Abstract
Gemtuzumab ozogamicin (GO) is an anti-CD33 antibody conjugated with calicheamicin (a cytotoxic antibiotic) that is used for the treatment of acute myeloid leukemia (AML). The relationship between the CD33 expression in leukemic cells and response to GO treatment has been controversial. We studied CD33 transcript and protein expression as well as polymorphisms in the CD33 gene in 22 uniformly treated pediatric AML patients and correlated the results with minimal residual disease (MRD) findings before and after GO. We found that a nonsynonymous coding change (Ala14Val) in CD33 was significantly associated with response to GO (P = .02) whereas CD33 transcript and protein expression were not (P > .2). The results suggest a novel mechanism of resistance to GO, one that may extend to other immunotoxins.
Introduction
Gemtuzumab ozogamicin (GO) consists of a humanized anti-CD33 monoclonal antibody linked to calicheamicin, a potent anti-tumor antibiotic. Because approximately 90% of patients with acute myeloid leukemia (AML) have myeloid blasts that express CD33 antigen,1 GO delivers calicheamicin is an attractive agent to treat AML cells. In the United States, GO is formulated and marketed as Mylotarg®, and is approved as monotherapy for patients greater than 60 years of age with a first relapse of AML who are ineligible for other cytotoxic therapy.2 A Medical Research Council (MRC) trial in younger adults indicated that the addition of GO (3 mg/m2 on day 1) to induction chemotherapy reduced the risk of relapse (37% vs. 52% at 3 years) without significantly increasing toxicity, thus resulting in a significantly better disease-free survival (DFS) in the GO arm (51% vs. 40% at 3 years).3
After binding to the CD33 antigen on the surface of myeloid cells, the GO-CD33 complex is internalized and calicheamicin is released. By binding to the DNA minor groove in a sequence specific manner, calicheamicin causes DNA double strand breaks, and cell killing.4 Thus, the binding of GO to cell surface CD33 and subsequent internalization is critical for its antileukemic effect.
In a recent study of patients with relapsed AML who received treatment with GO, significantly higher mean levels of CD33 protein expression and lower P-glycoprotein activity were observed in responders compared with nonresponders.5 However, other studies have failed to demonstrate a significant impact of CD33 expression of AML blasts on response to GO.6 Further, pharmacokinetic studies with GO have reported large inter-subject variability in both pediatric and adult patients.7;8 In the present study, we undertook a comprehensive analysis of CD33 expression, including polymorphisms in the CD33 gene and correlated the results to response to GO as measured by changes in minimal residual disease (MRD) levels.
Materials and Methods
Patients and treatment
Details of the multicenter AML02 trial have recently been presented.9 Briefly, patients were randomly assigned to receive high-dose or low-dose cytarabine (A), daunorubicin (D), and etoposide (E) as Induction I (combined therapy, ADE); all subsequent therapy is risk-adapted. Minimal residual disease (MRD) was monitored by flow cytometry as previously described. 10 MRD positivity was defined as 0.1% or more cells with a leukemia-associated immunophenotypes among bone marrow mononuclear cells. Initially, patients with no response (NR, ≥ 25% bone marrow blasts) to Induction I received low-dose ADE+GO (GO; 3 mg/m2) as Induction II, whereas all others received ADE. Because ADE + GO was effective in reducing tumor burden in patients with NR to Induction I and was well tolerated, the protocol was amended in February 2005 to expand the use of ADE + GO to patients with ≥ 1% MRD after Induction I. Five patients received ADE + GO because of NR to induction I, whereas 3 patients received ADE + GO because of MRD levels ≥ 1%. Patients who were MRD+ after two courses of induction therapy and who have not previously received GO, were given GO, 6 mg/m2 as a single agent, before consolidation chemotherapy or stem cell transplantation (n=14). The MRD levels were measured before (ranging from days 1 to 9) and after treatment (between days 11 to 20) with GO.10
The study design and investigations are approved by the St. Jude Children’s Research Hospital Institutional Review Board. Written informed consent and assent were obtained from patients, parents, or guardians, as appropriate, before enrollment in the study.
All 22 patients received GO, either as single agent (n=14) or in combination with ADE (n=8) are included in the present study.
CD33 gene sequencing
The CD33 gene has 7 exons that encode for a protein of 364 amino acids. We sequenced all 7 exons and the flanking intronic sequence in all 22 patients with AML who had received GO prior to August 2005 (Primer sequences are available on request). Amplification was carried out in a 1 × PCR buffer using 5 ng of DNA (obtained at diagnosis), as described earlier.11 Sequencing was carried out using an ABI Prism 3700 automated sequencer and PCR primers. Sequences were assembled using the Phred-Phrap-Consed package as described earlier.11
Statistical analysis
We used therapy-stratified permutation tests to explore the association of change in MRD with other variables. Each analysis stratified according to whether or not patients received ADE in combination with their GO therapy. An MRD-change variable was computed by subtracting post-GO MRD from pre-GO MRD and dividing this result by pre-GO MRD. Negative values indicate a decrease in MRD during GO treatment. To measure the association of MRD-change with clinical variables, we computed the Wilcoxon rank-sum or Kruskal-Wallis statistic within each stratum, and determined a weighted average of the statistics from the two strata. The weights were proportional to sample size. The association of MRD-change with genotype was measured with weighted average of Spearman correlation coefficients of MRD-change with the number of copies of a specific allele. The significance of the averaged statistics was determined by 10,000 stratified permutations of MRD-change variable. We used the classical Wilcoxon rank-sum test to examine the association of MRD-change with therapy (GO alone vs. ADE+GO). For each polymorphism, the classical Spearman correlation was used to examine the association of CD33 expression levels with the number of alleles. All tests are two sided; no multiple testing adjustments were used. The analysis was performed using an exact routine implemented in R software.
Results and Discussion
CD33 expression and response to GO
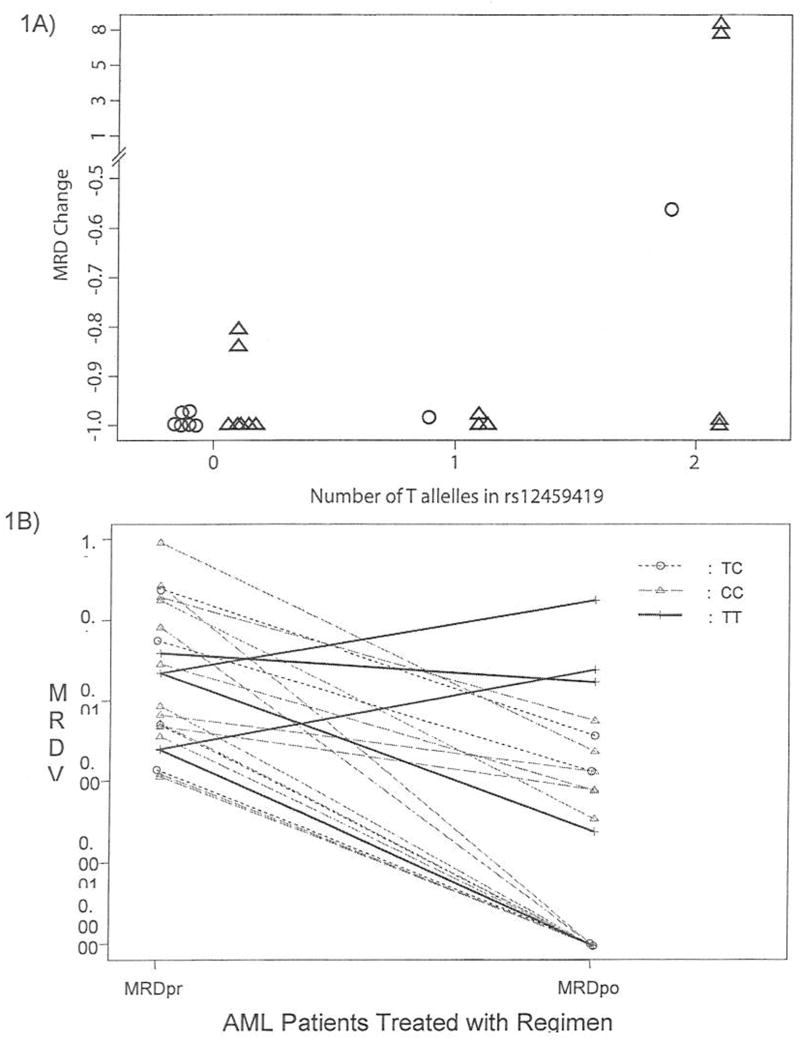
Among the 22 patients studied, 14 had less than 0.1% residual disease following treatment with GO, five had between 0.1% and 0.5% residual disease (Fig 1B). The median and upper quartiles of the MRD-change variable were −0.998 and −0.972, respectively. However, three patients had a very poor response. Two of them had an increase in MRD (patient #1, 0.0234 to 0.188, MRD change = 7.03; patient #2, 0.00264 to 0.0258, MRD change = 8.77) and one with slight decrease (0.0414 to 0.0181, MRD change = −0.56). The MRD change was not associated with initial induction arm, race, combination of GO with ADE, risk group, karyotype or FAB (P > 0.39 for each comparison).
Fig 1. Association of rs12459419 (Ala14Val) polymorphism with response to GO in pediatric patients with AML.
(A) Change in the MRD of patients with AML after treatment including GO, according to number of T alleles (Val) in rs12459419. ○ indicates measure of change in MRD for patients with AML who received ADE+GO; Δ indicates measure of change in MRD for patients with AML who received GO. P=0.026. (B) MRD levels in leukemic blasts pre and post treatment with GO containing regimens in 22 patients with AML. The lines joining the MRD levels of patients also depict the genotype for rs12459419 as shown in the inset.
Data on CD33 transcript expression as measured by Affymetrix 133 GeneChip in leukemic cells obtained at diagnosis were available for 15 patients. There was no significant association between MRD-change and CD33 expression (P = .80). Additionally, we did not observe significant evidence of an association of MRD-change with flow-cytometric measurements of the percentage of blasts expressing CD33 (P = .23). The three non-responders had similar expression of CD33 (measured in either assay). There was no significant relation between GO response and ABCB1 (Pgp1) transcript levels measured in the 15 patients studied for global gene expression by micro array (P=.55)
Single nucleotide polymorphisms in CD33
Sequencing of CD33 coding region in 22 AML patients identified 8 polymorphisms (Table 1). However, only Val14Ala, Arg69Gly, Arg304Gly, Ser305Pro, and a 3′UTR SNP occurred with a minimum allele frequency of greater than 10%.
We determined the association of the genotype of each of the CD33 SNPs with changes in MRD during treatment with GO. None of the SNPs were significantly associated with CD33 expression levels (P > .34 for each polymorphism). We found that the number of T alleles of rs12459419 has a significant positive association with the MRD-change variable (P = .024), with increasing number of T alleles associated with a poorer response to GO-containing regimens (either alone or in combination with ADE) (Fig. 1A). None of the other 7 polymorphisms were significantly associated with change in MRD during treatment with GO (P > .35 for each polymorphism). MRD levels in each patient before and after GO-containing regimens are shown in Fig. 1B. Out of 5 patients homozygous for rs12459419 (Val14Val), 2 demonstrated an increase in MRD and 1 had very little change in MRD (Fig. 1B). Interestingly, we observed that all 3 AML patients with poor MRD response to GO (MRD levels >0.01) were homozygous TT (Val14Val) for the rs12459419 variant (allele frequency 100%), whereas only 2 of 19 (11%) patients with response to GO (MRD levels < 0.01) were homozygous TT.
The C>T allele of rs12459419 results in an Ala (codon GCC) to Val (codon GTC) amino acid change at codon 14 in the protein. The Ala to Val change occurs on the surface of the protein and is present within the signal peptide (amino acid 1–17). Furthermore, the SIFT software (Sorting Intolerant From Tolerant, http://blocks.fhcrc.org/sift/SIFT.html] predicts the Val14Ala change to affect the protein function with a score of 0.04. SIFT predicts whether an amino acid substitution affects protein function based on sequence homology and the physical properties of the amino acids. This provides a plausible biological explanation for the observed association described above. This amino acid change might affect the CD33 targeting or compartmentalization, or protein structure, which could result in aberrant recognition by GO or internalization of the complex. In this regard, the mRNA level and percentage of CD33 positive cells do not associate with MRD-change in this study.
Although the sample size is small, this is the first report exploring and demonstrating an association between coding polymorphisms in CD33 with the response to GO. Further confirmation of these results in a larger and independent cohort of AML patients as well as molecular biological studies to functionally characterize these coding polymorphisms are needed and are planned in our laboratory.
Acknowledgments
We are grateful to Dr. Kristine Crews for providing her input in preparing this report, to Dr. Erin Schuetz for providing supporting lab materials and to Dr. Donald Samulack for scientific editing. This work was supported in part by the National Institutes of Health, National Institute of General Medical Sciences Pharmacogenetics Research Network and Database (U01GM61374; http://www.pharmgkb.org) under Grant U01 GM61393; by the Cancer Center Support Grant CA-21765; and by the American Lebanese Syrian Associated Charities (ALSAC).
References
- 1.Legrand O, Perrot JY, Baudard M, et al. The immunophenotype of 177 adults with acute myeloid leukemia: proposal of a prognostic score. Blood. 2000;96:870–877. [PubMed] [Google Scholar]
- 2.Bross PF, Beitz J, Chen G, et al. Approval summary: gemtuzumab ozogamicin in relapsed acute myeloid leukemia. Clin Cancer Res. 2001;7:1490–1496. [PubMed] [Google Scholar]
- 3.Alan K Burnett, Kell William J, Goldstone Anthony H, et al. The Addition of Gemtuzumab Ozogamicin to Induction Chemotherapy for AML Improves Disease Free Survival without Extra Toxicity: Preliminary Analysis of 1115 Patients in the MRC AML 15 Trial. Blood. 2006;108:13. [Google Scholar]
- 4.Ikemoto N, Kumar RA, Ling TT, et al. Calicheamicin-DNA complexes: warhead alignment and saccharide recognition of the minor groove. Proc Natl Acad Sci U S A. 1995;92:10506–10510. doi: 10.1073/pnas.92.23.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walter RB, Gooley TA, van dV V, et al. CD33 expression and P-glycoprotein-mediated drug efflux inversely correlate and predict clinical outcome in patients with acute myeloid leukemia treated with gemtuzumab ozogamicin monotherapy. Blood. 2007;109:4168–4170. doi: 10.1182/blood-2006-09-047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sievers EL. Efficacy and safety of gemtuzumab ozogamicin in patients with CD33-positive acute myeloid leukaemia in first relapse. Expert Opin Biol Ther. 2001;1:893–901. doi: 10.1517/14712598.1.5.893. [DOI] [PubMed] [Google Scholar]
- 7.Buckwalter M, Dowell JA, Korth-Bradley J, Gorovits B, Mayer PR. Pharmacokinetics of gemtuzumab ozogamicin as a single-agent treatment of pediatric patients with refractory or relapsed acute myeloid leukemia. J Clin Pharmacol. 2004;44:873–880. doi: 10.1177/0091270004267595. [DOI] [PubMed] [Google Scholar]
- 8.Dowell JA, Korth-Bradley J, Liu H, King SP, Berger MS. Pharmacokinetics of gemtuzumab ozogamicin, an antibody-targeted chemotherapy agent for the treatment of patients with acute myeloid leukemia in first relapse. J Clin Pharmacol. 2001;41:1206–1214. doi: 10.1177/00912700122012751. [DOI] [PubMed] [Google Scholar]
- 9.Rubnitz Jeffrey, Razzouk Bassem, Bowman Paul, et al. Improved Remission Induction Rate of Childhood AML: Preliminary Results of the AML02 Trial. Blood. 2005;106:275. [Google Scholar]
- 10.Coustan-Smith E, Ribeiro RC, Rubnitz JE, et al. Clinical significance of residual disease during treatment in childhood acute myeloid leukaemia. Br J Haematol. 2003;123:243–252. doi: 10.1046/j.1365-2141.2003.04610.x. [DOI] [PubMed] [Google Scholar]
- 11.Lamba JK, Crews K, Pounds S, et al. Pharmacogenetics of deoxycytidine kinase: identification and characterization of novel genetic variants. J Pharmacol Exp Ther. 2007;323:935–945. doi: 10.1124/jpet.107.128595. [DOI] [PubMed] [Google Scholar]