Summary
Hepatitis B virus (HBV) covalently closed circular DNA (CCC DNA) is the source of HBV transcripts and persistence in chronically infected patients. The novel aspect of this study was to determine the effect of RNA interference (RNAi) on HBV CCC DNA when administered prior to establishment of HBV replication or during chronic HBV infection. HBV replication was initiated in HepG2 cells by transduction with HBV baculovirus. Subculture of HBV expressing HepG2 cells at 10 days post-transduction generates a system in which HBV replication is ongoing and HBV is expressed largely from CCC DNA thus simulating chronic HBV infection. HepG2 cells were transduced with short hairpin RNA (shRNA) expressing baculovirus prior to initiation of HBV replication or during chronic HBV replication and the levels of HBV RNA, HBsAg, replicative intermediates (RI), extracellular (EC) and CCC DNA species were measured. HBsAg, HBV RNA and DNA levels were markedly reduced through day 8 whether cells were transduced with shRNA prior to or during a chronic infection; however, the CCC DNA species were only affected when shRNA was administered prior to initiation of infection. We conclude that RNAi may have therapeutic value for controlling HBV replication at the level of RI and EC DNA and for reducing establishment of CCC DNA during HBV infection. Our data support previous findings demonstrating the stability of HBV CCC DNA to antiviral therapy. This study also reports the development of a novel HBV baculovirus subculture system that can be used to evaluate antiviral effects on chronic HBV replication.
INTRODUCTION
More than 350 million individuals worldwide are chronically infected with hepatitis B virus (HBV) (Kane, 1995, Margolis, 1998) and are 100 times more likely to manifest hepatocellular carcinoma than uninfected individuals (Beasley, 1988). Current treatments for HBV include nucleos(t)ide analogues and interferon-alpha administration. Interferon-alpha has a low population response rate (~30 %) and adverse side effects (Ganem & Prince, 2004). Nucleoside analogues directly block viral replication by targeting the reverse transcriptase, but do not affect viral antigen load (Lau et al., 2000, Papatheodoridis et al., 2002). This limitation can induce a host immune response resulting in hepatocellular injury (Ganem & Prince, 2004). Another major limitation of nucleoside analogues is the development of drug resistant mutants (Melegari et al., 1998). Due to low efficacy and undesirable outcomes of these treatments, new therapies capable of targeting multiple aspects of the HBV viral life cycle are needed.
The HBV genome contains four overlapping open reading frames that encode the core, polymerase, surface, and X proteins as well as the pre-genomic RNA (pgRNA), the template for HBV genome replication (Ganem & Varmus, 1987). The unique arrangement of the HBV transcripts and the fact that replication occurs through an RNA intermediate make HBV an attractive target for RNA interference (RNAi) therapy.
RNAi is the process by which gene expression is silenced through the sequence-specific degradation of mRNA (Fire et al., 1998, Lau et al., 2001). RNAi is mediated by activation of the RNA-induced silencing complex through its association with 21–23 nucleotide small interfering RNAs (siRNA) derived from Dicer-processed long double-stranded RNAs (Hammond et al., 2000, Hammond et al., 2001). Several studies have demonstrated the effectiveness of synthetic siRNA, vector-generated siRNA, and short hairpin RNA (shRNA) in the inhibition of HBV replication and antigen production (Chen et al., 2006, Jia et al., 2006, McCaffrey et al., 2003, Moore et al., 2005, Shlomai & Shaul, 2003, Uprichard et al., 2005).
Retroviral vectors expressing shRNA sequences are capable of efficiently transducing hepatocytes and inducing long-term inhibition of HBV replication (Moore et al., 2005); however, successful treatment using retroviral vectors is dependent upon cell division and genomic integration. The baculovirus Autographa californica nuclear polyhedrosis virus can mediate gene delivery to a wide variety of mammalian cells (reviewed in (Kost & Condreay, 2002, Kost et al., 2005)). Recombinant baculoviruses are capable of delivering target genes to liver-derived cells with efficiencies greater than 90 % (Boyce & Bucher, 1996, Delaney & Isom, 1998, Hofmann et al., 1995). We previously reported the generation of a system for studying HBV replication in vitro, in which a HBV-expressing baculovirus efficiently delivers the HBV genome to HepG2 cells to initiate a productive infection wherein HBV transcripts, intracellular and secreted HBV antigens, HBV DNA replicative intermediates (RI), and HBV covalently closed circular DNA (CCC DNA) are produced and extracellular (EC) HBV virions are secreted (Abdelhamed et al., 2003, Abdelhamed et al., 2002, Delaney & Isom, 1998, Heipertz et al., 2007). In this system, HBV transcription is initially driven from input HBV baculovirus DNA; however, during the first several days post-transduction, the input baculovirus DNA declines to levels virtually undetectable by Southern blot analysis and HBV CCC DNA begins to accumulate inside the nucleus where it drives HBV transcription for at least 4 weeks post-transduction (Abdelhamed et al., 2002, Heipertz et al., 2007).
Recent studies demonstrated that a baculovirus construct expressing shRNAs targeting lamin A/C can be used to transduce HepG2 cells, resulting in knockdown of the target gene (Nicholson et al., 2005). This suggests that recombinant baculovirus is an efficient vector for delivery of shRNA to HepG2 cells. In this study, we generated a baculovirus expressing a HBV-specific shRNA, and tested its ability to inhibit HBV replication when delivered prior to the initiation of HBV replication established by the previously described HBV baculovirus system (Delaney & Isom, 1998). This study also reports the development of a novel subculture system, derived from the original HBV baculovirus system, that mimics a chronic HBV infection with HBV transcription driven from HBV CCC DNA, the primary source of HBV transcripts and causative agent of HBV persistence in patients. The subculture system was used to test the ability of the anti-HBV shRNA-expressing baculovirus to inhibit chronic HBV replication.
MATERIALS AND METHODS
Cell culture
HepG2 cells were cultured in minimal medium (MEM; Invitrogen) supplemented with 10 % heat-inactivated fetal bovine serum (FBS; HyClone) (MEM-FBS) and incubated at 37 °C in a humidified incubator at 5 % CO2 (Knowles et al., 1980). Sf21 insect cells were cultured at 28 °C without CO2 in a nonhumidified incubator and maintained in Grace’s insect medium supplemented with yeastolate (Mediatech, Inc.), lactalbumin hydrolase (Mediatech, Inc.) and 10 % FBS.
Generation of shRNA sequences and expression vectors
Potential shRNA sequences against various regions of the HBV genome were generated using Ambion’s Target Finder (www.ambion.com).
To create pSilencer plasmids, the following oligos were synthesized (Macromolecular Core Facility, Penn State College of Medicine): 5′-GATCCCGGTATGTTGCCCGTTTGTCTTCAAGAGAGACAAACGGGCAACATACCTTTTT TGGAAA-3′(sense) and 5′-AGCTTTTCCAAAAAAGGTATGTTGCCCGTTTGTCTCTCTTGAAGACAAACGGGCAACA TACCGG-3′(antisense) for pSiU6S; 5′-GATCCCGCCCCCACTGGCTGGGGCTTTTCAAGAGAAAGCCCCAGCCAGTGGGGGTT TTTTGGAAA-3′ (sense) and 5′-AGCTTTTCCAAAAAACCCCCACTGGCTGGGGCTTTCTCTTGAAAAGCCCCAGCCAGT GGGGGCGG-3′ (antisense) for pSiU6E1; 5′-GATCCCATTGGTCTGCGCACCAGCATTCAAGAGATGCTGGTGCGCAGACCAATTTTTT TGGAAA-3′ (sense) and 5′-AGCTTTTCCAAAAAAATTGGTCTGCGCACCAGCATCTCTTGAATGCTGGTGCGCAGAC CAATGG-3′ (antisense) for pSiU6E2X; 5′-GATCCCGCCTCCAAGCTGTGCCTTGTTCAAGAGACAAGGCACAGCTTGGAGGCTTTT TTGGAAA-3′ (sense) and 5′-AGCTTTTCCAAAAAAGCCTCCAAGCTGTGCCTTGTCTCTTGAACAAGGCACAGCTTGG AGGCGG-3′ (antisense) for pSiU6C. Oligos were annealed and ligated into pSlilencer 2.0-U6 BamHI and EcoRI sites according to manufacturer’s instructions (Ambion). pSilencer 2.0-U6 Negative Control (Ambion) was used as a negative control (pSiU6Control). DNA sequencing was used to verify all pSiU6 plasmids created. pSiU6 plasmids were digested with HindIII and EcoRI and the U6 promoter-shRNA sequence was subsequently cloned into the HindIII-EcoRI sites of pBlueBac4.5 (pBB4.5; Invitrogen) to generate pBBU6E1, pBBU6E2X, pBBU6C, and pBBU6Control.
Generation of HBV shRNA-expressing baculovirus
HBV baculovirus was produced as described (Delaney & Isom, 1998). This method was used to generate shRNA expressing baculovirus (Delaney & Isom, 1998). Briefly, pBBU6S and pBBU6Control were individually co-transfected along with linear AcMNPV baculovirus DNA into Sf21 cells using the BacNBlue transfection kit (Invitrogen), and baculoviruses were amplified in Sf21 cells from clones isolated by plaque assay according to manufacturer’s instructions. DNA from baculoviruses was extracted as described (O’Reilly, 1997), subjected to HindIII and EcoRI restriction enzyme digestion, and visualized by Southern blotting according to described methods (Sambrook, 1989) to determine which virus isolates contained intact U6-shRNA inserts. Positive baculoviruses were amplified in Sf21 suspension cultures, concentrated, and titered by end-point dilution as described (O’Reilly, 1997).
Co-transfection and baculovirus transduction
1.5 μg of pBBU6S, pBBU6E1, pBBU6E2X, pBBU6C, and pBBU6Control were individually co-transfected along with 1.5 μg of pBB4.5HBV1.3 into HepG2 cells 16–24 hrs after seeding in 60 mm plates using Effectene transfection reagent (Qiagen) according to manufacturer’s instructions. 1.5 μg of pBB4.5HBV1.3 transfected into HepG2 cells as described above served as a negative control. Cells were fed every other day and harvested on day 4 post-transfection for analysis of HBV RI. All transfections were carried out in duplicated and repeated in triplicate in independent experiments.
Baculovirus transduction of HepG2 cells was carried out as described (Abdelhamed et al., 2003, Delaney & Isom, 1998). Briefly, HepG2 cells were seeded at a confluency of 20–40 % in 60 mm plates 16–24 hrs prior to baculovirus transduction. The day of transduction, an average cell count was determined. The appropriate volume of high-titer baculovirus stock needed to obtain desired multiplicity of infection was calculated and diluted in 0.5 mL MEM-FBS. Baculovirus inoculum was applied drop-wise to HepG2 cells, allowed to absorb for 1 hr at 37 °C with gentle rocking every 15 minutes, and removed by gently washing cells two times with PBS. HepG2 cells were re-fed and maintained as described above.
HBV DNA extraction and Southern blot
Intracellular HBV RI and EC DNA were extracted from HepG2 cells as described (Abdelhamed et al., 2003, Abdelhamed et al., 2002). HBV RI and EC DNA were migrated in 1 % agarose gels and Southern blotted as described (Sambrook, 1989). Membranes were hybridized with probe generated from a full-length double-stranded HBV genome radiolabeled with [α-32P]dCTP using a Random Prime Labeling kit (Roche Diagnostics).
To isolate CCC DNA, HBV-infected HepG2 cells were harvested and pelleted by centrifugation (1000 × g for 5 minutes) in cold PBS. Pellets were lysed on ice for 20 minutes by the addition of 750 μL of cold PBS containing 0.005 % Nonidet P-40 (Roche). The cytoplasmic fraction was separated from the nuclear fraction by centrifugation at 1700 × g at 4 °C. HBV RI DNA was extracted from the cytoplasmic fraction as described (Abdelhamed et al., 2003, Abdelhamed et al., 2002). The nuclear pellet was resuspended in 400 μL of TE (50:1) and lysed using a modified previously described procedure (Zhang & Summers, 2000). Briefly, 300 μL of a solution containing 0.15 N NaOH and 6 % SDS was added to the nuclear pellet then, rotated for 10 minutes at room temperature, followed by incubation at 37 °C for 10 minutes to allow for the irreversible denaturation of cellular DNA. The alkaline lysate was neutralized with 200 μL of a 3 M KAc (pH 5) neutralization buffer and centrifuged at 21000 × g for 20 minutes at 4 °C. The supernatant was phenol and chloroform extracted, and nucleic acid was recovered by isopropanol precipitation with the addition of 10 mu;g tRNA. Precipitated nucleic acid was resuspended in TE (10:1), digested with RNase 100 μg/mL for 30 minutes at 37 °C, and analyzed by electrophoresis and Southern blotting as described above.
HBV RNA extraction and Northern blot
The single-step acid guanidinium method (Chomczynski & Sacchi, 1987) was utilized to extract total RNA from HepG2 cells. 10 μg of total RNA was analyzed by electrophoresis and Northern blotting. The membranes were hybridized with HBV and GAPDH-specific [α-32P]dCTP -radiolabeled probes.
Baculovirus DNA extraction and Southern blot
Total DNA was harvested from baculovirus transduced HepG2 cells at various time points post-transduction as described (Abdelhamed et al., 2002). Samples were analyzed by electrophoresis and Southern blotting. A [α-32P]dCTP -radiolabeled 332 bp fragment from pBB4.5 was used to specifically detect baculovirus DNA.
Subculture of HBV-infected HepG2 cells
HepG2 cells were infected with HBV-expressing baculovirus (MOI 100) according to methods previously described (Abdelhamed et al., 2003, Delaney & Isom, 1998). On day 10 post-baculovirus mediated HBV infection, HBV-infected HepG2 cells were subcultured 1:4 into 60 mm dishes.
Measurement of secreted HBsAg
Secreted HBsAg was measured from culture media collected 24hrs post-refeeding by ELISA using Auszyme Monoclonal kit (Abbott Laboratories) according to manufacturer’s instructions. Appropriate dilutions of culture media were conducted as necessary.
Data analysis and statistical significance
Northern and Southern blots were visualized with a PhosphorImager and RNA and DNA bands were quantified using ImageQuant (Molecular Dynamics). Significance was determined using a homoscedastic two-tailed t-test. p values ≤0.05 were considered significant.
RESULTS
Selection and screen of shRNA sequences
Various regions of the HBV genome were chosen as potential RNAi target areas and four shRNA sequences against these regions were generated using Ambion’s Target Finder (www.ambion.com). All four shRNA sequences target the HBV 3.5 kb RNA (Fig. 1a), which is the template for HBV genomic replication and also the mRNA for the HBV polymerase, HBV e antigen (HBeAg) and core protein. shRNAs U6S and U6E1 also target the 2.4 and 2.1 kb mRNA transcripts for the HBV surface antigens (HBsAg). U6E2X targets all HBV transcripts, including the HBV × antigen mRNA.
FIG. 1. Inhibition of HBV RI DNA by transfection with shRNA expressing plasmids.
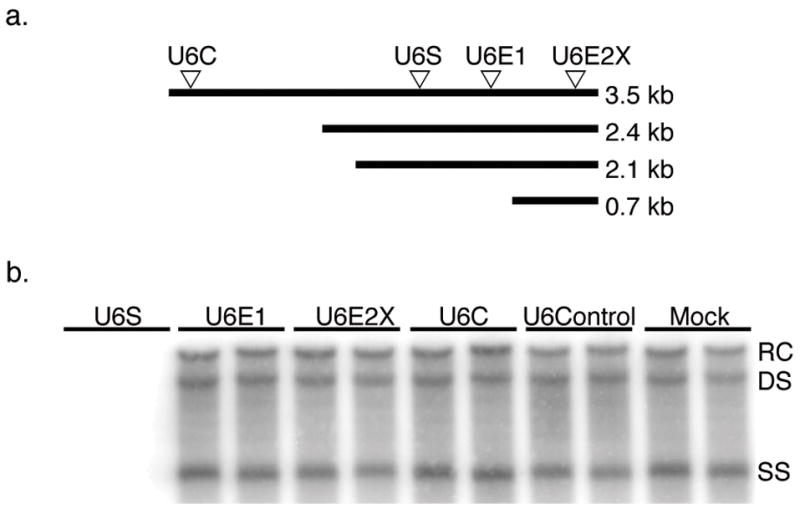
(a) Arrows represent location of RNAi target sites within the HBV transcripts. (b) HBV RI DNA was extracted from HepG2 cells four days post-cotransfection with a HBV expressing plasmid and either shRNA expressing plasmid pBBU6S, pBBU6E1, pBBU6E2X, pBBU6C or negative control pBBU6Control. Cells were also transfected with HBV expression plasmid alone (Mock). HBV RI DNA was visualized by Southern blotting. Bands indicate the relaxed circular (RC), double stranded (DS), and single stranded (SS) HBV RI DNA species.
To test the ability of each shRNA sequence to inhibit HBV replication in cell culture, a co-transfection assay was conducted. All four shRNA sequences were cloned into the baculovirus transfer vector, pBB4.5. HepG2 cells were co-transfected with pBB4.5HBV1.3, a greater than genome length HBV expression plasmid (Delaney & Isom, 1998) along with either pBBU6S, pBBU6E1, pBBU6E2X, pBBU6C, pBBU6Control or media alone (Mock). Four days post-co-transfection, cells were harvested and HBV RI were extracted and analyzed by Southern blotting (Fig. 1b). Of the four shRNA sequences tested, only U6S was capable of markedly inhibiting HBV RI levels. To our knowledge, the effects of U6E1 and U6E2X on HBV replication have not been evaluated. Previous studies showed that the U6C sequence caused a marked reduction in both HBV mRNA levels as well as HBsAg and HBeAg levels (Zhang et al., 2004). We did not investigate the effect of U6C on HBV mRNA or antigen expression, making it difficult to compare the two studies. In addition, different HBV systems as well as transfection methods were used which may account for the differences in effectiveness of the sequence. Although there is no known explanation for the failure of the other anti-HBV shRNA sequences tested, it is possible that there may be an inherent flaw in their design that affects proper strand incorporation or other biochemical factors necessary for function. U6S targets a specific region in the HBsAg reading frame that is conserved throughout all HBV genotypes and may contribute to its potency. The U6S sequence was selected for future experiments.
Pretreatment with shRNA expressing baculovirus
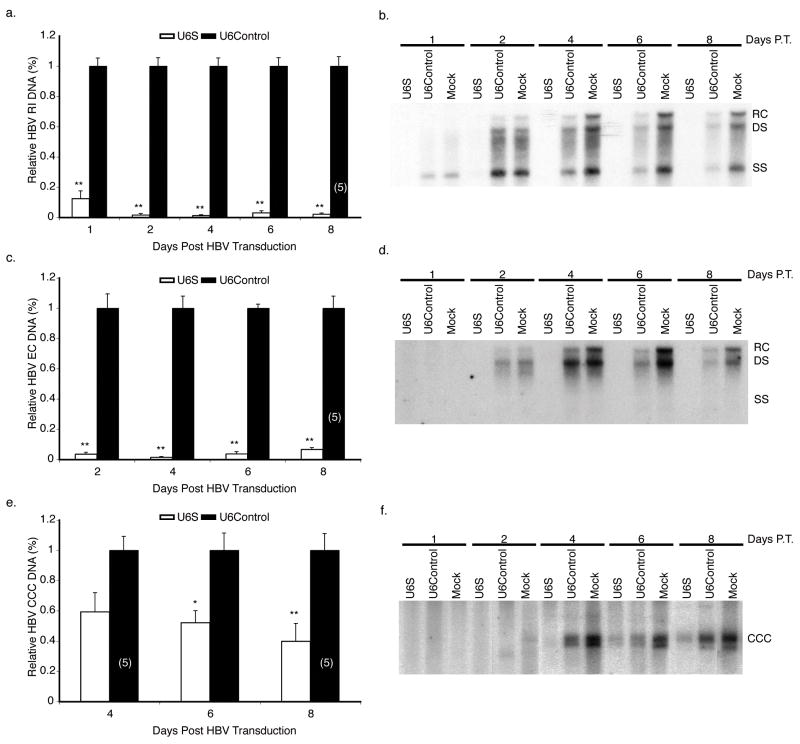
shRNA delivery is a major obstacle and determinant of RNAi efficacy. To overcome these issues, baculoviruses expressing U6S and U6Control were generated and their ability to prevent and inhibit HBV replication was examined. HepG2 cells were transduced with U6S or U6Control baculoviruses at a MOI of 100 pfu cell−1. 24 hrs later, HBV replication was initiated by transducing the cells with HBV baculovirus (MOI 100 pfu cell−1). Representative Southern blots for RI (Fig. 2b), EC (Fig. 2d), and CCC (Fig. 2f) DNAs from mock, U6Control, and U6S treated HepG2 cells transduced with HBV baculovirus are shown. It is important to note that when HepG2 cells are transduced with HBV baculovirus, RI at least with regard to single stranded HBV DNA is detectable by day 1, EC is not detectable until day 2, and CCC DNA is not detectable until day 4 post-transduction (Abdelhamed et al., 2002, Delaney & Isom, 1998, Heipertz et al., 2007). For this reason, quantitative data is provided beginning with day 1 for RI, day 2 for EC and day 4 for CCC DNAs. In these and other experiments (e.g. Fig. 5f), sometimes more than one band was observed at the expected CCC DNA position. We do not know the underlying reason; however, both bands were converted into a single 3.2 kb band upon treatment with the single-cutter enzyme Xho I (data not shown). Therefore, both bands were considered as CCC DNA and their sum was used to calculate the relative amount of CCC DNA in the samples. U6Control treatment caused some inhibition of HBV replication compared to mock treated cells; therefore, the effects of U6S on HBV replication, calculated using data from three independent experiments, are presented as a normalized mean relative to U6Control treated cells and not to mock (Fig. 2a, c, e). U6Control treatment is not toxic compared to mock treatment of cells and the U6Control nonspecific negative effect is most likely related to U6Control sequence expression. Pretreatment with U6S baculovirus significantly reduced the levels of HBV RI DNA by 97.9 ± 0.74 % (p = 3.25 E−08), EC DNA by 93.4 ± 1.10 % (p = 4.30 E−07), and CCC DNA by 60.2 ± 11.8 % (p = 0.005) compared to pretreatment with U6Control treated cells at day 8 post-initiation of HBV replication.
FIG. 2. Analysis of HBV DNA levels in HepG2 cells pretreated with shRNA-expressing baculovirus.
HBV RI (a, b), EC (c, d) and CCC (e, f) DNA were extracted from HepG2 cells transduced with the specified shRNA-expressing baculovirus at the indicated times post-initiation of HBV replication by a HBV-expressing baculovirus. In panels a, c, and e, results are standardized to U6Control treated cells and represented as means ± SEM values of three independent experiments. Representative Southern blots are shown in panels b, d, and f. n=6 unless otherwise indicated (n), p ≤ 0.05 considered statistically significant, * p ≤ 0.05, ** p ≤ 0.005. Relaxed circular (RC), double stranded (DS), single stranded (SS), and covalently closed circular (CCC) species of HBV DNA.
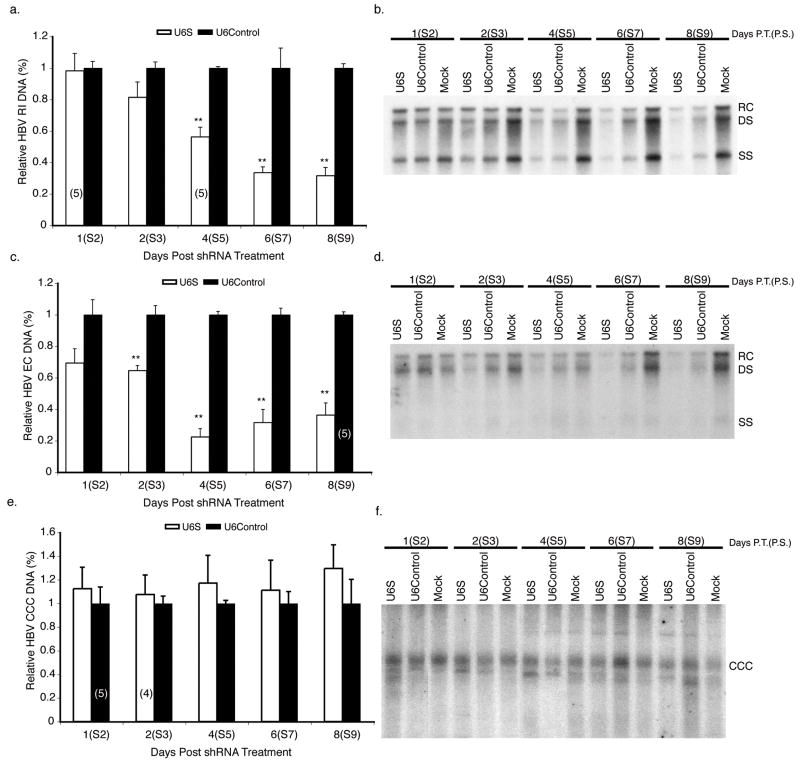
FIG. 5. Analysis of HBV DNA levels in subcultured HBV-infected HepG2 cells treated with shRNA-expressing baculovirus.
HBV RI (a, b), EC (c, d), and CCC (e, f) DNAs were extracted from HepG2 cells transduced with the specified shRNA-expressing baculovirus at the indicated times post-initiation of HBV replication with a HBV-expressing baculovirus. In panels a, c, and e, results are standardized to U6Control treated cells and represented as means ± SEM values of three independent experiments. Panels b, d, and f show representative Southern blots. n=6 unless otherwise indicated (n), p ≤ 0.05 considered statistically significant, * p ≤ 0.05, ** p ≤ 0.005. Bands indicate the relaxed circular (RC), double stranded (DS), single stranded (SS), and covalently closed circular (CCC) species of HBV DNA.
To ascertain the specificity of the shRNA for HBV transcripts, Northern blot analysis was conducted (Fig. 3a). U6S expressing baculovirus markedly reduced the 2.4/2.1 kb HBV transcripts as well as the 3.5 kb transcript by day 1 post-HBV baculovirus transduction (Fig. 3a). U6Control treatment had no effect on HBV RNA compared to mock. Results were normalized to the levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA and calculated using data obtained for U6S compared to U6Control from three independent experiments. The level of total HBV mRNA was significantly decreased by 84.1 ± 2.5 % (p = 3.80 E−10) at day 4 post-initiation of HBV replication compared to U6Control treated cells. In the U6S treated lanes of Fig. 3a, there appears to be a RNA species migrating between where the 2.4 and 2.1 kb bands are normally detected. Although we have not characterized this band, it was commonly detected in U6S treated samples and may be the product of differential splicing.
FIG. 3. Analysis of HBV mRNA levels in shRNA-expressing baculovirus pretreated HepG2 cells.
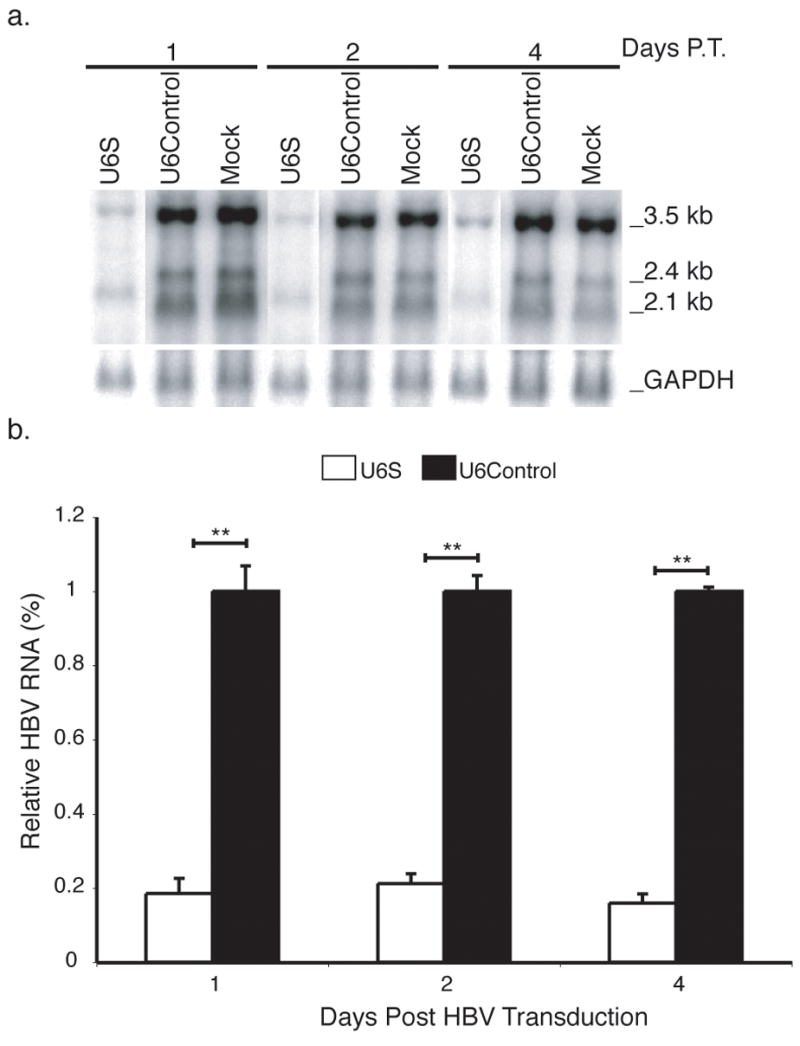
(a) Total RNA was harvested from shRNA pretreated HepG2 cells at the times indicated post-initiation of HBV replication by a HBV-expressing baculovirus and analyzed by Northern blotting. The positions of the major HBV transcripts and GAPDH are indicated. (b) Results are normalized to GAPDH, standardized to U6Control treated cells and represented as means ± SEM values of three independent experiments. n=6, p ≤ 0.05 considered statistically significant, * p ≤ 0.05, ** p ≤ 0.005.
The effect of U6S on HBsAg levels secreted into the culture media was determined. Results were calculated using data from three independent experiments and are presented as a normalized mean relative to U6Control treated cells. U6S expressing baculovirus significantly inhibited HBsAg levels compared to U6Control treated cells (Table 1).
Table 1.
Inhibition of HBsAg secretion in HepG2 cells by shRNA
| Relative HBsAg in culture medium (%)* |
|||
|---|---|---|---|
| Sample | Day 1 (P.T. HBV) | Day 2 (P.T. HBV) | Day 4 (P.T. HBV) |
| U6S | 1.13 ± 0.92† | 0.39 ± 0.26† | 0.28 ± 0.37† |
| U6Control | 100 ± 6.43 | 100 ± 6.13 | 100 ± 1.68 |
| Mock | 112.35 ± 15.43 | 89.74 ± 10.39 | 78.74 ± 15.95 |
Data represents mean ± SEM standardized to U6Control treated samples of three independent experiments performed in duplicate.
Statistical differences between U6S and U6Control were tested using Student’s t-test, P≤ 0.005.
HBV baculovirus/HepG2 subculture system
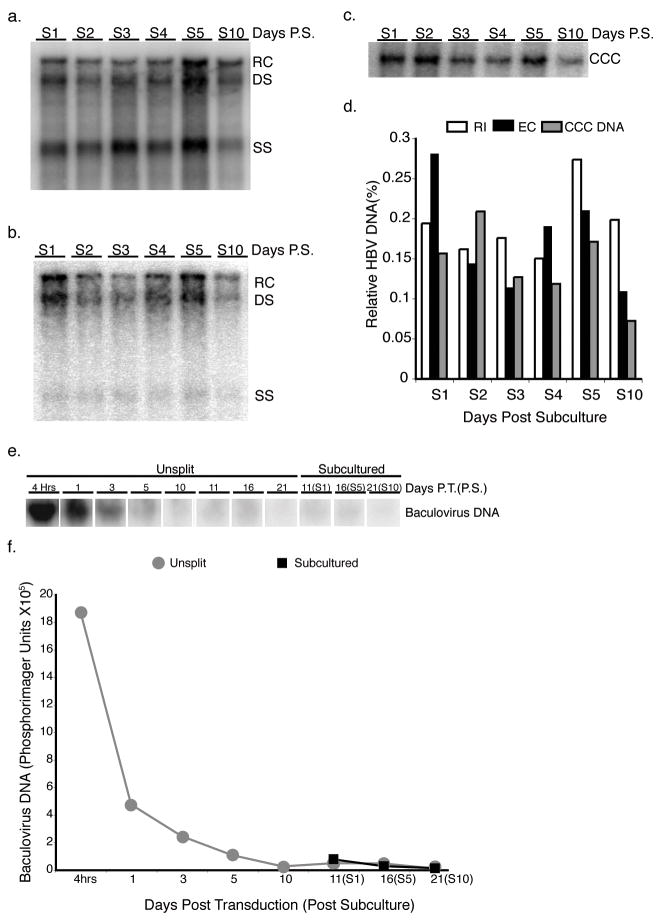
Long-term cell culture models of HBV chronic infection are limited and many rely on cell lines with integrated copies of the HBV genome. To test the ability of shRNA U6S expressing baculovirus to inhibit HBV replication under chronic conditions, we developed a novel system where HBV transcription occurs predominantly from CCC DNA. HepG2 cells were transduced with HBV baculovirus (MOI 100 pfu cell−1). At 10 days post-transduction, a time at which input baculovirus DNA is markedly reduced by Southern blot analysis and HBV transcription occurs from nuclear HBV CCC DNA (Heipertz et al., 2007), the HBV expressing HepG2 cells were subcultured at a 1:4 ratio. The subcultured cells continue to produce high levels of HBV RI (Fig. 4a), EC (Fig. 4b), and CCC (Fig. 4c) DNAs easily detectable by Southern blot analysis from day 1 until day 10 post-subculture. Southern blots for HBV RI, EC, and CCC DNAs from unsplit cells at day 10 post-transduction are not shown; however, data from PhosphorImager scans of these blots were used to calculate the relative levels of HBV DNA in subcultured cells compared to day 10 unsplit cells in Fig. 4d.
FIG. 4. Southern blot analysis of subcultured HBV-infected HepG2 cells and analysis of baculovirus DNA.
HBV RI (a), EC (b) and CCC (c) DNAs were extracted from HepG2 cells that were subcultured 1:4 10 days post-initiation of HBV replication with a HBV-expressing baculovirus at the time points indicated. (d) Quantification of the various HBV DNA species. Levels of HBV DNA in subcultured cells were standardized to day 10 unsplit cells. The relaxed circular (RC), double stranded (DS) and single stranded (SS) bands are indicated. Input baculovirus DNA was extracted from subcultured and unsplit HepG2 cells at the times indicated and analyzed by Southern blot. Image of Southern blot (e) and quantification of baculovirus input DNA (e).
To determine the levels of baculovirus DNA in the subculture system, HepG2 cells were transduced with HBV baculovirus (MOI 100 pfu cell−1) and total DNA was extracted at various time points post-transduction from both unsplit and subcultured cells. Baculovirus DNA levels markedly decrease through day 5 post-transduction and continue to decrease to virtually undetectable levels by day 10 post-transduction (Fig. 4e, f). Baculovirus DNA levels were essentially undetectable at day 21 post-transduction and day 10 post-subculture. These results agree with and extend previously published data (Heipertz et al., 2007).
shRNA mediated inhibition of a chronic HBV infection in vitro
We used the baculovirus subculture system to investigate the effect of U6S expressing baculovirus on HBV replication during a chronic infection. HepG2 cells were subcultured 1:4 at 10 days post-transduction with HBV baculovirus. 24 hrs post-subculture, the infected cells were transduced with U6S or U6Control baculovirus (MOI 100 pfu cell−1) and the inhibitory effect of U6S on HBV replication was examined. One of the strengths of the subculture system is that the HBV expressing HepG2 cells at 24 hrs after plating are at an appropriate cell density for transduction by a recombinant baculovirus, in this case, baculoviruses expressing U6S or U6Control. The inhibition of HBV replication by U6S was calculated using data from three independent experiments and is presented as a normalized mean relative to U6Control treated cells (Fig. 5a, c, e). U6S significantly inhibited HBV RI DNA levels by 68.4 ± 5.2 % (p = 3.81 E−07) and EC DNA levels by 63.7 ± 7.7 % (p = 4.42 E−05) at day 8 post-U6S transduction compared to cells transduced with U6Control (Fig. 5a, and c, respectively). No changes in HBV CCC DNA levels were observed when cells were transduced with U6S baculovirus (Fig. 5e, f).
Northern blot analysis was conducted to determine whether the observed inhibition in HBV RI and EC DNA levels was a consequence of U6S mediated degradation of HBV transcripts. U6S was capable of reducing the HBV 2.4/2.1 kb and 3.5 kb transcripts (Fig. 6a). Results were normalized to GAPDH levels. The effect of U6S on HBV mRNA levels was calculated from three independent experiments and presented as a normalized mean relative to U6Control treated cells (Fig. 6b). U6S significantly decreased total HBV mRNA levels by 81.1 ± 2.3 % (p = 6.90 E−05) at day 4 post-U6S transduction compared to U6Control treated cells (Fig. 6b).
FIG. 6. Northern blot analysis of HBV mRNA levels in shRNA-expressing baculovirus treated subcultured HBV-infected HepG2 cells.
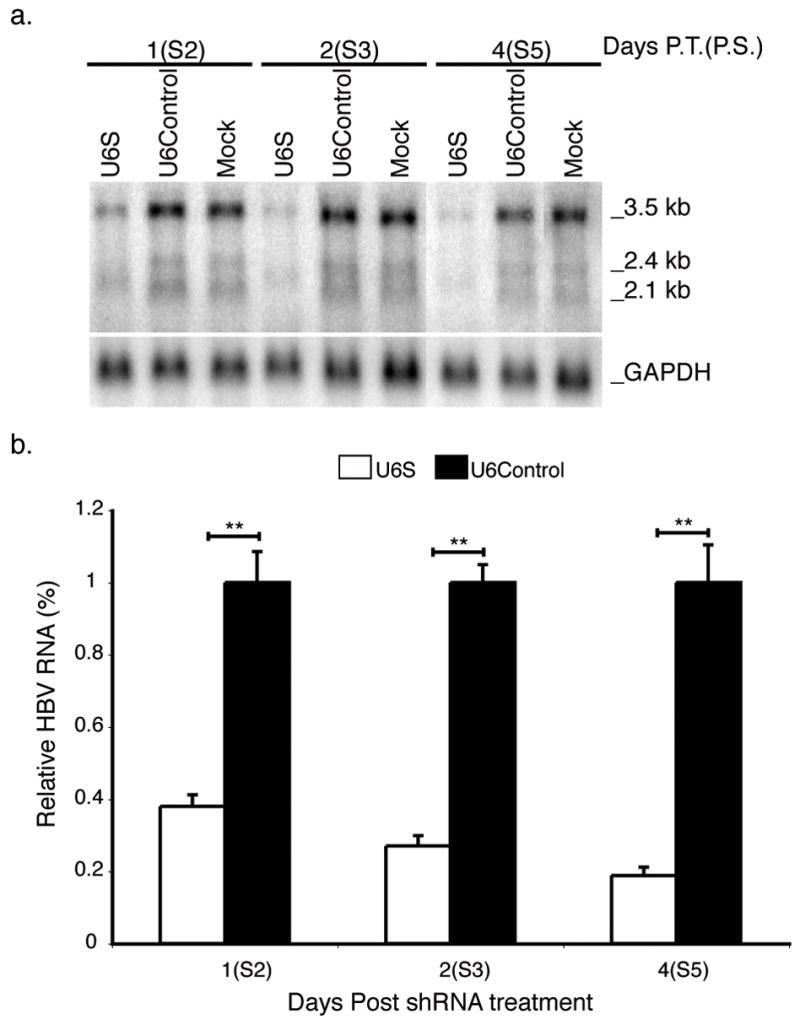
(a) Total RNA was harvested from subcultured HBV-infected HepG2 cells at the times indicated following transduction with shRNA-expressing baculovirus and analyzed by Northern blotting. The positions of the major HBV transcripts and GAPDH are indicated. (b) Results are normalized to GAPDH, standardized to U6Control treated cells and represented as means ± SEM values of three independent experiments. n=6, p ≤ 0.05 considered statistically significant, * p ≤ 0.05, ** p ≤ 0.005.
The effect of U6S on HBsAg levels secreted into the culture media was determined. Results were calculated using data from three independent experiments and are presented as a normalized mean relative to U6Control treated cells. U6S expressing baculovirus significantly inhibited HBsAg levels with time post-treatment compared to U6Control treated cells (Table 2).
Table 2.
Inhibition of HBsAg secretion in subcultured HepG2 cells by shRNA.
| Relative HBsAg in culture medium (%)* |
|||
|---|---|---|---|
| Sample | Day 2 (P.T. shRNA) | Day 4 (P.T. shRNA) | Day 6 (P.T. shRNA) |
| U6S | 42.78 ± 9.02† | 16.15 ± 4.22† | 5.38 ± 2.81† |
| U6Control | 100 ± 4.53 | 100 ± 3.75 | 100 ± 7.84 |
| Mock | 85.92 ± 13.15 | 110.09 ± 19.33 | 106.65 ± 17.06 |
Data represents mean ± SEM standardized to U6Control treated samples of three independent experiments performed in duplicate.
Statistical differences between U6S and U6Control were tested using Student’s t-test, P≤ 0.005.
DISCUSSION
Two different HBV/HepG2 cell model systems were used to evaluate the effects of a specific shRNA on HBV replication. To examine the effects on establishment of HBV replication, the original HBV recombinant baculovirus/HepG2 system we previously reported and characterized was used (Abdelhamed et al., 2003, Abdelhamed et al., 2002, Delaney & Isom, 1998, Heipertz et al., 2007). The HBV baculovirus system makes use of the fact that HepG2 cells can support high levels of HBV replication and the baculovirus can efficiently transduce HepG2 cells. The construct contains a 1.3 unit length HBV genome, strain ayw, and only HBV regulatory elements. HBV genetic information is delivered by the baculovirus to the HepG2 cell nucleus. The process of receptor mediated HBV infection, uncoating, and transport of the partially double-stranded DNA to the nucleus are bypassed, and as such, the system does not recapitulate the entry process of natural HBV replication. Within 10 hours after transduction, HBV transcripts and proteins are made and HBV capsids are found in the cytoplasm (Heipertz and Isom, unpublished data). Intracellular RI are detected during the first day post-transduction and EC and nuclear HBV CCC DNA are not detected before day 2 post-transduction. When HepG2 cells are transduced with a recombinant baculovirus containing a reporter gene under the control of a mammalian promoter, such as CMV promoter-driven LacZ, the expression of the gene product is transient, peaking at 48 to 72 hrs post-transduction and becoming undetectable by 5 to 6 days post-transduction. In contrast, when HepG2 cells are transduced with HBV baculovirus, HBV replication, including production of CCC DNA, is easily detectable for at least 30 to 35 days post-transduction (Abdelhamed et al., 2002). Input baculovirus DNA levels in transduced HepG2 cells decline rapidly between 4 and 24 hrs post-transduction and continue to decline becoming only minimally detectable by 5–10 days post-transduction. HBV transcription including production of HBV pgRNA occurs initially from the input HBV baculovirus DNA template. As CCC DNA accumulates, a switch occurs with HBV transcription being driven from CCC DNA by 5–10 days post-transduction. In the HBV baculovirus system, HBV capsids deliver newly synthesized HBV genomes back into the nucleus.
The present study examined the effects of an anti-HBsAg shRNA-expressing baculovirus on HBV replication. Four regions of the HBV genome were chosen as potential RNAi target areas. Treatment of cells with U6S, a sequence that targets the HBsAg, core, HBeAg, and polymerase mRNAs, as well as the HBV pgRNA resulted in significant suppression of viral CCC DNA when administered to cells 24 hrs prior to transduction with HBV baculovirus, although not to the same magnitude as was observed for RI and EC DNAs. These data demonstrated that U6S is capable of diminishing establishment of HBV replication in vitro. Although U6S in this study was developed independently, the powerful antiviral effects of this sequence have been previously reported and are comparable to those in this study. For example, a greater than 98 % inhibition of HBsAg and 80 % inhibition of HBV RNA were achieved when HepG2.2.15 cells were transduced with an adeno-associated virus shRNA expression vector (Moore et al., 2005). The present study is unique because it is the first study to evaluate the effects of this shRNA on HBV CCC DNA. The finding that pretreatment with U6S had a greater effect on RI or EC than CCC HBV DNAs was similar to the effects of lamivudine or L-FMAU on HBV replication using the HBV baculovirus system (Abdelhamed et al., 2003, Abdelhamed et al., 2002, Delaney et al., 1999).
Although the ideal goal is to develop an antiviral or combination of antiviral agents that will eliminate ongoing chronic HBV infection, there is value in identifying therapies that can block the initiation of HBV infection. Preventing recurrent HBV infection in liver transplantation patients is a vital component of successful therapy, but few effective treatment options are currently available (O’Grady et al., 1992, Todo et al., 1991). Sustained administration of HBV immunoglobulin and nucleoside analogues prior to and following liver transplantation has been shown to increase survival rates (Han et al., 2000, Markowitz et al., 1998, Zheng et al., 2006), but these agents can select for drug-resistant mutants (Melegari et al., 1998). The data reported in the current study indicate that pretreatment with anti-HBV shRNA U6S can delay or reduce establishment of replication in vitro. Unlike nucleoside analogues, which inhibit HBV only at the level of reverse transcription, the U6S sequence not only diminishes HBV DNA levels, but also inhibits the production of HBV RNA and protein (Cheng et al., 2005, Lau et al., 2000, Wu et al., 2005).
Many studies on HBV replication in vitro rely on cell lines containing integrated copies of the viral genome or cells transduced with HBV expression vectors. Although these systems are useful for studying the HBV life cycle and therapeutic intervention, they are incapable of modeling true chronic HBV infection. Because naturally occurring HBV transcription and replication are driven by nuclear HBV CCC DNA, it is crucial to assess the efficacy of anti-HBV agents in a system that mimics these conditions. We have modified the original HBV baculovirus system to include the usage of subculture at day 10 post-transduction to generate a HBV system in which transcription is driven from CCC DNA and HBV replication is ongoing. This is quite different from what is observed when HBV replication is initiated by transduction with HBV baculovirus where EC and CCC DNA are not detectable before day 2 post-transduction (Abdelhamed et al., 2002, Delaney & Isom, 1998). Using this model, we have demonstrated that although the U6S shRNA sequence significantly reduces HBV transcripts and inhibits HBV RI and EC DNA in chronically infected cells, it does not affect CCC DNA levels. Indeed these findings could be considered negative since the shRNA had no effect on preformed CCC DNA levels, but these data are highly important because they reemphasize the fact that HBV CCC DNA is resilient to antiviral attack, and support the concept that CCC DNA is extremely stable, once formed. In addition, U6S does not completely inhibit production of intercellular RI and virions and RI DNA-driven recycling; therefore, highly stable CCC DNA pools are replenished, thus maintaining a constant level of nuclear CCC DNA. It has been previously reported that transfection of HepG2.2.15 cells with vector-based siRNAs targeted to the HBV core nuclear localization signal either alone (Li et al., 2007) or in combination (Xin et al., 2008) inhibit HBV CCC DNA as measured by RT-PCR. It is not possible to compare these studies with our data due to differences in siRNA target sequences, in vitro HBV model systems, quantification, and methods of CCC DNA extraction; specifically, isolation of CCC DNA from the culture medium (Li et al., 2007) versus extraction of CCC DNA from the nuclear fraction of infected cells in our study.
Success at truly combating chronic HBV will require targeting multiple different stages in the HBV life cycle. The unique method of action and demonstrated efficacy of RNAi-based therapies make them an attractive complement to existing therapeutic regimes. Recombinant baculovirus vectors are valuable tools for research because of their ability to efficiently and reproducibly transduce cells in culture and modulate gene expression. They also have therapeutic potential. Baculovirus expressing the diptheria toxin under control of the glial fibrillary acidic protein promoter has been used successfully to inhibit the growth of glioma xenografts in rat brain (Wang et al., 2006). Several groups also have been able to elicit both humoral and cellular immune responses in vivo using recombinant baculovirus expression and display vectors (Abe et al., 2003, Kim et al., 2007, Strauss et al., 2007).
The goals of this study were to evaluate the use of RNAi technology on blocking the initiation of HBV replication and on inhibition of chronic ongoing HBV replication. In the process of addressing the latter question, the HBV baculovirus subculture system was established and its efficacy as a model for antiviral testing using an shRNA was evaluated. The HBV baculovirus system has specific advantages for drug studies because it is possible to measure the effect of antiviral agents on all aspects of the HBV life cycle including CCC DNA, and to do so quantitatively. HBV CCC DNA is present at sufficient levels in the original, as well as the subculture system, to be detected by Southern blot analysis. In addition, as demonstrated in this report, agents delivered by gene therapy can also be evaluated; specifically, the reseeding process after subculture makes it possible to reproducibly carry out a super-transduction with a second recombinant baculovirus expressing a DNA sequence of interest, in this case an shRNA. The HBV baculovirus subculture system has the potential to be used in the future to analyze the efficacy of nucleoside analogues, novel small molecule inhibitors, RNAi, or other agents alone or in combination on chronic HBV replication.
Acknowledgments
The molecular imaging and PhosphorImager analysis of Southern and Northern blots and the synthesis of oligodeoxnucleotides used in this study were carried out in the Macromolecular Core Facility of the Pennsylvania State University College of Medicine.
This work was supported, in part, by CA23931 to H.C.I. from the National Institutes of Health and also under a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds; the Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
Footnotes
Publisher's Disclaimer: This is an author manuscript that has been accepted for publication in Journal of General Virology, copyright Society for General Microbiology, but has not been copy-edited, formatted or proofed. Cite this article as appearing in Journal of General Virology. This version of the manuscript may not be duplicated or reproduced, other than for personal use or within the rule of 'Fair Use of Copyrighted Materials' (section 17, Title 17, US Code), without permission from the copyright owner, Society for General Microbiology. The Society for General Microbiology disclaims any responsibility or liability for errors or omissions in this version of the manuscript or in any version derived from it by any other parties. The final copy-edited, published article, which is the version of record, can be found at http://vir.sgmjournals.org, and is freely available without a subscription.
References
- Abdelhamed AM, Kelley CM, Miller TG, Furman PA, Cable EE, Isom HC. Comparison of anti-hepatitis B virus activities of lamivudine and clevudine by a quantitative assay. Antimicrob Agents Chemother. 2003;47:324–36. doi: 10.1128/AAC.47.1.324-336.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelhamed AM, Kelley CM, Miller TG, Furman PA, Isom HC. Rebound of hepatitis B virus replication in HepG2 cells after cessation of antiviral treatment. J Virol. 2002;76:8148–60. doi: 10.1128/JVI.76.16.8148-8160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe T, Takahashi H, Hamazaki H, Miyano-Kurosaki N, Matsuura Y, Takaku H. Baculovirus induces an innate immune response and confers protection from lethal influenza virus infection in mice. J Immunol. 2003;171:1133–9. doi: 10.4049/jimmunol.171.3.1133. [DOI] [PubMed] [Google Scholar]
- Beasley RP. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer. 1988;61:1942–56. doi: 10.1002/1097-0142(19880515)61:10<1942::aid-cncr2820611003>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Boyce FM, Bucher NL. Baculovirus-mediated gene transfer into mammalian cells. Proc Natl Acad Sci U S A. 1996;93:2348–52. doi: 10.1073/pnas.93.6.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Ko TM, Ma HI, Wu HL, Xiao X, Li J, Chang CM, Wu PY, Chen CH, Han JM, Yu CP, Jeng KS, Hu CP, Tao MH. Long-term inhibition of hepatitis B virus in transgenic mice by double-stranded adeno-associated virus 8-delivered short hairpin RNA. Gene Ther. 2006 doi: 10.1038/sj.gt.3302846. [DOI] [PubMed] [Google Scholar]
- Cheng TL, Chang WW, Su IJ, Lai MD, Huang W, Lei HY, Chang WT. Therapeutic inhibition of hepatitis B virus surface antigen expression by RNA interference. Biochem Biophys Res Commun. 2005;336:820–30. doi: 10.1016/j.bbrc.2005.08.173. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Delaney WEI, Isom HC. Hepatitis B virus replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology. 1998;28:1134–46. doi: 10.1002/hep.510280432. [DOI] [PubMed] [Google Scholar]
- Delaney WEt, Miller TG, Isom HC. Use of the hepatitis B virus recombinant baculovirus-HepG2 system to study the effects of (−)-beta-′,3′-dideoxy-3′-thiacytidine on replication of hepatitis B virus and accumulation of covalently closed circular DNA. Antimicrob Agents Chemother. 1999;43:2017–26. doi: 10.1128/aac.43.8.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118–29. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–93. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–50. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Han SH, Ofman J, Holt C, King K, Kunder G, Chen P, Dawson S, Goldstein L, Yersiz H, Farmer DG, Ghobrial RM, Busuttil RW, Martin P. An efficacy and cost-effectiveness analysis of combination hepatitis B immune globulin and lamivudine to prevent recurrent hepatitis B after orthotopic liver transplantation compared with hepatitis B immune globulin monotherapy. Liver Transpl. 2000;6:741–8. doi: 10.1053/jlts.2000.18702. [DOI] [PubMed] [Google Scholar]
- Heipertz RA, Jr, Miller TG, Kelley CM, Delaney WEt, Locarnini SA, Isom HC. In vitro study of the effects of precore and lamivudine-resistant mutations on hepatitis B virus replication. J Virol. 2007;81:3068–76. doi: 10.1128/JVI.02341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C, Sandig V, Jennings G, Rudolph M, Schlag P, Strauss M. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc Natl Acad Sci U S A. 1995;92:10099–103. doi: 10.1073/pnas.92.22.10099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia F, Zhang YZ, Liu CM. A retrovirus-based system to stably silence hepatitis B virus genes by RNA interference. Biotechnol Lett. 2006;28:1679–85. doi: 10.1007/s10529-006-9138-z. [DOI] [PubMed] [Google Scholar]
- Kane M. Global programme for control of hepatitis B infection. Vaccine. 1995;13(Suppl 1):S47–9. doi: 10.1016/0264-410x(95)80050-n. [DOI] [PubMed] [Google Scholar]
- Kim CH, Yoon JS, Sohn HJ, Kim CK, Paik SY, Hong YK, Kim TG. Direct vaccination with pseudotype baculovirus expressing murine telomerase induces anti-tumor immunity comparable with RNA-electroporated dendritic cells in a murine glioma model. Cancer Lett. 2007;250:276–83. doi: 10.1016/j.canlet.2006.10.022. [DOI] [PubMed] [Google Scholar]
- Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497–9. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- Kost TA, Condreay JP. Recombinant baculoviruses as mammalian cell gene-delivery vectors. Trends Biotechnol. 2002;20:173–80. doi: 10.1016/s0167-7799(01)01911-4. [DOI] [PubMed] [Google Scholar]
- Kost TA, Condreay JP, Jarvis DL. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat Biotechnol. 2005;23:567–75. doi: 10.1038/nbt1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau GK, Tsiang M, Hou J, Yuen S, Carman WF, Zhang L, Gibbs CS, Lam S. Combination therapy with lamivudine and famciclovir for chronic hepatitis B-infected Chinese patients: a viral dynamics study. Hepatology. 2000;32:394–9. doi: 10.1053/jhep.2000.9143. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Li GQ, Gu HX, Li D, Xu WZ. Inhibition of Hepatitis B virus cccDNA replication by siRNA. Biochem Biophys Res Commun. 2007;355:404–8. doi: 10.1016/j.bbrc.2007.01.163. [DOI] [PubMed] [Google Scholar]
- Margolis HS. Hepatitis B virus infection. Bull World Health Organ. 1998;76(Suppl 2):152–3. [PMC free article] [PubMed] [Google Scholar]
- Markowitz JS, Martin P, Conrad AJ, Markmann JF, Seu P, Yersiz H, Goss JA, Schmidt P, Pakrasi A, Artinian L, Murray NG, Imagawa DK, Holt C, Goldstein LI, Stribling R, Busuttil RW. Prophylaxis against hepatitis B recurrence following liver transplantation using combination lamivudine and hepatitis B immune globulin. Hepatology. 1998;28:585–9. doi: 10.1002/hep.510280241. [DOI] [PubMed] [Google Scholar]
- McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, Wieland SF, Marion PL, Kay MA. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639–44. doi: 10.1038/nbt824. [DOI] [PubMed] [Google Scholar]
- Melegari M, Scaglioni PP, Wands JR. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology. 1998;27:628–33. doi: 10.1002/hep.510270243. [DOI] [PubMed] [Google Scholar]
- Moore MD, McGarvey MJ, Russell RA, Cullen BR, McClure MO. Stable inhibition of hepatitis B virus proteins by small interfering RNA expressed from viral vectors. J Gene Med. 2005;7:918–25. doi: 10.1002/jgm.739. [DOI] [PubMed] [Google Scholar]
- Nicholson LJ, Philippe M, Paine AJ, Mann DA, Dolphin CT. RNA interference mediated in human primary cells via recombinant baculoviral vectors. Mol Ther. 2005;11:638–44. doi: 10.1016/j.ymthe.2004.12.010. [DOI] [PubMed] [Google Scholar]
- O’Grady JG, Smith HM, Davies SE, Daniels HM, Donaldson PT, Tan KC, Portmann B, Alexander GJ, Williams R. Hepatitis B virus reinfection after orthotopic liver transplantation. Serological and clinical implications. J Hepatol. 1992;14:104–11. doi: 10.1016/0168-8278(92)90138-f. [DOI] [PubMed] [Google Scholar]
- O’Reilly DR. Use of baculovirus expression vectors. Methods Mol Biol. 1997;62:235–46. doi: 10.1385/0-89603-480-1:235. [DOI] [PubMed] [Google Scholar]
- Papatheodoridis GV, Dimou E, Papadimitropoulos V. Nucleoside analogues for chronic hepatitis B: antiviral efficacy and viral resistance. Am J Gastroenterol. 2002;97:1618–28. doi: 10.1111/j.1572-0241.2002.05819.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Analysis and Cloning of Eukaryotic Genomic DNA. In: Nolan C, editor. Molecular Cloning. 2. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989. pp. 9.1–9.62. [Google Scholar]
- Shlomai A, Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764–70. doi: 10.1053/jhep.2003.50146. [DOI] [PubMed] [Google Scholar]
- Strauss R, Huser A, Ni S, Tuve S, Kiviat N, Sow PS, Hofmann C, Lieber A. Baculovirus-based vaccination vectors allow for efficient induction of immune responses against plasmodium falciparum circumsporozoite protein. Mol Ther. 2007;15:193–202. doi: 10.1038/sj.mt.6300008. [DOI] [PubMed] [Google Scholar]
- Todo S, Demetris AJ, Van Thiel D, Teperman L, Fung JJ, Starzl TE. Orthotopic liver transplantation for patients with hepatitis B virus-related liver disease. Hepatology. 1991;13:619–26. [PMC free article] [PubMed] [Google Scholar]
- Uprichard SL, Boyd B, Althage A, Chisari FV. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc Natl Acad Sci U S A. 2005;102:773–8. doi: 10.1073/pnas.0409028102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Li F, Yang Y, Guo HY, Wu CX, Wang S. Recombinant baculovirus containing the diphtheria toxin A gene for malignant glioma therapy. Cancer Res. 2006;66:5798–806. doi: 10.1158/0008-5472.CAN-05-4514. [DOI] [PubMed] [Google Scholar]
- Wu HL, Huang LR, Huang CC, Lai HL, Liu CJ, Huang YT, Hsu YW, Lu CY, Chen DS, Chen PJ. RNA interference-mediated control of hepatitis B virus and emergence of resistant mutant. Gastroenterology. 2005;128:708–16. doi: 10.1053/j.gastro.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XM, Li GQ, Jin YY, Zhuang M, Li D. Combination of small interfering RNAs mediates greater suppression on hepatitis B virus cccDNA in HepG2.2.15 cells. World J Gastroenterol. 2008;14:3849–54. doi: 10.3748/wjg.14.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XN, Xiong W, Wang JD, Hu YW, Xiang L, Yuan ZH. siRNA-mediated inhibition of HBV replication and expression. World J Gastroenterol. 2004;10:2967–71. doi: 10.3748/wjg.v10.i20.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Summers J. Low dynamic state of viral competition in a chronic avian hepadnavirus infection. J Virol. 2000;74:5257–65. doi: 10.1128/jvi.74.11.5257-5265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Chen Y, Liang T, Lu A, Wang W, Shen Y, Zhang M. Prevention of hepatitis B recurrence after liver transplantation using lamivudine or lamivudine combined with hepatitis B Immunoglobulin prophylaxis. Liver Transpl. 2006;12:253–8. doi: 10.1002/lt.20701. [DOI] [PubMed] [Google Scholar]