Abstract
Genetic mutations associated with Alzheimer’s disease (AD) in the Amyloid Precursor Protein (APP) gene specifically alter the production of the APP processing product, amyloid-β (Aβ) peptide, generated by β- and γ-secretases. The accumulation and deposition of Aβ is hypothesized to cause AD pathogenesis, leading to the debilitating neurological deficits observed in AD patients. However, it is unclear how processing of APP to generate Aβ corresponds with the age-dependent pattern of brain-regional neurodegeneration common in AD. We have previously shown that overexpression of BACE1, the primary β-secretase gene, in mice expressing an AD mutant form of APP leads to significantly elevated regional Aβ levels, which coincide with the regional pattern of Aβ deposition. In the current study, we have used our genomic-based β-secretase transgenic mice to determine how BACE1 regulates the spatial and temporal pattern of Aβ production throughout post-natal development. Specifically, we observed unique differences in the brain-regional expression pattern between neonatal and adult BACE1 transgenic mice. These alterations in the BACE1 expression profile directly corresponds with age-related differences in regional Aβ production and deposition. These studies indicate that modulation of BACE1 expression leads to dramatic alterations in APP processing and AD-like neuropathology. Furthermore, our studies provide further evidence that BACE1 plays a major role in the regulation of the APP processing pathway, influencing the age-dependent onset of AD pathogenesis.
Keywords: Alzheimer’s disease, β-Secretase, Amyloid-β, Genomic-based
1. Introduction
Amyloid-β (Aβ) peptides, the primary constituent of senile plaques in Alzheimer’s disease (AD), are generated from the sequential cleavages of Amyloid Precursor Protein (APP) by β- and γ-secretases. The generation and deposition of Aβ peptides in the AD brain is hypothesized to be the causative phenomenon in AD pathogenesis. This hypothesis, known as the amyloid cascade, is corroborated genetically by the presence of familial AD mutations in the APP gene [10,13,29] that shift the processing of the APP holoprotein towards β-secretase cleavage, resulting in the increase of Aβ levels accompanied by a decrease of the non-amyloidogenic and non-pathogenic p3 peptide generated by the other APP holoprotein secretase, known as α-secretase[5,11]. Aβ metabolism can also be altered by many additional pathways including Aβ degradation, clearance, transport and aggregation [29]. Nevertheless, the accumulation and aggregation of Aβ progresses in an age-related brain-regional pattern [3], but it is unknown how this pattern of age-dependent Aβ production and deposition is regulated. Thus, the spatial and temporal control of APP expression and its processing in the brain may have significant implications for Aβ production and deposition.
Expression studies have shown that APP expression in the rodent brain is increased in early embryonic and neonatal animals relative to adult animals, where specific brain regions, particularly the hippocampus and cortex, exhibit the highest expression [24,26]. Moreover, the coordinated expression of α-secretase, β-secretase, and APP throughout the brain [26] indicates the significance for strict regulation of APP processing. Thus, the competition for the APP holoprotein substrate by β- and α-secretases is a critical step in Aβ production and, potentially, the pathogenesis of AD.
Modulation of the activity of these APP secretases will provide insight into the competitive balance of the APP processing pathway. With the identification of BACE1 [36,37,40], it was determined that β-secretase cleavage can be, in fact, the rate-limiting event for Aβ production. In addition, overexpression of BACE1 in APP transgenic mice results in increased β-secretase cleavage products particularly increased Aβ levels [2,4,28] as well as altered amyloid deposition pathology [4,28,38]. Specifically, we have previously reported an altered brain-regional deposition profile in our double BACE1xAPP transgenic mice. Intriguingly, the brain regions exhibiting the most severe amyloid-β pathology correspond with both significantly elevated Aβ levels and highest BACE1 protein expression [4]. Our previous studies implicate BACE1 as a regulator of brain-regional Aβ production and deposition in vivo.
To further understand how BACE1 regulates APP processing in vivo, we have analyzed our genomic-based BACE1 transgenic mice for age-dependent and regional alterations in Aβ production. Our previous studies, comparing the cDNA-based Tg2576 and our genomic-based R1.40 transgenic, have demonstrated that brain-regional processing of the human Swedish mutant APP transgene differs by transgene type [21]. Furthermore, as we have previously shown, characterizing human APP expression and processing under the control of its native regulatory elements removes biases involving the spatial and temporal regulation of the APP transgene [19].
Here, we report the unique profile of BACE1 expression and brain-regional Aβ production in early post-natal and adult animals. We observe altered brain-regional levels of BACE expression and APP expression, which may control the production of Aβ by brain region at both post-natal ages and adult ages. Although Aβ levels are higher in young animals, the brain-regional and age-dependent increase of Aβ in adult animals is due to elevated regional BACE1 expression, particularly in the cortex. Most notably, increased human BACE1 expression corresponds with regional alterations in amyloid deposition. These findings provide key insights into the regulation of the APP processing pathway and provide evidence for the role of BACE1 as a primary regulator of age-related regional Aβ production throughout the mammalian lifespan.
2. Materials and methods
2.1. Animals and genetic crosses
All animals were handled according to official guidelines (IACUC). Animals were bred on a C57BL/6J background. BACE1 BAC transgenic animals [4], were crossed to either hemizygous or homozygous R1.40 YAC APP transgenic animals [19]. R1.40 APP transgene homozygosity was determined by fluorescence in situ hybridization, as described previously [17]. Progeny of these crosses were sacrificed at post-natal day 7 and 60 for biochemical and immunohistochemical analysis. For Aβ deposition analysis, animals were aged 13 and 18 months before being sacrificed. Bace1 knockout tissues were kindly provided by Lucia Pastorino (Mount Sinai School of Medicine) [30].
2.2. Tissue procurement
For post-natal ages day 7 and 60, whole brains were removed and immediately dissected along the midline. For brain region studies, brains were dissected on a filter soaked with phosphate buffer saline (PBS) while chilling on wet ice. Each brain was dissected into the following regions: olfactory bulb, cerebellum, hippocampus, and cortex. Each hemi-brain or brain region was immediately frozen on dry ice and transferred to -80 °C freezer until use for biochemical analysis described below. For immunohistochemical studies, animals were anesthetized with Avertin (0.02 cc/gm of body weight) and transcardially perfused with ice-cold phosphate buffer (0.1 M, pH 7.4) followed by 4% paraformaldehyde. After perfusion, whole brains were immersion-fixed in 4% paraformaldehyde for >24 h and then cryo-protected by immersion in a sucrose gradient of 10, 20, and 30% sucrose. Sagittal sections were cut on a sliding microtome and stored at -80 °C.
2.3. Protein extraction
Protein was isolated from frozen transgenic hemi-brain or brain region tissues by polytron homogenization using a PowerGen 125 homogenizer (Fisher, Pittsburgh, PA) with 5mm × 95 mm or 7 mm × 95 mm generator probes, depending on buffer volume needed for tissue homogenization. For Western blot analysis, tissues were homogenized in 1% CHAPS solution in PBS with 1x protease inhibitors [pepstatin, 1 μM; leupeptin, 4.5 μg/ml; aprotinin, 30 μg/ml; 4-(2-aminoethyl)-benzenesulfonyl fluoride (AEBSF), 1 mM] as previously described [20]. Total protein concentration was determined by the BCA assay (Pierce, Rockford, IL). For Aβ ELISA analysis, tissues were homogenized in 5 M Guanidine-HCl in 50 mM Tris-HCl, pH 8.0 as previously described [15].
2.4. Western blot analysis and quantitation
Total proteins were separated by electrophoresis and transferred onto Immobilon-P transfer membranes (Millipore, Billerica, MA) for antibody detection. All protein blots were probed with anti-rabbit HRP-conjugated (Amersham Biosciences, Piscataway, NJ) secondary antibody and detected by chemiluminescence (ECL, Amersham Biosciences, Piscataway, NJ). For analysis of BACE1 protein expression, equal amounts of total protein from brain tissue extracts were resolved on 8% Tris-Glycine gels (Invitrogen Life Technologies, Carlsbad, CA) and detected with rabbit polyclonal antibody 00/6 (kindly provided by G. Evin, Ph.D., University of Melbourne). The relative level of human BACE1 protein expression was determined by interpolating signal intensity captured by a fluorescence imager (Flour-S-Max Imaging Machine, Bio-Rad, Hercules, CA) relative to a standard curve of nontransgenic Bace1 expression, as described previously [4]. For analysis of APP C-terminal fragments, 20 μg of total protein were resolved on 4-12% Bis-Tris gradient gels (Invitrogen Life Technologies, Carlsbad, CA). Brain tissue extracts were blotted with 369, a polyclonal antibody raised against the C-terminus of APP (kindly provided by S. Gandy, M.D., Ph.D., Thomas Jefferson U.). APP C-terminal fragments were quantified by comparing the relative ratio of CTF-β to the total amount of C-terminal fragments generated for each animal, as described previously [4,20].
2.5. Sandwich ELISAs for Aβ
Brain extracts (either hemi-brain or brain region) animals were analyzed for levels of Aβ peptides. Protein extracts were analyzed for levels of Aβ peptides using Aβ1-40 ELISA (Biosource International, Camarillo, CA), as previously described [4]. The values were read using fluorometric plate reader Wallac 1420 multilabel counter (Perkin-Elmer, Wellesly, MA). Aβ1-40 concentrations were determined based on sample values relative to the serially diluted standards representing a standard curve of known Aβ1-40 concentration. Each sample was analyzed in triplicate and sample values are expressed as pmole Aβ1-40/gram brain tissue weight.
2.6. Immunohistochemistry
The methods for Aβ immunostaining were performed as previously described by Kulnane and Lamb [17] and stained with mAb 6E10. Sections were prepared according to standard methods using Vectastain ABC Kit (Vector Laboratories, Burlingame, CA) with the chromogen 3,3-diaminobenzidine tetrahydrochloride (DAB) and counter-stained with hematoxylin.
2.7. Statistical analysis
Two-tailed t-tests were utilized for statistical analyses when variances for each sample population were equal. Two-tailed t-test with Welch’s correction was used to adjust for unequal variances between sample populations. One-way ANOVA was utilized to compare BACE1 expression across brain regions. In these cases, Tukey’s post hoc test was utilized to assess multiple comparisons across several sample populations. All data were analyzed by GraphPad Prism 4.0 statistical software (GraphPad Software, Inc., San Diego, CA, 1998).
3. Results
3.1. Human BACE1 and APP processing by whole brain: neonatal versus adult mice
Knowing human BACE1 expression alters APP processing and Aβ production in the cDNA-based Swedish mutant APP transgenic (Tg2576) model [4], we wanted to determine how human BACE1 expression alters APP processing in a genomic-based APP transgenic model, the YAC APP R1.40 line [19]. The YAC APP transgenic line expresses the Swedish mutant human APP ≈ 2-fold higher than endogenous mouse App under the control of its native regulatory elements [19]. The genomic-based R1.40 APP line makes fewer assumptions about the spatial and temporal regulation of APP expression and processing. Thus, we crossed our human BACE1 BAC transgenic line to the YAC APP R1.40 transgenic line to characterize the regulation of APP processing over the course of the mouse lifespan.
Based on evidence that BACE1 gene expression is elevated in embryonic and post-natal animals [26], we compared young post-natal and adult mouse brain for differences in BACE1 protein expression and Aβ production. By Western blot, we confirmed human BACE1 protein expression is elevated in post-natal day 7 (P7) mice compared to adult mice (Fig. 1A). We determined human BACE1 expression is more than 2-fold higher in P7 transgenic animals compared to post-natal day 60 (P60) adult transgenics (Fig. 1B) (two-tailed t-test, p-value <0.0001).
Fig. 1.
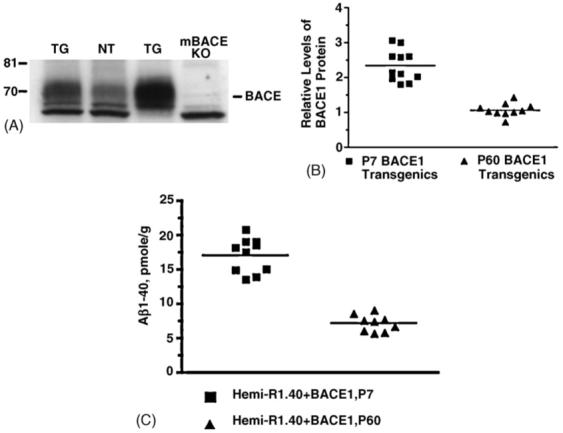
Human BACE1 expression and Aβ production in neonatal and adult BACE1 transgenic mice. (A) Western blot analysis of mouse and human BACE1 in 1% CHAPS brain extracts. Twenty micrograms of total protein from adult BACE1 transgenic in lane 1 (TG), adult nontransgenic control in lane 2 (NT), 1-week-old BACE1 transgenic in lane 3 (TG), and mouse Bace1 KO in lane 4 (mBACE KO) mice were run on 8% Tris-glycine gels, transferred to polyvinylidene difluoride membrane, and blotted with C-terminal antibody BACE-00/6. On the left for each gel are sizes of molecular weight markers in kilodaltons. (B) BACE1 protein expression was quantified by comparing the amount of BACE1 protein in post-natal day 7 (P7) BACE1 transgenics (n = 11) and post-natal day 60 (P60) BACE1 transgenics (n = 10) brain tissue extracts relative to a standard curve of Bace1 expression using a fluorescence imager for capture of chemiluminescent signal. Human BACE1 is expressed ∼2-fold higher in P7 transgenic animals compared to P60 transgenic animals (two-tailed t-test with Welch’s correction, p-value < 0.0001). (C) Brain extracts from BACE1/R1.40 YAC APP double transgenic animals were analyzed by Aβ1-40 ELISA. P7 double transgenic animals (n = 10) had significantly higher levels of Aβ1-40 compared to P60 double transgenic animals (n = 9) (two-tailed t-test with Welch’s correction, p-value < 0.0001).
To examine the impact of human BACE1 overexpression on Aβ metabolism in these double APPxBACE1 transgenics, the levels of Aβ peptide in whole brain extracts were measured by ELISA (Fig. 1C). We found that Aβ1-40 levels were significantly higher in P7 BACE1 and APP double transgenics compared to P60 BACE1 and APP double transgenics (two-tailed t-test, p-value <0.0001). Thus, Aβ production in whole brain extracts is significantly higher in neonatal mice than adult mice.
3.2. BACE1 protein expression by brain region
We have previously documented that brain regional differences in transgene-derived BACE1 expression was similar to the pattern of endogenouse Bace expression and was correlated with increased Aβ production and deposition [4]. To further examine the profile of BACE1 expression by brain region and its relationship to APP processing, we analyzed protein expression in early post-natal and adult BACE1 transgenic extracts by Western blot. The pattern of BACE1 protein expression differs in P7 transgenic mice (Fig. 2A) from that of adult transgenic mice (Fig. 2C). In the young P7 transgenics, BACE1 expression in the cortex and hippocampus is higher than expression in the cerebellum (ANOVA, p-value <0.01) (Fig. 2B). In the P60 adult transgenics, however, BACE1 expression in the hippocampus is the lowest-expressing brain region, while expression in the cortex remains elevated. Cortical BACE1 expression is significantly higher compared to cerebellum (ANOVA, p-value <0.05) and hippocampus (ANOVA, p-value <0.001), while olfactory bulb expression is only significantly higher than hippocampus (ANOVA, p-value < 0.001) (Fig. 2D).
Fig. 2.
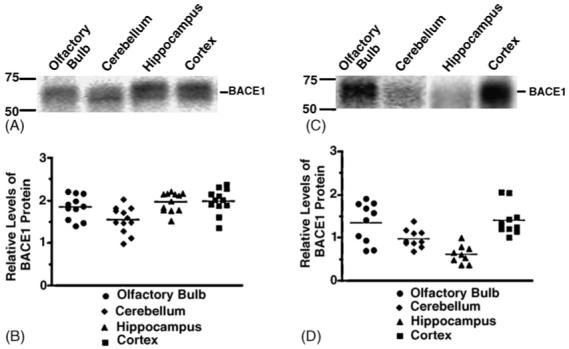
Human BACE1 protein expression by brain region in neonatal and adult BACE1 transgenic mice. (A and C) Western blot analysis of human BACE1 in 1% CHAPS extracts from olfactory bulb, cerebellum, hippocampus, and cortex brain regions in P7 BACE1 transgenics (A) and P60 BACE1 transgenics (C) were run on 8% Tris-glycine gels, transferred to polyvinylidene difluoride membrane, and blotted with C-terminal antibody BACE-00/6. (B and D) BACE1 protein expression was quantitated using Western blot analysis by comparing the amount of BACE1 protein relative to a standard curve of Bace1 expression using a fluorescence imager to capture chemiluminescent signal. For P7 BACE1 transgenics (B), human BACE1 expression is higher in the cortex and hippocampus than in the cerebellum (ANOVA, p-value <0.01). For the P60 BACE1 transgenics (D), olfactory bulb BACE1 expression is higher than hippocampus (ANOVA, p-value <0.001); expression in the cortex is higher than in cerebellum (ANOVA, p-value <0.05) and hippocampus (ANOVA, p-value <0.001).
3.3. APP processing by brain region: neonatal versus adult mice
To examine the potential consequences of altered regional BACE1 expression on APP processing, we analyzed APP expression by Western blot (Fig. 3A). First, based on relative intensity, we observed increased levels of the 100 kDa holoprotein expression at young ages compared to APP levels at adult stages (Fig. 3A, left).
Fig. 3.
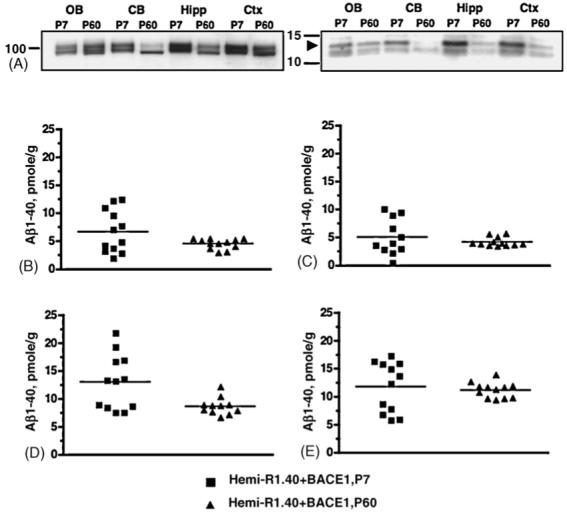
Amyloid-β metabolism by brain region in neonatal and adult BACE1 and APP double transgenic mice. (A) Western blot analysis of APP holoprotein and APP C-terminal fragments in 1% CHAPS brain region extracts from P7 and P60 BACE1/R1.40 YAC APP double transgenic animals. Twenty micrograms of total protein from olfactory bulb (OB), cerebellum (CB), hippocampus (Hipp), and cortex (Ctx) were run on 4-12% Bis-tris gradient gels, transferred to polyvinylidene difluoride membrane, and blotted with APP C-terminal polyclonal antibody 369. Shown are APP holoprotein (left) and C-terminal fragments (right) at a longer exposure timepoint. On the left of each gel are sizes of molecular weight makers in kilodaltons. Arrowhead marks 14 kDa CTF-β fragment. (B-E) Brain region extracts were analyzed by Aβ1-40 ELISA. P7 double transgenics were compared to P60 double transgenics for total Aβ1-40 levels in olfactory bulb (B), cerebellum (C), hippocampus (D), and cortex (E). Hippocampal Aβ1-40 is higher in P7 double transgenic animals compared to P60 double transgenics (two-tailed t-test with Welch’s correction, p-value = 0.018).
Although APP expression is generally elevated, we observed noticeably increased APP levels by brain region, particularly in the cortex and hippocampus, at this stage. Elevated APP holoprotein levels are accompanied by high levels of CTF-β, the precursor to Aβ (Fig. 3A, right) in the same brain regions. Consistent with in situ hybridization studies [26], these protein expression studies indicate that, along with increased APP holoprotein expression, APP processing is altered in early post-natal mice.
To examine the impact of human BACE1 and APP expression on Aβ metabolism by brain region in early post-natal and adult mice, the levels of Aβ peptide in brain region extracts were analyzed by ELISA (Fig. 3, B-E). Similar to the altered regional pattern of APP expression by Western blot, we observed a selective increase of Aβ levels by brain region, particularly in the hippocampus, in the young BACE1 and APP double transgenics compared to adult BACE1 and APP double transgenics (two-tailed t-test, p-value = 0.018). Increased regional APP expression levels in young animals correspond with increased Aβ levels, indicating the significance of APP expression in the APP processing pathway. While these studies demonstrate the impact of BACE1 and APP expression on Aβ production levels, they have not addressed the age-related phenomenon of Aβ metabolism commonly observed in AD.
3.4. Age-dependent brain regional Aβ production
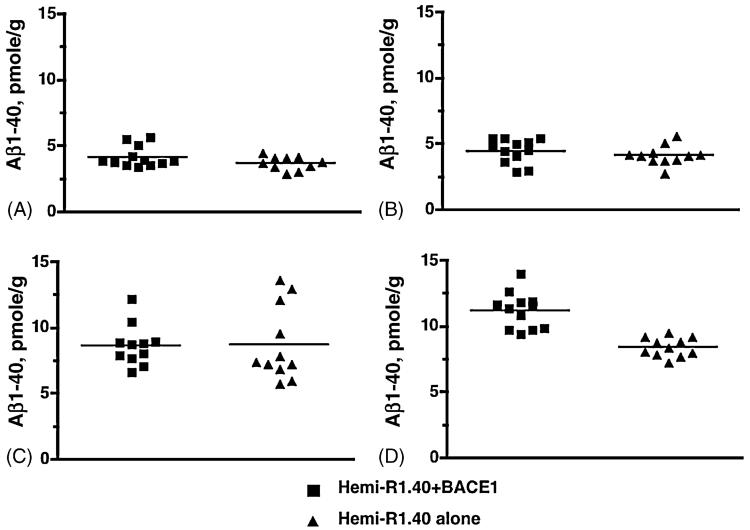
To examine age-dependent alterations in APP processing due to BACE1 expression, we compared Aβ levels in animals expressing both APP and BACE1 versus animals expressing APP alone. First, we analyzed Aβ1-40 levels in whole brain extracts by ELISA. Double transgenics for BACE1 and APP exhibit no significant differences in whole brain for Aβ production compared to APP transgenics alone (data not shown). Knowing regional BACE1 expression significantly alters Aβ levels [4], we investigated whether Aβ production differed by brain region in 2-month-old mice. Adult mice expressing human APP and human BACE1 did not exhibit any differences of Aβ1-40 levels compared to age-matched APP transgenics alone in brain region extracts for olfactory bulb, cerebellum, or hippocampus (Fig. 4A-C). However, double transgenics expressing human APP and human BACE1 did exhibit significantly higher levels of Aβ1-40 compared to age-matched APP transgenics alone in cortex, an increase of ∼ 25% (Fig. 4D) (two-tailed t-test, p-value <0.0001). These results indicate significantly elevated BACE1 protein expression in the cortex may directly influence cortical Aβ production in adult animals but not in younger animals.
Fig. 4.
Impact of BACE1 expression on Aβ production by brain region. Aβ1-40 ELISA analysis of brain region extracts from P60 BACE1/R1.40 YAC APP double transgenic mice and age-matched R1.40 YAC APP single transgenic mice. Total Aβ1-40 levels were analyzed in olfactory bulb (A), cerebellum (B), hippocampus (C), and cortex (D). BACE1/R1.40 transgenic mice have significantly higher Aβ1-40 levels in the cortex (D) compared to R1.40 transgenics alone (two-tailed t-test with Welch’s correction, p-value <0.0001).
3.5. Age-dependent regional alterations of Aβ deposition
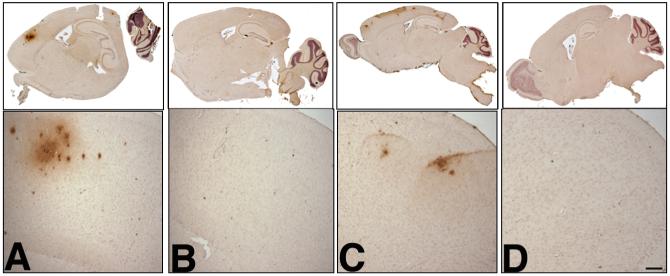
This age-related regional increase in Aβ1-40 levels is particularly striking when BACE1xAPP double transgenic animals are aged further. To examine the impact of altered regional BACE1 expression and Aβ production on Aβ deposition, we analyzed the Aβ immunohistochemical profile of BACE1xAPP double transgenic animals compared to age-matched single APP transgenics alone. Consistent with increased cortical BACE1 expression and Aβ production, Aβ deposits only developed in the cortex of double transgenics at both 13 months and 18 months (Fig. 5A and C) whereas age-matched APP transgenic controls did not develop any alterations in Aβ deposition (Fig. 5B and D). Furthermore, although Aβ pathology is strictly limited to the cortex, deposits spread throughout the primary and secondary motor cortices [31] from 13-months to the 18-month time-point, indicating an age-related expansion of amyloid deposition. This altered deposition profile indicates age-related differences in brain regional BACE1 expression influences the pattern and progression of AD-like neuropathology.
Fig. 5.
Amyloid-β deposition profile in aged BACE1xAPP animals. Brains from 13-month (A, B) and 18-month (C, D) old animals were fixed in 10% formalin, embedded in paraffin, and sectioned on a sagittal plane (10 μm thick). Scattered sections were analyzed by staining using standard protocols [17] with mAb 6E10, which detects amino acids 1-17 in the Aβ region. Shown are whole brain scans (upper) and frontal cortex brain sections (lower) of animals transgenic for both BACE1 and homozygous R1.40 YAC APP at 13 months (A) and 18 months (C). Shown are whole brain scans (upper) and frontal cortex brain sections (lower) of animals transgenic for homozygous R1.40 YAC APP alone at 13 months (B) and 18 months (D). Scale bar in D = 100 μm.
4. Discussion
According to the amyloid hypothesis, the generation of Aβ from APP is the primary pathogenic event in Alzheimer’s disease. The correlation of genetic mutations in early-onset AD pedigrees, particularly APP and the PSEN genes, with increased Aβ levels indicates amyloidogenic processing of APP may indeed be causative [10,13,23,29]. These and other mutations [33] that result in elevated Aβ production, carried out by β-secretase and γ-secretase, provide insight into how modulation of APP processing may be instrumental in AD pathogenesis. However, it is still unclear how the spatial and temporal patterns of APP processing correlate with the age-related profile of Aβ production and neuropathology observed in AD. Along these lines, increased regional activity of BACE1 [9], as the primary β-secretase gene, in the AD brain indicates BACE1 may play a major role in regulating the pattern of Aβ production and, ultimately, AD pathogenesis.
In hopes of understanding the influence of BACE1 expression on APP processing, we report that the profile of brainregional expression of human BACE1 in our β-secretase transgenic mice alters the temporal and spatial pattern of Aβ production. Specifically, we observe alterations in brain regional BACE1 protein expression as the animals age from early post-natal stages to adulthood. Interestingly, increased hippocampal BACE1 and APP expression are significantly elevated in young animals compared to adult animals. However, the regional pattern of BACE1 expression, specifically in the cortex, selectively influences regional Aβ production in adult animals, which ultimately results in distinct patterns of AD-like neuropathology. Our studies provide evidence that BACE1 regulates age-related Aβ production and deposition.
Understanding how the regulation of the APP processing pathway influences Aβ production has become a crucial area of investigation. As with most biochemical pathways, the expression and activity of the enzyme are as critical as the availability of the enzyme’s substrate with respect to the amount of product generated. In the APP processing pathway, the relative expression of APP coupled with the expression and activity of the APP secretases will likely dictate the amount of Aβ generated. Similar to previous studies [1,24,26], we observe increased APP protein expression in early post-natal animals relative to adult animals. It has been suggested that increased APP levels in young animals is due to its potential role in the developing brain [18]. However, it is not known why BACE1 protein levels are also more than 2-fold higher in younger animals. The increased BACE1 and APP protein levels lead to significant increases in Aβ levels at this young age compared to adult animals.
In our studies, elevated Aβ levels in animals co-expressing genomic transgenes of human BACE1 and human APP are primarily a result of an age-related regional distribution of both APP and BACE1 protein (refer to Table 1). Intriguingly, increased APP holoprotein levels in young animals is accompanied by selective brain-regional increases in APP processing products, namely the Aβ precursor peptide, CTF-β.
Table 1.
Summary of alterations in brain-regional BACE1 expression and APP processing
| Brain region | BACE1 expression |
HOLO-APP expression |
CTF-β levels |
Aβ levels |
Aβ deposition | ||||
|---|---|---|---|---|---|---|---|---|---|
| P7 | P60 | P7 | P60 | P7 | P60 | P7 | P60 | ||
| Olfactory bulb | +++ | +++ | + | ++ | ++ | + | ++ | ++ | None |
| Cerebellum | ++ | ++ | ++ | + | ++ | + | + | + | None |
| Hippocampus | ++++ | + | ++++ | ++ | ++++ | + | ++++ | +++ | None |
| Cortex | ++++ | ++++ | ++++ | ++ | ++++ | + | ++++ | ++++ | Positive |
(+) indicates relative levels of expression or protein processing products. P7 corresponds to post-natal day 7 animals; P60 corresponds to post-natal day 60 animals.
Along these lines, cortical and hippocampal Aβ peptides are almost 2-fold higher than Aβ levels in cerebellum and olfactory bulb. Although BACE1 expression is significantly higher in the cortex and hippocampus at this age, only Aβ levels in the hippocampus are increased relative to adult animals. One of the possible explanations for this is that the only brain region in adult animals expressing human BACE1 with increased Aβ levels is the cortex, where BACE1 protein expression is the highest. Our studies suggest that as APP expression reduces with age, BACE1 can significantly influence APP processing, possibly indicating that coordinated expression levels of BACE1 and APP alters the APP processing pathway.
However, the coordinated expression for BACE1 and APP in each of the brain regions in adult animals does not necessarily reflect the pattern of APP processing intermediates, namely CTF-β (Table 1). Although adult animals have higher Aβ levels, CTF-β levels are markedly lower compared to neonatal animals, particularly in the cortex. These discrepancies in APP processing and Aβ metabolism are consistent with previous studies analyzing the impact of reduced Bace1 expression on APP processing [25] as well as analysis of different APP transgenic mouse models [18] and suggest that APP processing is not determined solely by the expression levels of BACE1 and APP.
Several studies have shown that BACE1 activity can be regulated at a variety of different levels, including signaling through acetylcholine receptors [41], interaction with reticulon [12] or PAR-4 proteins [39] and alteration by cellular cholesterol levels [6,32,35] all of which could impact the unique profile of APP processing in the brain regions of our BACE1xAPP double transgenic animals. By stimulation of expression, protein-protein interactions, or alterations in the cellular microenvironment, each of these mechanisms can differentially regulate the activity of BACE1, resulting in a unique pattern of Aβ production and accumulation.
Degradation and clearance of Aβ from the brain is another possible mechanism by which Aβ levels are regulated. Among many proteases, recent studies have built strong evidence for the ability of two metalloproteases, neprilysin (NEP) and insulin degrading enzyme (IDE), to hydrolyze Aβ and reduce amyloid plaque burden in mice [8,14,22,27]. Similarly, two receptor-associated proteins, low-density lipoprotein receptor-related protein (LRP) and the receptor for advanced glycation end products (RAGE), are instrumental in Aβ transport from the brain to the periphery for degradation [7,16,28,34]. Since we do not observe Aβ plaque accumulation in young animals, it might be possible that Aβ clearance is more efficient at this age. The changes we observe in the regional distribution of Aβ levels as BACE1 transgenic animals age indicates Aβ degradative activity may also differ by brain region. Analyzing age-related differences in activity of known Aβ clearance genes by brain region would likely provide insight into the balance between Aβ clearance versus Aβ accumulation.
Finally, the age-related brain-regional differences in Aβ production we observe is particularly significant due to the altered regional deposition profile in the aged BACE1xAPP double transgenics. As has been described previously [21], YAC APP single transgenics only start to develop Aβ deposits in the cortex at 13 months of age, while deposits are not observed in the hippocampus until ∼20 months, indicating that endogenous Bace1 levels influence the onset and pattern of Aβ deposition. Similarly, as we have previously shown in another APPxBACE1 transgenic model [4], our studies implicate further that brain-regional BACE1 expression in aged, depositing animals corresponds with significant alterations in the regional deposition profile. The cortex-specific deposition profile in the BACE1 and YAC APP double transgenic animals indicates human BACE1 regional expression may influence the onset and site of Aβ deposition. These studies suggest that age-dependent brain-regional fluctuations in BACE1 and APP protein expression can ultimately have a dramatic impact on regional Aβ deposition (Table 1).
In summary, we have characterized unique patterns of Aβ production in our BACE1xAPP double transgenic animals. Although Aβ levels are generally increased in young animals due to elevated levels of both BACE1 and APP, the pattern of BACE1 overexpression alone significantly impacts brain regional Aβ production in an age-dependent manner. Although APP processing by brain region in adult animals of our YAC APP transgenic model has been analyzed previously [21], these studies demonstrate the functional consequences of β-secretase overexpression on regional Aβ production at different ages. These studies demonstrate how BACE1 and APP expression modulate age-related Aβ metabolism and provide invaluable insight into the regulation of the APP processing pathway. Our studies lead to interesting questions regarding the regulation of BACE1 expression and activity throughout aging and disease. Further studies examining the temporal and spatial patterns of BACE1 activity regulation and Aβ production might offer a crucial link underlying the mechanism of BACE1’s role in Aβ deposition and AD pathogenesis.
Acknowledgements
We thank S. Gandy for the generous gift of the 369 antibody and G. Evin for the generous gift of the 00/6 antibody. We would also like to thank Lucia Pastorino and Joseph Buxbaum for the generous gift of Bace1 knockout tissues, and thank Nick Varvel and Amanda Bringard for technical assistance. This work was supported by National Institutes of Health Grant AG14451, an Alzheimer’s Association grant, and an American Health Assistance Foundation Grant (to B.T.L.) as well as support from the University Alzheimer Center (AG08012) and the Ireland Cancer Center (CA43703). M.J.C. was supported in part by National Institutes of Health Training Grant GM08056-21.
References
- [1].Basha MR, Wei W, Bakheet SA, Benitez N, Siddiqi HK, Ge YW, et al. The fetal basis of amyloidogenesis: exposure to lead and latent overexpression of amyloid precursor protein and beta-amyloid in the aging brain. J Neurosci. 2005;4(25):823–9. doi: 10.1523/JNEUROSCI.4335-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bodendorf U, Danner S, Fischer F, Stefani M, Sturchler-Pierrat C, Wiederhold KH, et al. Expression of human beta-secretase in the mouse brain increases the steady-state level of beta-amyloid. J Neurochem. 2002;80(5):799–806. doi: 10.1046/j.0022-3042.2002.00770.x. [DOI] [PubMed] [Google Scholar]
- [3].Braak H, Braak E. Frequency of stages of Alzheimer-related lesions in different age categories. Neurobiol Aging. 1997;18(4):351–7. doi: 10.1016/s0197-4580(97)00056-0. [DOI] [PubMed] [Google Scholar]
- [4].Chiocco MJ, Kulnane LS, Younkin L, Younkin S, Evin G, Lamb BT. Altered amyloid-beta metabolism and deposition in genomic-based beta-secretase transgenic mice. J Biol Chem. 2004;279(50):52535–42. doi: 10.1074/jbc.M409680200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Citron M, Oltersdorf T, Haass C, McConlogue L, Hung AY, Seubert P, et al. Mutation of the beta-amyloid precursor protein in familial Alzheimer’s disease increases beta-protein production. Nature. 1992;360(6405):672–4. doi: 10.1038/360672a0. [DOI] [PubMed] [Google Scholar]
- [6].Cordy JM, Hussain I, Dingwall C, Hooper NM, Turner AJ. Exclusively targeting beta-secretase to lipid rafts by GPI-anchor addition up-regulates beta-site processing of the amyloid precursor protein. Proc Natl Acad Sci USA. 2003;100(20):11735–40. doi: 10.1073/pnas.1635130100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Deane R, Du Yan S, Submamaryan RK, LaRue B, Jovanovic S, Hogg E, et al. RAGE mediates amyloid-beta peptide transport across the blood-brain barrier and accumulation in brain. Nat Med. 2003;9(7):907–13. doi: 10.1038/nm890. [DOI] [PubMed] [Google Scholar]
- [8].Farris W, Mansourian S, Leissring MA, Eckman EA, Bertram L, Eckman CB, et al. Partial loss-of-function mutations in insulin-degrading enzyme that induce diabetes also impair degradation of amyloid beta-protein. Am J Pathol. 2004;164(4):1425–34. doi: 10.1016/s0002-9440(10)63229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fukumoto H, Cheung BS, Hyman BT, Irizarry MC. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch Neurol. 2002;59(9):1381–9. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- [10].Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349(6311):704–6. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- [11].Haass C, Hung AY, Selkoe DJ, Teplow DB. Mutations associated with a locus for familial Alzheimer’s disease result in alternative processing of amyloid beta-protein precursor. J Biol Chem. 1994;269(26):17741–8. [PubMed] [Google Scholar]
- [12].He W, Lu Y, Qahwash I, Hu XY, Chang A, Yan R. Reticulon family members modulate BACE1 activity and amyloid-beta peptide generation. Nat Med. 2004;10(9):959–65. doi: 10.1038/nm1088. [DOI] [PubMed] [Google Scholar]
- [13].Hendriks L, van Duijn CM, Cras P, Cruts M, Van Hul W, van Harskamp F, et al. Presenile dementia and cerebral haemorrhage linked to a mutation at codon 692 of the beta-amyloid precursor protein gene. Nat Genet. 1992;1(3):218–21. doi: 10.1038/ng0692-218. [DOI] [PubMed] [Google Scholar]
- [14].Iwata N, Tsubuki S, Takaki Y, Shirotani K, Lu B, Gerard NP, et al. Metabolic regulation of brain Abeta by neprilysin. Science. 2001;292(5521):1550–2. doi: 10.1126/science.1059946. [DOI] [PubMed] [Google Scholar]
- [15].Johnson-Wood K, Lee M, Motter R, Hu K, Gordon G, Barbour R, et al. Amyloid precursor protein processing and A beta42 deposition in a transgenic mouse model of Alzheimer disease. Proc Natl Acad Sci USA. 1997;94(4):1550–5. doi: 10.1073/pnas.94.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kang DE, Pietrzik CU, Baum L, Chevallier N, Merriam DE, Kounnas MZ, et al. Modulation of amyloid beta-protein clearance and Alzheimer’s disease susceptibility by the LDL receptor-related protein pathway. J Clin Invest. 2000;106(9):1159–66. doi: 10.1172/JCI11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kulnane LS, Lamb BT. Neuropathological characterization of mutant amyloid precursor protein yeast artificial chromosome transgenic mice. Neurobiol Dis. 2001;8(6):982–92. doi: 10.1006/nbdi.2001.0446. [DOI] [PubMed] [Google Scholar]
- [18].Lahiri DK, Nall C, Chen D, Zaphiriou M, Morgan C, Nurnberger JI., Sr Developmental expression of the beta-amyloid precursor protein and heat-shock protein 70 in the cerebral hemisphere region of the rat brain. Ann N Y Acad Sci. 2002;965:324–33. doi: 10.1111/j.1749-6632.2002.tb04174.x. [DOI] [PubMed] [Google Scholar]
- [19].Lamb BT, Call LM, Slunt HH, Bardel KA, Lawler AM, Eckman CB, et al. Altered metabolism of familial Alzheimer’s disease-linked amyloid precursor protein variants in yeast artificial chromosome transgenic mice. Hum Mol Genet. 1997;6(9):1535–41. doi: 10.1093/hmg/6.9.1535. [DOI] [PubMed] [Google Scholar]
- [20].Lehman EJ, Kulnane LS, Gao Y, Petriello MC, Pimpis KM, Younkin L, et al. Genetic background regulates beta-amyloid precursor protein processing and beta-amyloid deposition in the mouse. Hum Mol Genet. 2003;12(22):2949–56. doi: 10.1093/hmg/ddg322. [DOI] [PubMed] [Google Scholar]
- [21].Lehman EJ, Kulnane LS, Lamb BT. Alterations in beta-amyloid production and deposition in brain regions of two transgenic models. Neurobiol Aging. 2003;24(5):645–53. doi: 10.1016/s0197-4580(02)00153-7. [DOI] [PubMed] [Google Scholar]
- [22].Leissring MA, Farris W, Chang AY, Walsh DM, Wu X, Sun X, et al. Enhanced proteolysis of beta-amyloid in APP transgenic mice prevents plaque formation, secondary pathology, and premature death. Neuron. 2003;40(6):1087–93. doi: 10.1016/s0896-6273(03)00787-6. [DOI] [PubMed] [Google Scholar]
- [23].Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995;269(5226):973–7. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- [24].Loffler J, Huber G. Beta-amyloid precursor protein isoforms in various rat brain regions and during brain development. J Neurochem. 1992;59(4):1316–24. doi: 10.1111/j.1471-4159.1992.tb08443.x. [DOI] [PubMed] [Google Scholar]
- [25].Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P, et al. Mice deficient in BACE1, the Alzheimer’s beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci. 2001;4(3):231–2. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- [26].Marcinkiewicz M, Seidah NG. Coordinated expression of beta-amyloid precursor protein and the putative beta-secretase BACE and alpha-secretase ADAM10 in mouse and human brain. J Neurochem. 2000;75(5):2133–43. doi: 10.1046/j.1471-4159.2000.0752133.x. [DOI] [PubMed] [Google Scholar]
- [27].Marr RA, Rockenstein E, Mukherjee A, Kindy MS, Hersh LB, Gage FH, et al. Neprilysin gene transfer reduces human amyloid pathology in transgenic mice. J Neurosci. 2003;23(6):1992–6. doi: 10.1523/JNEUROSCI.23-06-01992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mohajeri MH, Saini KD, Nitsch RM. Transgenic BACE expression in mouse neurons accelerates amyloid plaque pathology. J Neural Transm. 2004;111(3):413–25. doi: 10.1007/s00702-003-0057-z. [DOI] [PubMed] [Google Scholar]
- [29].Mullan M, Crawford F, Axelman K, Houlden H, Lilius L, Winblad B, et al. A pathogenic mutation for probable Alzheimer’s disease in the APP gene at the N-terminus of beta-amyloid. Nat Genet. 1992;1(5):345–7. doi: 10.1038/ng0892-345. [DOI] [PubMed] [Google Scholar]
- [30].Pastorino L, Ikin AF, Lamprianou S, Vacaresse N, Revelli JP, Platt K, et al. BACE (beta-secretase) modulates the processing of APLP2 in vivo. Mol Cell Neurosci. 2004;25(4):642–9. doi: 10.1016/j.mcn.2003.12.013. [DOI] [PubMed] [Google Scholar]
- [31].Paxinos G, Keith BJ. The mouse brain in stereotaxic coordinates. IInd ed. Academic Press; San Diego: 2001. [Google Scholar]
- [32].Riddell DR, Christie G, Hussain I, Dingwall C. Compartmentalization of beta-secretase (Asp2) into low-buoyant density, noncaveolar lipid rafts. Curr Biol. 2001;11(16):1288–93. doi: 10.1016/s0960-9822(01)00394-3. [DOI] [PubMed] [Google Scholar]
- [33].Rocchi A, Pellegrini S, Siciliano G, Murri L. Causative and susceptibility genes for Alzheimer’s disease: a review. Brain Res Bull. 2003;61(1):1–24. doi: 10.1016/s0361-9230(03)00067-4. [DOI] [PubMed] [Google Scholar]
- [34].Shibata M, Yamada S, Kumar SR, Calero M, Bading J, Frangione B, et al. Clearance of Alzheimer’s amyloid-ss(1-40) peptide from brain by LDL receptor-related protein-1 at the blood-brain barrier. J Clin Invest. 2000;106(12):1489–99. doi: 10.1172/JCI10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sidera C, Parsons R, Austen B. Post-translational processing of beta-secretase in Alzheimer’s disease. Proteomics. 2005;5(6):1533–43. doi: 10.1002/pmic.200401185. [DOI] [PubMed] [Google Scholar]
- [36].Sinha S, Anderson JP, Barbour R, Basi GS, Caccavello R, Davis D, et al. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature. 1999;402(6761):537–40. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- [37].Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, et al. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286(5440):735–41. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- [38].Willem M, Dewachter I, Smyth N, Van Dooren T, Borghgraef P, Haass C, et al. beta-site amyloid precursor protein cleaving enzyme 1 increases amyloid deposition in brain parenchyma but reduces cerebrovascular amyloid angiopathy in aging BACE x AP [V717I] double-transgenic mice. Am J Pathol. 2004;165(5):1621–31. doi: 10.1016/s0002-9440(10)63419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xie J, Guo Q. PAR-4 is involved in regulation of beta-secretase cleavage of the Alzheimer amyloid precursor protein. J Biol Chem. 2005;280(14):13824–32. doi: 10.1074/jbc.M411933200. [DOI] [PubMed] [Google Scholar]
- [40].Yan R, Bienkowski MJ, Shuck ME, Miao H, Tory MC, Pauley AM, et al. Membrane-anchored aspartyl protease with Alzheimer’s disease beta-secretase activity. Nature. 1999;402(6761):533–7. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- [41].Zuchner T, Perez-Polo JR, Schliebs R. Beta-secretase BACE1 is differentially controlled through muscarinic acetylcholine receptor signalling. J Neurosci Res. 2004;77(2):250–7. doi: 10.1002/jnr.20152. [DOI] [PubMed] [Google Scholar]