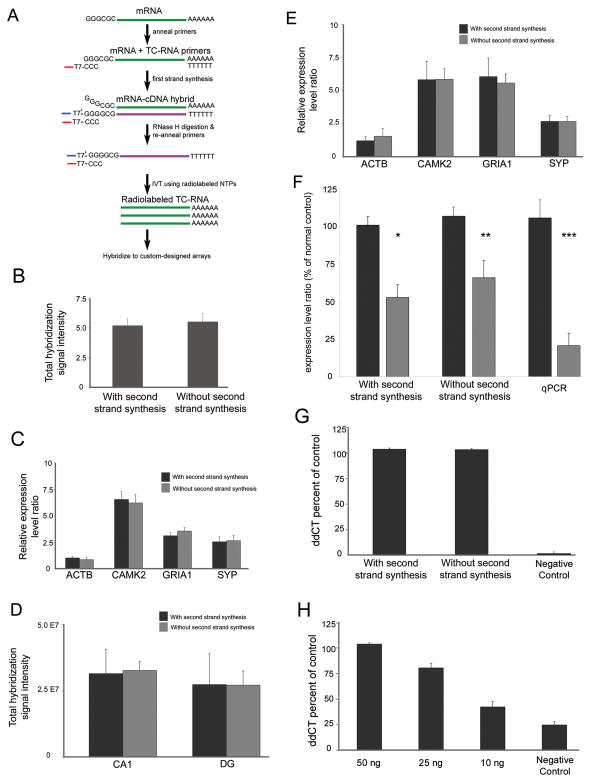
Figure 1. Comparison of TC RNA amplification methodologies.
A. Schematic representation of TC RNA amplification without second strand synthesis. The mRNA to be amplified (green line) and the TC primer serve as templates for the first strand synthesis with poly d(T) acting as a primer. First strand cDNA consists of three portions, the 5′ end comprised of the poly d(T), the mRNA complementary portion in the middle (purple line), and the 3′ end is comprised of the TC primer complementary to the cDNA (denoted as TC′). The TC′ portion hybridizes with the TC primer present in the reaction and forms a double stranded region without need for further second strand synthesis. Since the TC primer contains the T7 bacteriophage transcription promoter sequence, double stranded TC primer regions provide a functional RNA synthesis promoter for IVT and subsequent robust RNA amplification.
B. Histogram indicating no significant differences between the original TC RNA amplification method with second strand synthesis and TC RNA amplification without second strand synthesis in terms of total hybridization signal intensity ± standard deviation. Depicted assays were performed with 50 ng of mouse brain RNA using the original protocol (n = 6) and modified protocol (n = 8).
C. Comparison of amplification protocols using 50 ng of mouse brain RNA on specific gene expression levels. Histogram demonstrating no significant differences in expression levels in representative genes with varying levels of hybridization signal intensity. Key: ACTB, beta actin; CAMK2, calcium/calmodulin-dependent protein kinase II alpha; GRIA1, AMPA glutamate receptor 1 subunit; SYP, synaptophysin.
D. Similar to B, no differences in total hybridization signal intensity are observed between amplification protocols using RNA extracted from 50 cells per reaction obtained from normal control human postmortem CA1 neurons and DG granule cells.
E. Consistent with C, no differences are observed within representative individual genes following TC RNA amplification using the original or modified protocols in CA1 neurons acquired via LCM.
F. SYP expression levels are down regulated in CA1 neurons obtained from AD brains compared to age-matched normal controls using the original TC RNA amplification protocol (single asterisk denotes p < 0.01) and the TC RNA amplification without second strand synthesis methodology (double asterisk denotes p < 0.03). Consistent with the microarray analysis, significant down regulation of SYP expression was observed via qPCR in AD hippocampus as compared to age-matched controls (triple asterisk denotes p < 0.002).
G. Histograms depicting no significant differences in GRIA1 qPCR products across TC RNA amplification protocols in relation to a negative control using 50 ng of input mouse brain RNA.
H. GRIA1 qPCR products at three low concentrations (10 ng, 25 ng, and 50 ng) following TC RNA amplification without second strand synthesis. Note the linear nature of the amplification above the negative control.
A new adaptation of the TC RNA amplification procedure is presented that enables robust RNA amplification without the need for second strand synthesis, a cost-saving and potential yield-preserving method compatible with homogeneous cell analysis by laser capture microdissection (LCM) and subsequent microarray analysis. Herein, TC RNA amplification without second strand synthesis is compared to the original TC RNA amplification protocol using RNA extracted from mouse brain and hippocampal CA1 neurons and dentate gyrus granule cells obtained via LCM from human postmortem brain tissues to demonstrate viability of the modification.
Materials and Methods
Tissue and RNA accession
Animal protocols have been approved by the Institutional Animal Care and Use Committee (IACUC) of the Nathan Kline Institute/NYU School of Medicine and postmortem tissue accession were in full accordance with NIH guidelines. Wild-type C57BL/6 mice were euthanized by cervical dislocation and brains stored at −80°C. Cortex was weighed and approximately 100 mg was utilized for RNA extraction. Trizol (Invitrogen, Carlsbad, CA) was added at 10X (w/v) and tissue was homogenized. RNA was then extracted with chloroform and precipitated utilizing isopropanol and resuspended in 18.2 mega Ohm RNase-free water (Nanopure Diamond, Dubuque, IA). RNA purity and concentration was analyzed utilizing the total RNA Nano procedure by bioanalysis (2100, Agilent Technologies). Hippocampal CA1 neurons were harvested from postmortem human brains as described previously (Ginsberg and Che, 2005). Briefly, 50 individual neurofilament-immunoreactive CA1 neurons and 50 individual neurofilament-immunoreactive dentate gyrus (DG) granule cells were microaspirated via LCM per reaction from 6 um thick paraffin embedded tissue sections (n=8 normal aged controls) and (n=6 subjects with AD) (Table I). A total of 3–5 samples per cell type per case were collected and analyzed for both RNA amplification procedures (e.g., original TC RNA amplification protocol and TC RNA amplification without second strand synthesis).
Table I.
Case demographics
| Clinical Diagnosis |
|||
|---|---|---|---|
| AD (N=11) | CTR (N=9) | ||
| Age at death (yrs): mean ± sd (range) | 81.4±4.6 (72–92) | 78.3±11.7 (66–98) | p = 0.23a |
| Number (%) of males: | 4 (36.4%) | 4 (44.4%) | p = 0.57b |
| PMI (hrs): mean ± sd (range) | 11.3 ± 4.5 (4.5–17) | 9.1 ± 4.9 (4–17) | p = 0.72a |
Kruskal-Wallis test
Fisher’s exact test
No significant differences were observed between the conditions in terms of average age, number of males or postmortem interval (PMI). Brains were accrued from the Rush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, IL and the Penn Alzheimer’s Center, University of Pennsylvania School of Medicine, Philadelphia PA. All of the tissue samples were harvested using the same methods and procedures. Neuropathological designations for the diagnosis of AD were based on the NIA Reagan and CERAD criteria (Hyman and Trojanowski, 1997; Mirra et al., 1991). Assessment of demographic variables between conditions was assessed by the Kruskal-Wallis test (age and PMI) and Fisher’s exact test (percentage of males) as described previously (Ginsberg et al., 2006a; Ginsberg et al., 2006b). Only brains confirmed to have abundant RNAs by acridine orange histofluorescence were selected for study (Ginsberg et al., 1997). A total of 8 AD and 6 control brains were used in the microarray study. A total of 5 AD and 5 control brains were used in the qPCR study (tissue from 2 AD brains and 2 control brains were used for both the microarray and qPCR studies). This study was performed in accordance with IRB guidelines administrated by the Rush University Medical Center, University of Pennsylvania School of Medicine, and the Nathan Kline Institute/NYU School of Medicine.
TC RNA amplification
The TC RNA amplification protocol developed in this laboratory (Che and Ginsberg, 2004) has been modified to obviate the need for second strand cDNA synthesis altogether. The original and new TC RNA amplification protocols are available at http://cdr.rfmh.org/2005/Ginsberg_protocol1.html. Briefly, RNAs were reverse transcribed in the presence of poly d(T) primer (100 ng/ul) and TC primer (100 ng/ul) in 1X first strand buffer (Invitrogen), 2 ug of linear acrylamide (Applied Biosystems, Foster City, CA), 10 mM dNTPs, 100 uM DTT, 20 U of SuperRNase Inhibitor (Applied Biosystems) and 200 U of reverse transcriptase (Superscript III, Invitrogen). Single stranded cDNAs were digested by adding the following and then placed in a thermal cycler: 10 mM Tris (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, and 10 U RNase H (Invitrogen) in a final volume of 100 ul. RNase H digestion step at 37 ºC, 30 minutes; denaturation step 95 ºC, 3 minutes; primer re-annealing step 60 ºC, 5 minutes. Samples were purified by column filtration (Montage, Millipore, Billerica, MA). Column reservoirs were filled with 300 μl of 18.2 mega Ohm RNase-free water and the cDNA reaction was then added to the reservoir. The columns were then spun at 1000 × g for 15 minutes. To recover the cDNA, 20 μl of 18.2 mega Ohm RNase-free water was added to the columns, and the columns were inverted into clean microfuge tubes and spun at 1000 × g for 2 minutes. The volume of the samples was measured and adjusted to 19 μl by speed vacuum and resuspension in 18.2 mega Ohm RNase-free water. Hybridization probes were synthesized by IVT using 33P incorporation in 40 mM Tris (pH 7.5), 6 mM MgCl2, 10 mM NaCl, 2 mM spermidine, 10 mM DTT, 2.5 mM ATP, GTP and CTP, 100 uM of cold UTP, 20 U of RNase inhibitor, 2 KU of T7 RNA polymerase (Epicentre, Madison, WI), and 120 uCi of 33P -UTP (Perkin-Elmer, Boston, MA) (Ginsberg, 2008). The reaction was performed at 37°C for 4 hours. Radiolabeled TC RNA probes were hybridized to custom-designed cDNA arrays without further purification.
Custom-designed cDNA array platforms and data analysis
Array platforms consisted of 1 μg of linearized cDNA purified from plasmid preparations adhered to high-density nitrocellulose (Hybond XL, GE Healthcare, Piscataway, NJ). Each cDNA and/or expressed sequence-tagged cDNA (EST) was verified by sequence analysis and restriction digestion. cDNA clones and ESTs from mouse, rat, and human were employed. Approximately 576 cDNAs/ESTs were utilized on the current array platform. Arrays were prehybridized (2 hours) and hybridized (14–16 hours) in a solution consisting of 6X SSPE, 5X Denhardt’s solution, 50% formamide, 0.1% sodium dodecyl sulfate (SDS), and denatured salmon sperm DNA (200 μg/ml) at 42 °C in a rotisserie oven (Che and Ginsberg, 2004; Ginsberg, 2005). Following hybridization, arrays were washed sequentially in 2X SSC/0.1% SDS, 1X SSC/0.1% SDS and 0.5X SSC/0.1% SDS for 15 minutes each at 37°C. Arrays were placed in a phosphor screen for 24 hours and developed on a phosphor imager (GE Healthcare). All array phosphor images were adjusted to the same brightness and contrast levels for data acquisition and analysis. Hybridization signal intensity was determined utilizing ImageQuant software (GE Healthcare). Statistical procedures for custom-designed microarray analysis have been described in detail previously (Ginsberg, 2005, 2008). Briefly, the arrays were compared to negative control arrays performed utilizing the respective protocols without any starting RNA (Ginsberg, 2008). Expression of TC amplified RNA bound to each linearized cDNA (approximately 576 cDNAs/ESTs on the array) minus background was then expressed as a ratio of the total hybridization signal intensity of the array (a global normalization approach). Global normalization effectively minimizes variation due to differences in the specific activity of the synthesized probe as well as the absolute quantity of probe present (Eberwine et al., 2001; Ginsberg, 2008). Data analyzed in this manner does not allow the absolute quantitation of mRNA levels. However, an expression profile of relative changes in mRNA levels was generated. Relative changes in total hybridization signal intensity and percentage of cDNA clones above negative control were analyzed by one-way analysis of variance (ANOVA) with post-hoc analysis (Neumann-Keuls test; level of significance was set at p < 0.05) and correction for multiple observations for individual comparisons.
Real-time quantitative PCR (qPCR)
qPCR was performed in triplicate on microdissected frozen hippocampal tissue samples subjected to the RNA amplification procedures as described previously (Ginsberg and Che, 2005). TaqMan hydrolysis probes designed against AMPA glutamate receptor subunit 1 (GRIA1) (Mn00514377_m1), beta actin (ACTB; Mm00447557_m1), synaptophysin (SYP; Hs00300531_m1), and the housekeeping gene glyceraldehyde-3 phosphate dehydrogenase (GAPDH; Hs00266705_g1) were employed (Applied Biosystems). Samples were assayed on a real-time PCR cycler (7900HT) in 96-well optical plates covered with optical caps as per the manufacturer instructions (Applied Biosystems) as described previously (Ginsberg, 2008); (Ginsberg and Che, 2005). Standard curves and cycle threshold (Ct) were generated using serial dilution standards obtained from total mouse brain RNA (GRIA1, ACTB, and GAPDH) and human brain RNA (SYP and GAPDH). The ddCT method was employed to determine relative gene level differences with GAPDH qPCR products used as a control (ABI, 2004; Ginsberg and Mirnics, 2006). A total of 3–4 independent samples per condition were run in triplicate for the qPCR assessments. Negative controls consisted of the reaction mixture without input RNA. Alterations in PCR product synthesis were analyzed by one-way ANOVA with post-hoc analysis (Neumann-Keuls test; level of statistical significance was set at p < 0.05).
Results and Discussion
A representative comparison of the original and modified TC RNA amplification methods is presented in Figure 1. Using mouse brain RNA extracts (1 ng, 10 ng, 25 ng, and 50 ng), the TC RNA amplification procedure without second strand synthesis provided robust hybridization signal intensity similar to results published previously by our group using a second strand synthesis step (Che and Ginsberg, 2004). The similarity between procedures is evidenced for total hybridization signal intensity (Fig. 1B) as well as individual genes with low, moderate, and high relative expression levels including ACTB, calcium/calmodulin-dependent protein kinase II alpha (CAMK2), GRIA1, and SYP (Fig. 1C). Hybridization signal intensity levels are depicted for representative custom-designed arrays with and without second strand synthesis in Figure 2. Similar results indicated that levels of total hybridization signal intensity on the custom-designed array (Fig 1D) as well as individual genes (Fig. 1E) did not differ statistically between TC RNA amplification procedures for human CA1 neurons and DG granule cells acquired via LCM. Additional experiments were performed to assess the fidelity of the new modification on the ability to detect expression level differences across diagnostic conditions. Specifically, microarray analysis of CA1 neurons acquired via LCM from normal control and AD brains indicated a significant down regulation of the synaptic-related marker SYP in AD with the original TC RNA amplification protocol (52.6% ± 8.6 decrease as percentage of normal aged control) or without second strand synthesis second strand synthesis (65.7% ± 11.5 decrease as percentage of normal aged control) at a similar level of high statistical significance (Fig. 1F). Validation of the observed SYP down regulation via microarray analysis was performed with frozen human hippocampus using qPCR. Pronounced down regulation was found (82.2% ± 10.2 decrease as percentage of normal aged control), similar in direction (down regulation) and percentage of decrease to the microarray assessments with the two distinct TC RNA amplification protocols (Fig 1F). A greater decrease in SYP expression in AD using the qPCR technique may indicate greater sensitivity of this method for quantitation of one primer set (Bustin, 2000), or could also indicate that down regulation of SYP gene expression occurs in multiple cell types within the hippocampal formation (e.g., CA3 pyramidal neurons and dentate gyrus granule cells in addition to CA1 pyramidal neurons profiled via LCM for the present microarray analysis). Importantly, down regulation of SYP gene expression and protein levels has been observed by our group and others via several independent genomic and protein-based techniques within the AD postmortem human hippocampal formation (Callahan et al., 1999; Ginsberg et al., 2000; Gutala and Reddy, 2004; Knobloch and Mansuy, 2008), further validating the current technical and research observations.
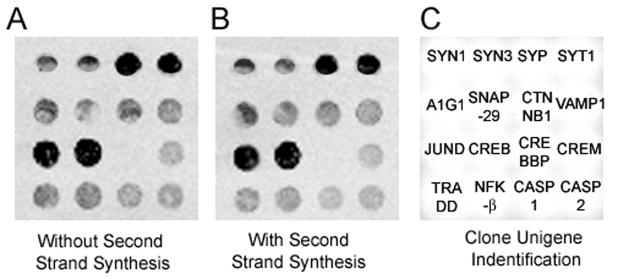
Figure 2. Comparison of the two TC RNA amplification procedures.
A. Representative custom-designed array displaying hybridization signal intensity derived from mouse brain RNA (50 ng input RNA sample) following the original TC RNA amplification procedure.
B. An identical section of a custom-designed array as in A, using the TC RNA amplification procedure without second strand synthesis. Note the similarity between the two arrays.
C. Unigene/NCBI clone identification for each feature on the custom designed array.
Key: SYN1, synapsin 1; SYN3, synapsin 3; SYP, synaptophysin; SYT1, synaptotagmin I; A1G1, adapter-related protein complex 1 gamma 1 subunit (gamma-adaptin); SNAP-29, synaptosomal-associated protein, 29kD; CTNNB1, beta catenin; VAMP1, vesicle-associated membrane protein 1; JUND, jun D proto-oncogene; CREB, cAMP responsive element binding protein; CREBBP, CREB binding protein; CREM, CAMP responsive element modulator; TRADD, Tumor necrosis factor receptor 1A-associated death domain; NFK-β, nuclear factor kappa-β; CASP1, caspase 1
Additional validation of the present microarray results was performed via qPCR with probes against GRIA1 (Figs. 1G, H) and ACTB (not shown) using input RNA from mouse brain following the two RNA amplification procedures. The original and modified TC RNA amplification methods did not differ statistically in their ability to detect both ACTB and GRIA1 at 10 ng, 25 ng, and 50 ng of input RNA with high specificity above negative control samples, and notable linearity of amplicon products at the 3 concentrations (Fig. 1H). The present qPCR-based ddCT expression levels are commensurate with previous observations using the TC RNA amplification method compared to native RNA levels in unamplified samples (Che and Ginsberg, 2004; Ginsberg and Che, 2005), further demonstrating the viability of the TC RNA amplification methodology without second strand synthesis.
Minute amounts of mRNAs within individual cells make RNA amplification methods requisite for functional genomics approaches when single cell and/or population cell resolution is desired, particularly if a microaspiration method such as LCM is employed. The process by which single cell genomic is performed, namely LCM, RNA amplification, microarray analysis, and subsequent qPCR validation is time consuming and expensive, and fraught with many sites where experimenter error and/or sample degradation can take place. The present report demonstrates that a modification of the original TC RNA amplification method is a highly effective, reproducible, and cost-effective advance that is compatible with single cell gene expression and subsequent microarray analysis and related downstream genetic analyses.
Acknowledgments
Support for this project comes from NIH grants AG09466, AG17617, and NS48447 and the Alzheimer’s Association. We thank Irina Elarova and Shaona Fang for expert technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- ABI. Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR. Applied Biosystems Product Guide; 2004. pp. 1–60. [Google Scholar]
- Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–93. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- Callahan LM, Vaules WA, Coleman PD. Quantitative decrease in synaptophysin message expression and increase in cathepsin D message expression in Alzheimer disease neurons containing neurofibrillary tangles. J Neuropathol Exp Neurol. 1999;58:275–87. doi: 10.1097/00005072-199903000-00007. [DOI] [PubMed] [Google Scholar]
- Che S, Ginsberg SD. Amplification of transcripts using terminal continuation. Lab Invest. 2004;84:131–7. doi: 10.1038/labinvest.3700005. [DOI] [PubMed] [Google Scholar]
- Dafforn A, Chen P, Deng G, Herrler M, Iglehart D, Koritala S, Lato S, Pillarisetty S, Purohit R, Wang M, Wang S, Kurn N. Linear mRNA amplification from as little as 5 ng total RNA for global gene expression analysis. BioTechniques. 2004;37:854–7. doi: 10.2144/04375PF01. [DOI] [PubMed] [Google Scholar]
- Eberwine J, Kacharmina JE, Andrews C, Miyashiro K, McIntosh T, Becker K, Barrett T, Hinkle D, Dent G, Marciano P. mRNA expression analysis of tissue sections and single cells. J Neurosci. 2001;21:8310–4. doi: 10.1523/JNEUROSCI.21-21-08310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. Analysis of gene expression in single live neurons. Proc Natl Acad Sci U S A. 1992;89:3010–4. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD. RNA amplification strategies for small sample populations. Methods. 2005;37:229–37. doi: 10.1016/j.ymeth.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD. Transcriptional profiling of small samples in the central nervous system. Methods Mol Biol. 2008;439:147–58. doi: 10.1007/978-1-59745-188-8_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Che S. Expression profile analysis within the human hippocampus: Comparison of CA1 and CA3 pyramidal neurons. J Comp Neurol. 2005;487:107–18. doi: 10.1002/cne.20535. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Che S. RNA amplification in brain tissues. Neurochem Res. 2002;27:981–92. doi: 10.1023/a:1020944502581. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Che S, Counts SE, Mufson EJ. Shift in the ratio of three-repeat tau and four-repeat tau mRNAs in individual cholinergic basal forebrain neurons in mild cognitive impairment and Alzheimer’s disease. J Neurochem. 2006a;96:1401–8. doi: 10.1111/j.1471-4159.2005.03641.x. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Che S, Wuu J, Counts SE, Mufson EJ. Down regulation of trk but not p75 gene expression in single cholinergic basal forebrain neurons mark the progression of Alzheimer’s disease. J Neurochem. 2006b;97:475–87. doi: 10.1111/j.1471-4159.2006.03764.x. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Crino PB, Lee VM-Y, Eberwine JH, Trojanowski JQ. Sequestration of RNA in Alzheimer’s disease neurofibrillary tangles and senile plaques. Ann Neurol. 1997;41:200–9. doi: 10.1002/ana.410410211. [DOI] [PubMed] [Google Scholar]
- Ginsberg SD, Hemby SE, Lee VM-Y, Eberwine JH, Trojanowski JQ. Expression profile of transcripts in Alzheimer’s disease tangle-bearing CA1 neurons. Ann Neurol. 2000;48:77–87. [PubMed] [Google Scholar]
- Ginsberg SD, Mirnics K. Functional genomic methodologies. Prog Brain Res. 2006;158:15–40. doi: 10.1016/S0079-6123(06)58002-1. [DOI] [PubMed] [Google Scholar]
- Goff LA, Bowers J, Schwalm J, Howerton K, Getts RC, Hart RP. Evaluation of sense-strand mRNA amplification by comparative quantitative PCR. BMC Genomics. 2004;5:76. doi: 10.1186/1471-2164-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutala RV, Reddy PH. The use of real-time PCR analysis in a gene expression study of Alzheimer’s disease post-mortem brains. J Neurosci Methods. 2004;132:101–7. doi: 10.1016/j.jneumeth.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hyman BT, Trojanowski JQ. Consensus recommendations for the postmortem diagnosis of Alzheimer disease from the National Institute on Aging and the Reagan Institute Working Group on diagnostic criteria for the neuropathological assessment of Alzheimer disease. J Neuropathol Exp Neurol. 1997;56:1095–7. doi: 10.1097/00005072-199710000-00002. [DOI] [PubMed] [Google Scholar]
- Iscove NN, Barbara M, Gu M, Gibson M, Modi C, Winegarden N. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat Biotechnol. 2002;20:940–3. doi: 10.1038/nbt729. [DOI] [PubMed] [Google Scholar]
- Kacharmina JE, Crino PB, Eberwine J. Preparation of cDNA from single cells and subcellular regions. Methods Enzymol. 1999;303:3–18. doi: 10.1016/s0076-6879(99)03003-7. [DOI] [PubMed] [Google Scholar]
- Knobloch M, Mansuy IM. Dendritic spine loss and synaptic alterations in Alzheimer’s disease. Mol Neurobiol. 2008;37:73–82. doi: 10.1007/s12035-008-8018-z. [DOI] [PubMed] [Google Scholar]
- Kurn N, Chen P, Heath JD, Kopf-Sill A, Stephens KM, Wang S. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin Chem. 2005;51:1973–81. doi: 10.1373/clinchem.2005.053694. [DOI] [PubMed] [Google Scholar]
- Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 1999;27:1558–60. doi: 10.1093/nar/27.6.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, Vogel FS, Hughes JP, van Belle G, Berg L. The Consortium to Establish a Registry for Alzheimer’s Disease (CERAD). Part II Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology. 1991;41:479–86. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- Nygaard V, Hovig E. Options available for profiling small samples: a review of sample amplification technology when combined with microarray profiling. Nucleic Acids Res. 2006;34:996–1014. doi: 10.1093/nar/gkj499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Buness A, Huber W, Volz J, Kioschis P, Hafner M, Poustka A, Sultmann H. Systematic analysis of T7 RNA polymerase based in vitro linear RNA amplification for use in microarray experiments. BMC Genomics. 2004;5:29. doi: 10.1186/1471-2164-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanGelder R, von Zastrow M, Yool A, Dement W, Barchas J, Eberwine J. Amplified RNA (aRNA) synthesized from limited quantities of heterogeneous cDNA. Proc Natl Acad Sci U S A. 1990;87:1663–7. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM. High-fidelity mRNA amplification for gene profiling. Nat Biotechnol. 2000;18:457–9. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- Zhumabayeva B, Diatchenko L, Chenchik A, Siebert PD. Use of SMART-generated cDNA for gene expression studies in multiple human tumors. BioTechniques. 2001;30:158–63. doi: 10.2144/01301pf01. [DOI] [PubMed] [Google Scholar]