Abstract
Congenital deficiency in the WRN protein, a member of the human RecQ helicase family, gives rise to Werner syndrome, a genetic instability and cancer predisposition disorder with features of premature aging. Cellular roles of WRN are not fully elucidated. WRN has been implicated in telomere maintenance, homologous recombination, DNA repair, and other processes. Here I review the available data that directly address the role of WRN in preserving DNA integrity during replication and propose that WRN can function in coordinating replication fork progression with replication stress-induced fork remodeling. I further discuss this role of WRN within the context of damage tolerance group of regulatory pathways and redundancy and cooperation with other RecQ helicases.
Keywords: Werner Syndrome, RecQ helicase, human cell culture, replication stress, damage tolerance
Introduction
The WRN gene is mutated in Werner syndrome (WS), a rare autosomal recessive disorder associated with premature aging (progeria) and predisposition to cancer [1,2]. Progeroid features of Werner syndrome include early onset of type II diabetes mellitus, atherosclerosis, cataracts, skin atrophy, graying and loss of hair, and osteoporosis. WRN is one of five members of the human RecQ helicase gene family, two other genes of which, BLMand RECQL4, are mutated in Bloom syndrome (BS) and a subset of Rothmund Thompson syndromes (RTS), respectively [3,4]. These syndromes share the cancer predisposition feature with WS, but do not exhibit such a pronounced progeroid component. Interestingly, the cell types and lineages susceptible to carcinogenesis in these three syndromes are somewhat different. WS carries an enhanced risk of neoplasms of mesenchymal origin, BS elevates the risk of the whole sporadic neoplasm spectrum, and RTS exhibits increased prevalence of osteosarcomas (reviewed in [5], also [6]). The molecular underpinnings of these differences are not yet established and await a systematic comparison of cellular phenotypes of the human RecQ helicases.
Recent work has revealed that the role of WRN in human pathogenesis may be broader than envisaged before, and goes beyond heritable disease. The WRNgene is inactivated by methylation in a large fraction of common sporadic epithelial malignancies such as colon cancer, in otherwise WRN-positive individuals [7]. Understanding molecular functions of WRN thus becomes a critical task relevant both to the study of human aging and the study of sporadic carcinogenesis.
Many areas of research into the functions of the human WRN RecQ helicase have been reviewed extensively (for the latest update, see [8–10]). Here, I will focus on the role of human WRN in DNA replication and specifically in replication fork metabolism during stress, as caused by damage to DNA. I will revisit the data that address this problem, put these in the context of current concepts of replication stress response, and evaluate whether a “fork-centric” view of WRN function helps arrive at a better understanding of WRN role in human cells.
Is S phase prolonged in WRN-deficient cells?
Slow population growth in culture [11] was one of the first phenotypes observed in patient-derived, WRN-deficient human fibroblasts maintained in ambient oxygen (20%), and it was evident that at least two factors could be contributing to it. The labeling index – a percentage of cells that incorporated a pulse of 3H-thymidine per specified unit of time – was lower in WS cells, indicating that they entered S phase less frequently or a larger fraction of them ceased to proliferate. In addition, when an S phase subpopulation (identified as incorporating 3H-thymidine) was followed over time by monitoring appearance of 3H-labeled mitoses, it became evident that those WS cells that entered the division cycle, spent about 10–20% more time between two consecutive mitoses. Moreover, analysis of these data according to [12], led to a conclusion that S phase was extended by about 30% in these WS fibroblasts [13].
More recently, Poot et al. used immortalized lymphoblasts grown in low oxygen to analyze WS cell cycle [14]. The authors used a technique of continuous BrdU labeling followed by Hoechst/EthBr staining and FACS, which allows distinguishing cells that divided one, two, or three times after the start of labeling [15]. Applying cell cycle modeling to the data (with specific assumptions such as a normal distribution of the probability of entering division), has yielded a conclusion that the only two statistically significant differences between a set of WRN−/−cells and an unrelated set of controls is, on average, a 39% extension of S phase and an increased fraction of cells permanently arrested in S phase. Later, Rodriguez-Lopes et al. counted mitotic indices of a non-isogenic pair of normal and WS primary fibroblasts grown at ambient oxygen, and found that in WS cells S phase was extended by up to 30%, the overall cell cycle by about 40%, and in addition to that, the fraction of dividing cells was lower [16].
We have looked at the cell cycle kinetics in pedigree matched immortalized lymphoblast or unrelated transformed fibroblast WS cells grown in ambient oxygen (JS unpub.). WRN-deficiency consistently correlated with a lower fraction of dividing cells. We assessed the duration of S and G2/M phases by following division of cells synchronized in late G1 by mimosine (with and without BrdU labeling). FACS profiles revealed both cell type-specific and between line-specific variability. Two out of three pairs of pedigree matched lymphoblasts exhibited slight (less than 10%) delay in S and/or G2/M phases in the absence of WRN, and only one out of two WS fibroblast lines was slower, albeit dramatically, in S phase than an unrelated WRN+/+control.
We thus used acute retroviral depletion of WRN to generate isogenic pairs of WRN+ and WRN- cells and found that WRN depletion from primary fibroblasts retarded S and/or G2/M progression and markedly reduced the fraction of dividing cells under standard growth conditions (ambient oxygen) [17,18]. Extension of S and/or G2/M phases in WRN-depleted primary fibroblasts was suppressed by lowering oxygen tension (Dhillon et al., submitted). This is consistent with the observation that reducing oxygen tension partially suppresses the growth rate defect seen in WS primary fibroblasts [19].
In contrast, WRN depletion from SV40 transformed fibroblasts led to a WRN-dependent delay of cell division only when these cells were subjected to replication stress, for instance, DNA damage during S phase The fraction of proliferating cells was not significantly affected by WRN depletion from SV40 transformed fibroblasts [18].
In summary, the data suggest that the fraction of dividing cells can be lower in WRN-deficient cell cultures, and in addition to that, cells that commit to a round of division, can take more time completing it. However, both of these phenotypes appear to be modulated by cell type, transformation status, and growth conditions, and in particular oxygen tension. It is an open question whether the lower dividing fraction and the extended S and/or G2/M of WRN-deficient cells are linked or independent phenotypes, and in fact Szekely et al. have proposed a separate role for WRN in counteracting oxidative damage in G1 [20]. Below I will focus on the data addressing S phase extension in WS cells.
What is the mechanism of the S phase extension in the absence of WRN?
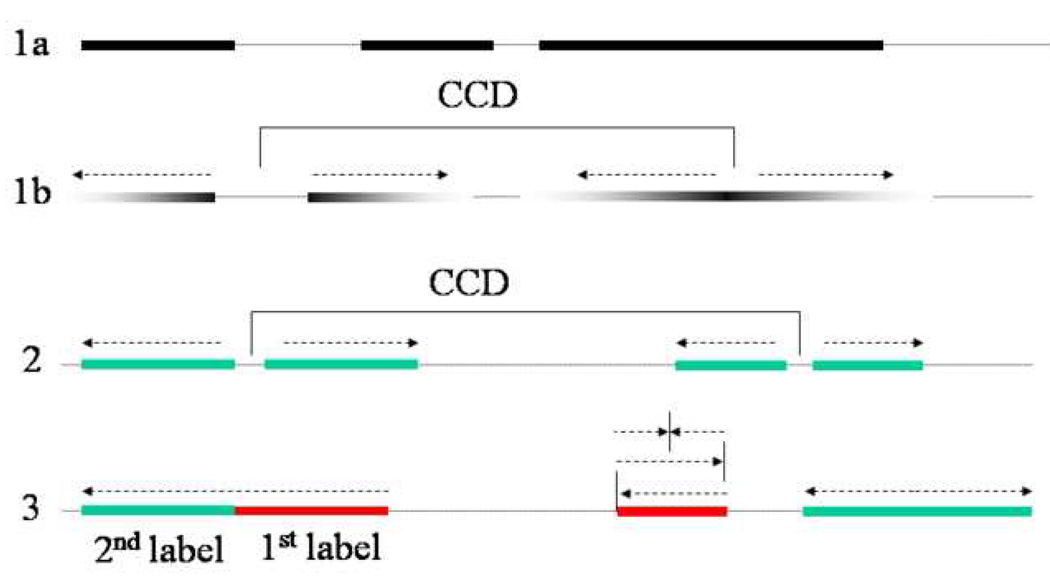
In the earliest experiments, sedimentation in alkaline sucrose gradients was used to assess the rate of DNA chain growth in WS and normal primary fibroblasts. This rate was found either retarded in WS cells [21,22] or indistinguishable from control [13]. Takeuchi et al. used a more sensitive approach of DNA fiber autoradiography to ask what aspect of S phase was altered in WS fibroblasts [23]. Cells were pulse-labeled with 3H-thymidine for up to one hour, lysed, and the released DNA fibers were spread on glass, layered with photo-emulsion and exposed. Replication was visualized as series of labeled tracks (Figure 1, fiber 1a). The length distributions of these tracks for each labeling time and the derived rate of DNA chain elongation were identical in WS and normal cells. The authors also addressed whether inter-replicon distance was affected in WS cells. A “hot” pulse of 3H thymidine was followed by a “warm” chase with a label of lower specific activity. Fork direction could then be determined from track appearance: signal intensity at the leading end of a track would be diminishing gradually (Figure 1, fiber 1b). Closely spaced pairs of tracks left by divergent forks would then identify a replication bubble growing out of an origin of replication at its center of symmetry. From this, one can measure replicon center-to-center distances (CCD), which would correspond to the distance between centers of symmetry of two consecutive bubbles (Figure 1, fiber 1b). In the study in question, the actual measurements were specified as “between adjacent tracks with a tandem array of at least two sets of tracks,” which could have been a distance between converging forks, not the CCD. Both the mean and mode of the obtained length distribution were significantly higher in WS cells than in controls, leading to the conclusion that the decreased frequency of initiation was the cause of the prolonged S phase in WS cells. However, the replicon density measurement obtained in this study is not conclusive (see above), and may be affected by replication bubble asymmetry as well as by skewed distribution of cells within S phase. A definitive measure of CCDs will require collecting more extensive data on non-synchronized as well as synchronized cells, at specified times in S phase.
Figure 1. Analysis of initiation and elongation of replication using pulse labeling of DNA with subsequent stretching.
1A) 3H thymidine labeling allows measuring lengths of labeled tracks in stretched DNA after exposure to photo emulsion. 1B) A high specific activity pulse of 3H thymidine followed by a low specific activity chase provides information about direction of replication fork movement. Replicon center to center distance (CCD) can be measured as a distance between centers of symmetry of pairs of divergent forks. 2) Synchronization of cells early in S phase by aphidicolin and pulsing with BrdU 5 minutes after release from aphidicolin visualizes replication bubbles in stretched DNA as pairs of tracks with small gaps of unlabeled DNA in between. CCD would correspond to a distance between these small gaps. 3) Double labeling with consecutive pulses of IdU and CldU allows distinguishing ongoing forks, terminated forks and newly fired forks, and shows direction of movement of ongoing forks.
In the meantime, a more recent analysis carried out by Rodriguez-Lopez and coauthors [16] led to the oppositeconclusion, e.g. that the elongation in WS primary fibroblasts was impaired, while the initiationof replication was normal. The authors’ experimental design offered a more rigorous assessment of fork initiation and progression. Primary fibroblasts were arrested in early G1 by serum starvation, then released and incubated with aphidicolin, a DNA polymerase inhibitor, for 1 hour. This two-step procedure enriched for origin firing events and ensured that WS cells and controls were in the same position, at the very beginning of S phase. Five minutes after aphidicolin was removed, cells were pulsed with BrdU. Labeled DNA was stretched and BrdU-labeled replication tracks were visualized by immunofluorescence. The only difference observed between WS and wild type cells was a 25% reduction of mean lengths of tracks.
Importantly, the authors could determine that this difference arose from slowness or inactivation of a subset of forksrather than from a uniform elongation rate decrease. This was possible because the experimental design allowed identifying, which tracks came from the same origin of replication. As long as cells are at the beginning of S phase and the labeling pulse is short, a pair of tracks separated by a very small gap can be regarded as two divergent forks belonging to the same replication bubble (Figure 1, fiber 2). When such track pairs were examined, more pairs (up to 75%) showed unequal lengths in WS cells than in controls (15%), suggesting that in many replication bubbles one of the forks was abnormally slow or stalled prematurely. Finally, CCDs could be estimated between centers of bubbles, and though the data presented were for a low nof 10, it appeared that CCDs were not higher in WS cells compared to controls, at least for the early S phase. It should be noted that aphidicolin, as an agent that stalls forks, could have enhanced the fork abnormalities observed in WS cells. However, the authors repeated some of the experiments without aphidicolin, and a moderate difference between mean track lengths of wild type and WRN−/− cells was still detectable (L. Cox, pers. comm.).
We recently analyzed replication fork elongation in WRN-depleted cells and isogenic controls [18], using double labeling with CldU and IdU to identify ongoing forks (Figure 1, fiber 3). To enrich for S phase cells, the cultures were arrested in late G1 by mimosine, released, and incubated for about 9 hours prior to labeling. As mentioned before, SV40 transformed fibroblasts are less sensitive to WRN absence than primary fibroblasts, and we could observe WRN-dependent fork defects in these cells only when we introduced replication stress by treatment with MMS or HU. This, together with double labeling, offered an advantage of an internal control to compare pre-stress and in-stress fork elongation, ruling out random sample heterogeneity or DNA processing artifacts. The lengths of pre-damage segments in ongoing forks were comparable between WRN-depleted cells and controls, however, after DNA alkylation damage by MMS WRN-depleted cells had an increased fraction of forks (for example 50% in WRN-depleted vs. 21% in control) that became dramatically slow (e.g. their tracks had post- MMS segments that were ≥5 times shorter than their pre-MMS segments). Interestingly, a similar phenotype was observed in WRN-depleted cells treated with HU. Both in control and WRN-depleted cells, a similar proportion of the forks that were elongating before HU was able to resume DNA synthesis immediately after HU was removed, but in WRN-depleted cells, unlike in controls, many of these forks were extremely slow.
In conclusion of this section, the data obtained thus far by others and us suggest that WRN deficient cells are impaired in replication fork elongation, though the magnitude of this impairment depends on the degree of replication stress. Notably, even a short treatment with aphidicolin, presence of BrdU, oxidative damage due to growth in ambient oxygen, metabolic perturbations due to serum starvation or mimosine addition, or oncogene overexpression, may apply mild stresses on replication, and elicit detectable WRN-dependent differences in fork progression. In turn, the initial observation of equivalent fork rates in WRN-deficient and control cells made by Takeuchi et al. can be explained by minimized replication stress [23].
Are all forks equal when it comes to WRN?
The replication track data described above indicate that WRN deficiency increases heterogeneity of fork population, eliciting appearance of a subset of forks that move very slowly or stop prematurely. A simple, quantitative interpretation of this data is that forks may be differentially sensitive to stress and the loss of WRN affects the most sensitive forks first.
The existing data argue in favor of differential sensitivity of forks to stress. Certain loci may naturally cause forks to slow down and additional stress can then disable such a fork more readily. Some loci, like rDNA [24], may have features that prevent efficient activation of the checkpoint response when a fork is stalled at them [25], thus such a fork is not protected by the fork-stabilizing functions of the checkpoint and can collapse [26–28]
The sequence composition of DNA that is being replicated is considered a major determinant of the differential sensitivity of forks to stress. Unusually A–T or G-C rich sequences and repeated sequences may pose problems to replication since they can distort DNA and/or form various secondary structures [29,30]. Telomeres, common fragile sites, minisatellites, trinucleotide and other microsatellite repeats, and rDNA all fall into this category [31–33]. Though not directly demonstrated (with the exception of rDNA, see above), indirect evidence suggests that during replication these sequences may be enriched for slow or stalled forks. In particular, these sequences can preferentially develop strand breaks (suggestive of fork collapse) or expansions-contractions (suggestive of fork pausing), in response to low level of exogenous replication stress or to deficiency in replication stress response proteins (ATR, Chk1, [34,35], and others) or replication accessory helicases such as Rrm3 [36] or Dna2 [37].
WRN has long been implicated in telomere maintenance [8], and Crabbe et al. proposed that WRN facilitates replication of the telomeric lagging strand DNA [38]. Using CO-FISH, these authors showed that in the absence of WRN, a subset of telomeres lacked their nascent lagging strands after a round of BrdU labeling (due to a unidirectional replication of telomeres the leading and lagging strand assignments are always the same and the lagging strand is synthesized off the G-rich strand of the telomeric repeat). Caburet et al. used DNA combing to observe a higher proportion of rearranged, palindromic arrays of rDNA units in WRN-deficient cells [39]. FRA16B, a rare AT rich repeat-based fragile site is one of the hot spots of translocations in WRN-deficient fibroblasts [40], and FRA16B has a replication pausing activity in a plasmid context in mammalian cells (Maria Krasilnikova, JS, unpub.). Moreover, a recent work by Pirzio et al. demonstrated that several common fragile sites are expressed at a higher level, e.g. develop more strand breaks in WRN-null or WRN helicase mutant fibroblasts [41], and an (AT)n sequence present in one of these sites, FRA16D, has a strong fork pausing activity when introduced into yeast [42]. Thus, enough data suggest that the sequences that may be difficult to replicate and are most sensitive to replication stress are also the ones preferentially affected in WRN-deficient cells. This is consistent with the view that WRN is required to facilitate fork progression during replication stress.
An alternative interpretation of the above data is that WRN is simply a sweepase, a non-replicative helicase recruited to the fork to clear unwanted secondary structures. Sweepases such as Rrm3 are well characterized in yeast, and in their absence forks pause more frequently on natural pause sites such as rDNA and telomeric DNA [36]. In fact a sweepase role has been proposed for WRN at telomeres: it may unwind telomeric D-loops (t-loops) [8], and/or unwind G-quadruplexes that form on a G-rich telomeric repeat strand [38,43]. However, since the proportion of forks affected by replication stress in the absence of WRN can vary and appears to be greater than what telomeric replication accounts for, it is reasonable to assume that WRN responsibilities are broader than sweeping a fixed number of secondary structures. A more flexible, dynamic view of WRN implies that it mitigates a common consequence of a secondary structure and replication stress – a slow or stalled fork.
A mechanistic model for WRN role in replication elongation
Emerging evidence suggests that DNA replication fork progression rate is “soft-coupled” rather than “hard-wired”: e.g. that fork progression is actively regulated, rather than being a simple product of DNA polymerization rate. This is best illustrated by findings that fork progression rates can be fasterin mutant than they are in wild type cells. For example, in chicken DT40 cells, absence of the RAD51 paralog XRCC3 increasesthe normally depressed rate of fork progression on BPDE- or CDDP-damaged DNA [44,45]. Similarly, when in mammalian cells fork progression is depressed by camptothecin (CPT) damage, inhibition or depletion of CHK1, a global mediator of the S phase checkpoint response, reverses this decrease, with forks progressing as if no damage was present [46]. CDDP generates inter- and intra-strand crosslinks and CPT traps topoisomerase I (TopoI) on DNA, presenting a protein-DNA adduct or a nick to approaching forks. Both compounds would be expected to impede fork progression, however, the above data suggest that the picture is not that simple. It is possible that the observed global slow-down of forks after CPT or other drugs is imposed on forks in-transby the checkpoint, and slows forks regardlessof whether they have a lesion in front of them [47]. Alternatively, a fork may be able to traverse lesions, though it will leave gaps in replicated DNA [48]. If so, then the observed slowing of forks may be a result of an in-ciscoupling of replication with repair in order to prevent gaps.
The increased slowing of forks during replication stress in the absence of WRN could therefore be attributed to either an in-trans or in-cis mechanism. WRN absence could upregulate the S phase checkpoint and cause an exaggerated fork slowing response in-trans. This interpretation can explain why such different stress agents as HU and MMS can elicit a similar response in WRN-depleted cells. However, it is not consistent with the observation that not all forks are equally affected by WRN absence. Also, CHK1 activation does not appear to be higher in WRN-depleted cells (JS unpub).
A more mechanistically attractive view is that WRN-dependent fork modulation is an in-cismechanism that couples fork progression with management of gaps and lesions. Three models of this mechanism have been proposed, and are partially supported by experimental data: fork regression via a Chicken foot (CF), and template switching via a double Holliday junction (HJ, [49,50]) or a hemicatenane (HC, [51,52]). All three make use of helicase, endo- and exonuclease, and strand melting/annealing activities such as branch migration and strand exchange, to reversibly and quickly switch between daughter/mother to daughter/daughter and mother/mother duplexes (Figure 2, see the legend for details and the DNA Repair special issue Replication Fork Repair Processes (2007) for further reading). Bypassing replication-blocking lesions and preventing gaps are some of the services such remodeling can provide, but other uses such as protecting the ends of the daughter strands, are possible. Forks slowed by HU or MMS can therefore undergo the same remodeling.
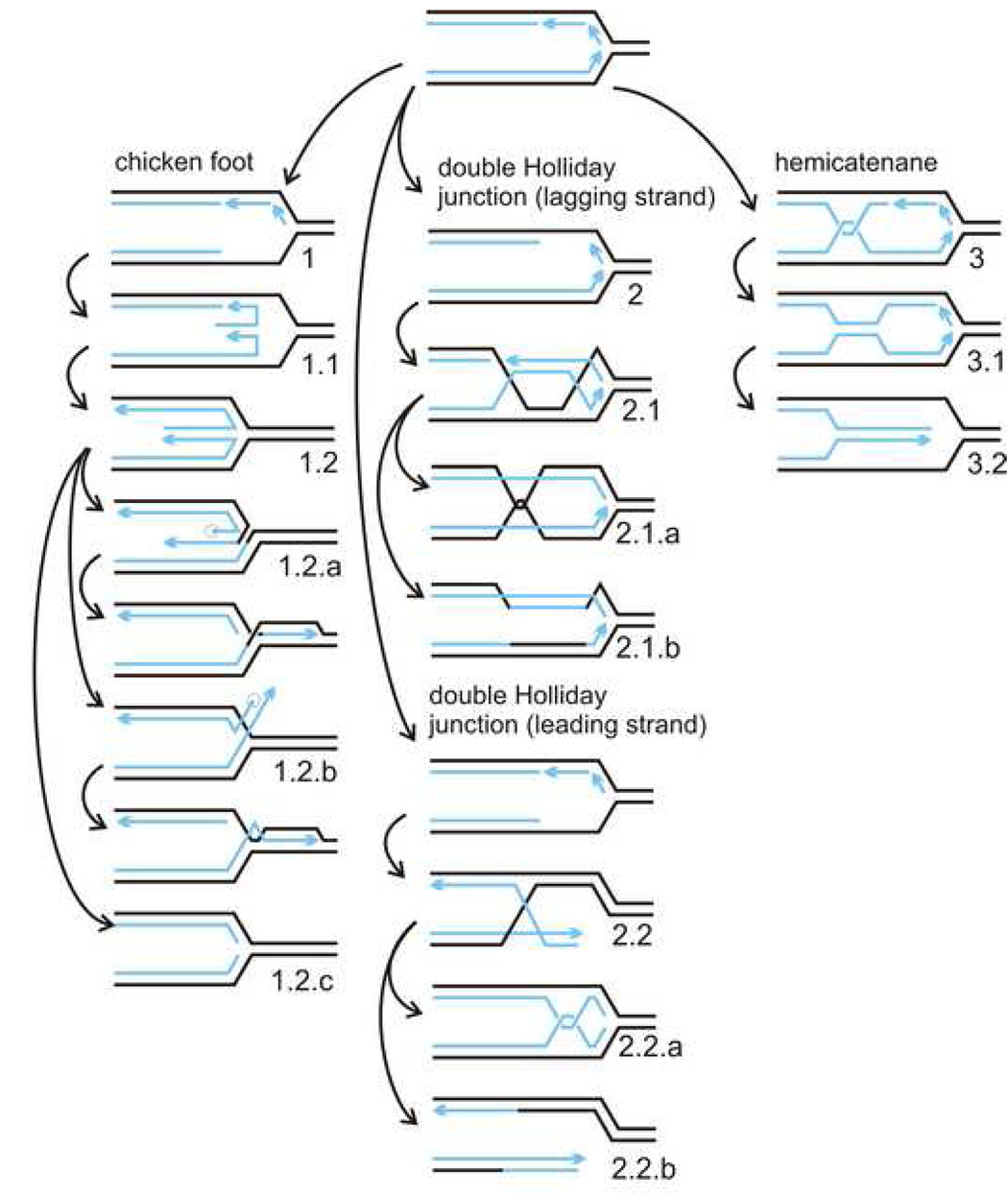
Figure 2. Pathways of replication fork remodeling, via chicken foot (CF), double Holliday junction (double HJ) and hemicatenane (HC) intermediates.
Further processing of these structures can include more than one subpathway (denoted 1.1, 1.2, etc.). Note that the pathway 2.2. can lead to undesirable double strand break intermediates depending on how the double HJ is resolved. Chicken Foot (CF, 1): CF can be simply reversed by unwinding or branch migration, or further processed in one out of three ways: 1.2.a. Resolution with subsequent end processing and strand invasion. 1.2.b. End processing with subsequent strand invasion. 1.2.c. Complete exonucleolytic degradation. Double HJ (2): 2.1. Stand exchange. 2.1.a. Dissolution through branch migration and decatenation. 2.1.b. Resolution. 2.2. Note that resolution applied when leading strand synthesis is blocked can lead to double strand breaks, 2.2.b. Hemicatenane (HC, 3): HC can be reversed by branch migration and decatenation. Alternatively, it can expand into a daughter/daughter duplex (3.1.) and then a chicken foot (3.2.).
In the fork regressionmodel two daughter strands anneal beginning from the fork-proximal ends in, and mother strands snap together, creating a three-pronged “chicken foot”, CF (Figure 2, pathway 1). In the double Holliday junction(HJ) model annealing of daughter strands occurs internally (Figure 2, pathway 2) by nucleation of a RAD51 filament on single-stranded stretches of mother strands (perhaps assisted by BRCA2, BRCA1, and hUBC13 [53,54]), and a strand exchange that results in mother/mother and daughter/daughter duplexes [55]. In the hemicatenane(HC) model, an intrinsically occurring topological linkage between daughter strands that branch migrates behind the fork, can promote daughter strand pairing when fork progression is impeded (Figure 2, pathway 3). Expansion of a daughter/daughter strand duplex can then convert the structure into a CF or HJ.
The following important considerations should be mentioned before I discuss potential contributions of WRN to fork remodeling. First, all three models postulate the intermediates that could be reverted back to the original fork configuration either directly, or via further remodeling along more than one pathway (Figure 2). These pathways can be complex, multi-step, and “costly” processes. Depending on the specific situation, for example the position of a blocking lesion on a leading or lagging strand, some of these pathways may be unproductive, leading to double-strand breaks or dead-end configurations (Figures 2, pathways 1.2.a and 2.2.b, also Figure 3). Thus there should exist mechanisms that ensure the choice of an optimal pathway [56]. Second, it is easy to envision that daughter/daughter duplexes or extended hemicatenanes, while ensuring that replication-blocking lesions are bypassed, will slow or stop overall fork progression. Unless they are reversed properly and timely, fork progression may remain impeded. Third, all three models provide for error-free maintenance of the fork. A mutagenic alternative is the trans-lesion synthesis (TLS) pathway [57]. Involvement of TLS can slow fork progression rates, as shown in a recent study [58]. TLS may also work in conjunction with other pathways of fork remodeling: the TLS polymerase eta has been implicated in template switching as it may be recruited to extend the recessed 3’ end in a daughter/daughter duplex [59–61].
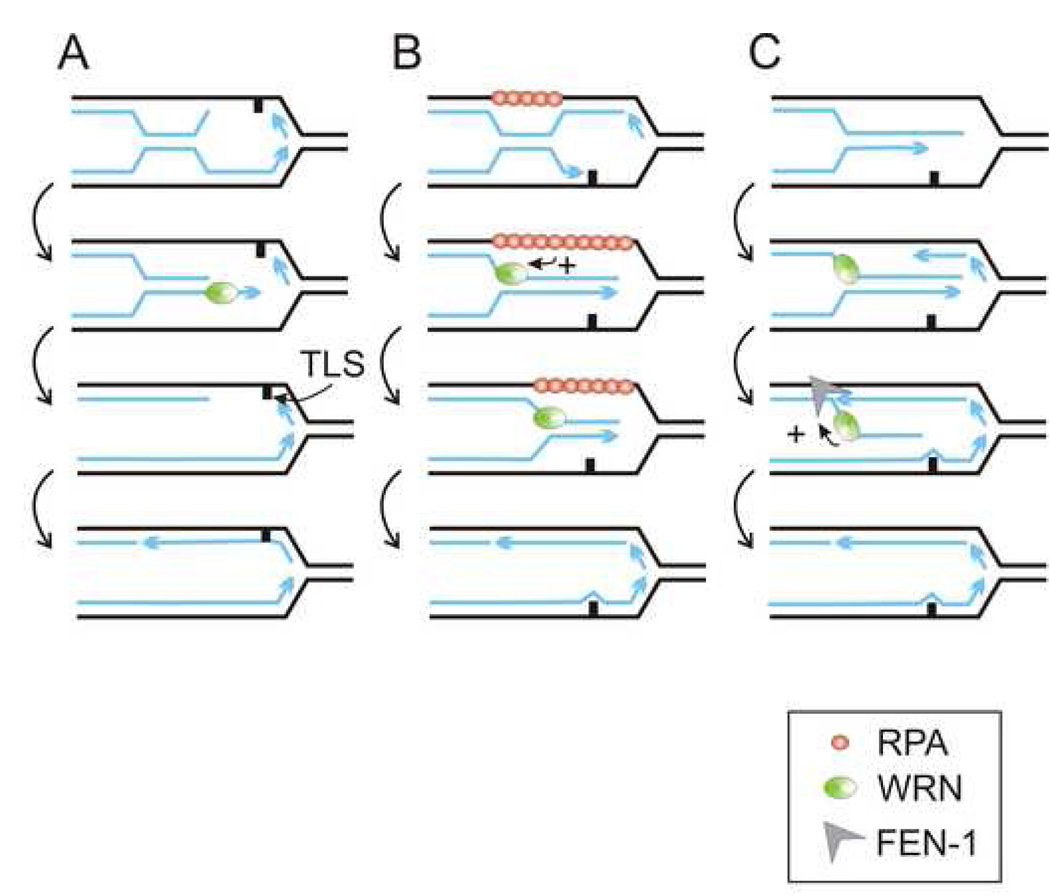
Figure 3. Possible scenarios of WRN function in coordinating fork progression with damage repair via control over daughter/daughter duplex expansion and/or half-life.
A) An unproductive daughter/daughter duplex with the 3’ overhang is unwound to redirect damage bypass towards translesion synthesis (TLS). B) An extension of a daughter/daughter duplex leads to exposure of ssDNA regions of mother strands (for simplicity, only one of the strands is shown coated with RPA). Accumulation of RPA stimulates helicase activity of WRN to limit propagation of daughter/daughter duplex and restore an original fork conformation. C) Lagging strand synthesis in the presence of a daughter/daughter duplex can lead to formation of long flaps. WRN can prevent their formation by limiting half-life of a daughter/daughter duplex, or stimulate FEN-1 to cleave such flaps once they are formed.
In vitrobiochemical activities of WRN are consistent with its participation in any of the outlined models. WRN exhibits 3’–5’ exonuclease [62–64], 3’–5’ helicase [65,66], ATPase [67], and strand melting/pairing activities [68], which can be modulated by other repair and replication proteins. WRN can regress model forks [69,70], and also reverse regressed forks, as it can branch migrate [71] and unwind HJs [72]. WRN can unwind or degrade the invading strand of a D-loop [73] or a recessed 3’ end of a leading strand [70]. WRN can also increase processivity of TLS polymerases [74], pol beta [75] and pol delta [76].
It is quite conceivable that rather than having a singular enzymatic or regulatory role, WRN may help maintain a balance between many opposing processes at the fork and direct them towards the most efficient route. As mentioned above, one important aspect of fork remodeling is limiting expansion and reducing half-life of a daughter/daughter duplex. Figure 3 shows three specific examples of “balancing acts” that can depend on WRN. First, WRN can counteract formation of unproductive daughter/daughter duplexes with recessed 5’ ends (Figure 3A). Second, WRN can limit propagation and half-life of any daughter/daughter duplex (Figure 3B). Here, a specific WRN-mediated feedback loop can be envisaged, where extension of a daughter-daughter duplex coincides with extension of single-stranded stretches on mother strands. Recruitment of RPA to ssDNA, and/or its displacement by RAD51 and thus an increase of the local concentration of RPA could stimulate helicase activity of WRN [77,78] on the daughter/daughter duplex, and this can generate a negative feedback to limit further propagation of this duplex. Finally (Figure 3C), WRN can optimize Okazaki fragment maturation in forks subjected to remodeling. If new Okazaki fragments are generated while the daughter/daughter duplex persists, unwinding of the latter may result in the appearance of long flaps on the lagging strand, which FEN-1 may be less able to cleave [79]. If long flaps include repeat DNA, formation of hairpins can further aggravate Okazaki fragment maturation and cause repeat instability [80]. Again, WRN function of limiting expansion of the daughter/daughter duplex could be safeguarding against such a development. In addition, WRN can directly stimulate nucleolytic activity of FEN-1 on long flaps [81,82]. FEN-1 could be recruited by WRN to the ss/ds DNA junction of the flap [83] generated as WRN unwinds the daughter/daughter duplex. WRN can also recruit EXO1 to degrade long flaps [84]. The DNA2 endonuclease is capable of cutting long flaps, and yeast dna2-1mutant is HU and MMS sensitive. This agrees with the idea that daughter/daughter duplexes and thus long flaps are more abundant in forks upon HU or MMS treatment. Consistent with this, expression of WRN in yeast dna2-1cells suppresses their HU and MMS sensitivity [85].
In conclusion of this section, I have described different but not mutually exclusive models of fork restructuring in response to DNA damage or replication arrest and proposed that WRN is well suited to balance remodeling processes at the fork, one specific example of which is controlling propagation of daughter/daughter duplexes.
WRN and damage tolerance pathways
The view presented above places WRN within the damage tolerancegroup of pathways. In yeast, these pathways have been associated with regulatory mono-, polyubiquitination, and sumoylation of proteins [86,87]. These modifications are carried out respectively, by the Rad6/Rad18 complex, Rad6/Rad18 in conjunction with Rad5/Ubc13/Mms2, and Ubc9 complexes (Ubc9/Siz1 for sumoylation of PCNA, Ubc9/Mms21 for sumoylation of Sgs1). These proteins have orthologs in higher eukaryotes and at least some of their functions appear to be conserved in these organisms, for example, regulatory mono- and polyubiquitination of PCNA [87,88]. Recent genetic and biochemical evidence supports regulatory roles for polyubiquitination and sumoylation in fork remodeling. Yeast Rad5 promotes fork regression into a CF in vitro[89], and UBC13 is required for formation of RAD51 filaments in DT40 cells [54], suggesting that polyubiquitination may promote formation of daughter/daughter duplexes. On the other hand, sumoylation of yeast PCNA limits nucleation of Rad51 filaments by recruiting Srs2 [90,91]. Also, sumoylation of the yeast RecQ helicase Sgs1 is thought to counteract propagation of daughter/daughter duplexes by regulating hemicatenane and double HJ dissolution activity of the Sgs1/TopoIIIalpha/RmiI complex [92].
Three lines of evidence suggest that WRN is also targeted by these regulatory circuits. First, WRN binds Ubc9 and is sumoylated in mice [93] and humans [94]. Second, in DT40 cells genetic evidence suggests that WRN is downstream of the RAD18-dependent circuit [95]. Third, WRN interacts with WRNIP1 protein, a homolog of yeast DNA damage tolerance protein Mgs1 [96–98]. WRNIP1 is a polyubiquitin-binding protein [99], while Mgs1 may inhibit Rad18/Rad6 activity when no damage is present [100,101] by stimulating turnover of ubiquitinated proteins [99]. Hence, all these lines of evidence place WRN downstream of regulatory pathways that use sumoylation and polyubiquitination to control the half-life of daughter/daughter duplexes in fork remodeling.
WRN, BLM and the question of RecQ helicase redundancy
The fact that WRN is not an essential gene has long suggested that WRN function may be partially redundant with the function of other proteins, and first and foremost, other RecQ helicases. Among the latter, a likely candidate is BLM, the RecQ helicase associated with Bloom syndrome [3]. DNA fiber autoradiography studies implicated BLM in replication elongation [102,103], and this association has been maintained by subsequent research [104]. WRN and BLM associate with each other, share many binding partners, and are active on many of the same model substrates in vitro, however, they have at least three distinguishing features. First, only WRN possesses 3’–5’ exonuclease activity. Second, WRN and BLM have different topoisomerase partners. WRN interacts with TopoI [105,106], a major eukaryotic type IB topoisomerase present at the fork, which relaxes both negative and positive supercoils [107]. BLM is in a complex with the type IA topoisomerase TopoIIIalpha [108], which relaxes only negative supercoils but can efficiently decatenate DNA [107]. Finally, BLM but not WRN strips RAD51 off DNA and inhibits formation of a D-loop in vitro when presented with an active RAD51-ssDNA filament, RPA, and naked dsDNA [109,110]. All these differences make it likely that BLM and WRN act at the fork in at least partially non-overlapping or complementary fashion. BLM, in complex with Topo3alpha and hRMII, is thought to dissolve daughter/daughter strand double HJs or hemicatenanes [111,112]. WRN on the other hand, could degrade daughter leading strands and unwind daughter/daughter duplexes in HJs and CFs as proposed above. Since formation of a CF decreases positive supercoiling that accumulates in front of the fork and reversal of a CF restores it, by removing positive supercoils, WRN partner TopoI could facilitate CF reversal by WRN.
Recent replication track analyses have identified potential non-redundancy in the in vivofunction of WRN and BLM at the replication fork. After 2–8 hours in HU, resumption of replication from stalled forks is reduced to about 50% of control in BLM null cells [113] but is comparable to control in WRN-depleted cells [18]. Although checkpoint-dependent global inhibition of origin firing is disrupted in BLM null cells after HU (ibid.), our preliminary data suggest that this is not the case for WRN. Albeit incomplete, these data may suggest an intriguing pattern of complementary roles for the two RecQ helicases. A definitive answer awaits a more extensive comparison of the replication phenotypes of WRN- and/or BLM-deficient cells within the same experimental setup.
Wrapping it all up
In this review I proposed that human WRN participates in replication fork remodeling to coordinate fork progression with the removal of DNA lesions or structural obstacles. I suggested specific scenarios where WRN controls propagation of daughter/daughter strand duplexes through its concerted exonuclease, helicase and strand pairing activities. The absence of WRN leads to increased half-life and/or expansion of daughter/daughter duplexes, which slows fork progression. Several lines of data are consistent with the idea that these functions of WRN in fork remodeling can be regulated by polyubiquitination and/or sumoylation.
The requirement for WRN in the cell appears to be strongly driven by replication stress. I propose that under “normal” growth conditions, the only forks likely to be compromised and thus to require WRN, are those that replicate telomeres, since these sequences possess several “high-risk” features: telomeres are repetitive, they can form secondary structures, and they are replicated unidirectionally off a single fork and will remain unreplicated if this fork is disabled. Thus, telomere defects are a consistent, well recognized feature of WRN-deficient cells. When these cells are challenged with more stress, additional deficiencies are revealed. These can reflect replication problems in other susceptible areas, such as fragile sites, satellite repeats, or any genomic segments that replicate unidirectionally. Finally, systemic deregulation of replication, by genotoxic drugs or oncogenes such as hyperactive Myc [114], can elicit severe S phase defects in WRN- cells [18,115] (C. Grandori, pers. comm.)
Improper remodeling of forks in the absence of WRN may result in DNA breakage and thus activation of the DNA damage response branch of the cell cycle checkpoint. In agreement with this, chromosomal rearrangements [116], breaks [115,117], and persistent gamma H2AX foci [17,20] are more frequent in WRN-deficient cells; also a higher percentage of WRN-deficient primary fibroblasts persist in very late S and/or G2/M phases of the cell cycle during normal growth and after replication stress [18]. Carryover of the unrepaired damage into the next G1 and persistence of checkpoint signaling can explain why more WRN-deficient cells remain in G1, and why this phenotype appears to depend in part on the factors that strongly contribute to the G1 checkpoint, such as p53 and Rb [18,20,118].
The emphasis on WRN activity at the fork does not imply that WRN cannot be additionally recruited to perform similar functions postreplicatively or outside S and G2 phases. Gapped DNA can persist after passage of a fork and may be repaired by pairing of daughter strands according to the template switching in double HJ or hemicatenane models [48]. Similarly, telomeric D-loops (t-loops) and RNA-DNA hybrid R-loops emerging during transcription [119] can become WRN substrates in G1. The “fork-centric” view of WRN function does not aim to limit WRN roles to replication alone, only to remind us that the type of a substrate that WRN appears to act upon can be exceedingly common, abundant, and tractable during S phase, and comprises a replication fork. A systematic characterization and quantitative comparison of in vivo replication phenotypes of WRN, BLM, and other protein factors implicated in fork remodeling will result in a deeper understanding of WRN function and of the pathogenesis of Werner syndrome and other DNA metabolism disorders.
Acknowledgments
I would like to thank Drs. R. Monnat, C. Grandori, W. Feng, and G. Alvino for critical reading of the manuscript and Dr. A. Carr for encouragement and support; also Drs. C. Grandori and L. Cox for communicating unpublished data. Thanks are due to A. Hackman for help with graphics. The author’s work on the WRN helicase is made possible by the Werner Program Project grant P01 CA77872 from NCI to R. Monnat and the Seattle Cancer and Aging Program grant P20 CA103728-04, subaward 000618167 from NCI to the author.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement:
The author declares that there are no conflicts of interest.
Literature cited
- 1.Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner's syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore) 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 3.Ellis NA, Groden J, Ye T-Z, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 4.Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 5.Mohaghegh P, Hickson ID. DNA helicase deficiencies associated with cancer predisposition and premature ageing disorder. Hum. Mol. Genet. 2001;10:741–746. doi: 10.1093/hmg/10.7.741. [DOI] [PubMed] [Google Scholar]
- 6.Wang LL, Gannavarapu A, Kozinetz CA, Levy ML, Lewis RA, Chintagumpala MM, Ruiz-Maldanado R, Contreras-Ruiz J, Cunniff C, Erickson RP, Lev D, Rogers M, Zackai EH, Plon SE. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. Journal of the National Cancer Institute. 2003;95:669–674. doi: 10.1093/jnci/95.9.669. [DOI] [PubMed] [Google Scholar]
- 7.Agrelo R, Cheng W-H, Setien F, Ropero S, Espada J, Fraga MF, Herranz M, Paz MF, Sanchez-Cespedes M, Artiga MJ, Guerrero D, Castells A, von Kobbe C, Bohr VA, Esteller M. Epigenetic inactivation of the premature aging Werner syndrome gene in human cancer. Proceedings of the National Academy of Sciences. 2006;103:8822–8827. doi: 10.1073/pnas.0600645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Opresko PL. Telomere ResQue and preservation--Roles for the Werner syndrome protein and other RecQ helicases. Mechanisms of Ageing and Development. 2008;129:79–90. doi: 10.1016/j.mad.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Kudlow BA, Kennedy BK, Monnat RJ. Werner and Hutchinson-Gilford progeria syndromes. mechanistic basis of human progeroid diseases. 2007;8:394–404. doi: 10.1038/nrm2161. [DOI] [PubMed] [Google Scholar]
- 10.Cheng W-H, Muftuoglu M, Bohr VA. Werner syndrome protein: Functions in the response to DNA damage and replication stress in S-phase. Experimental Gerontology. 2007;42:871–878. doi: 10.1016/j.exger.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Salk D, Bryant E, Hoehn H, Johnston P, Martin GM. Growth characteristics of Werner syndrome cells in vitro. Advances in Experimental Medicine & Biology. 1985;190:305–311. doi: 10.1007/978-1-4684-7853-2_14. [DOI] [PubMed] [Google Scholar]
- 12.Kruse PF, Patterson MK, editors. Tissue Culture. New York: Academic Press; 1973. A. Macieira-Coelho Cell cycle analysis; pp. 412–422. [Google Scholar]
- 13.Takeuchi F, Hanaoka F, Goto M, Yamada M, Miyamoto T. Prolongation of S phase and whole cell cycle in Werner's syndrome fibroblasts. Exp Gerontol. 1982;17:473–480. doi: 10.1016/s0531-5565(82)80009-0. [DOI] [PubMed] [Google Scholar]
- 14.Poot M, Hoehn H, Runger TM, Martin GM. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Experimental Cell Research. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- 15.Rabinovitch PS, Kubbies M, Chen YC, Schindler D, Hoehn H. BrdU-Hoechst flow cytometry: a unique tool for quantitative cell cycle analysis. Exp Cell Res. 1988;174:309–318. doi: 10.1016/0014-4827(88)90302-3. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Lopez A, Jackson D, Iborra F, Cox L. Asymmetry of DNA replication fork progression in Werner's syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- 17.Dhillon KK, Sidorova J, Saintigny Y, Poot M, Gollahon K, Rabinovitch PS, Monnat RJ. Functional role of the Werner syndrome RecQ helicase in human fibroblasts. Aging Cell. 2007;6:53–61. doi: 10.1111/j.1474-9726.2006.00260.x. [DOI] [PubMed] [Google Scholar]
- 18.Sidorova JM, Li N, Folch A, Monnat RJ., Jr The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle. 2008;7:796–807. doi: 10.4161/cc.7.6.5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis T, Wyllie FS, Rokicki MJ, Bagley MC, Kipling D. The Role of Cellular Senescence in Werner Syndrome: Toward Therapeutic Intervention in Human Premature Aging. Ann NY Acad Sci. 2007;1100:455–469. doi: 10.1196/annals.1395.051. [DOI] [PubMed] [Google Scholar]
- 20.Szekely AM, Bleichert F, Numann A, Van Komen S, Manasanch E, Ben Nasr A, Canaan A, Weissman SM. Werner Protein Protects Nonproliferating Cells from Oxidative DNA Damage. Mol. Cell. Biol. 2005;25:10492–10506. doi: 10.1128/MCB.25.23.10492-10506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujiwara Y, Higashikawa T, Tatsumi M. A retarded rate of DNA replication and normal level of DNA repair in Werner's syndrome fibroblasts in culture. J Cell Physiol. 1977;92:365–374. doi: 10.1002/jcp.1040920305. [DOI] [PubMed] [Google Scholar]
- 22.Fujiwara Y, Kano Y, Ichihashi M, Nakao Y, Matsumura T. Abnormal fibroblast aging and DNA replication in the Werner syndrome. Adv Exp Med Biol. 1985;190:459–477. doi: 10.1007/978-1-4684-7853-2_23. [DOI] [PubMed] [Google Scholar]
- 23.Takeuchi F, Hanaoka F, Goto M, Akaoka I, Hori T, Yamada M, Miyamoto T. Altered frequency of initiation sites of DNA replication in Werner's syndrome cells. Hum Genet. 1982;60:365–368. doi: 10.1007/BF00569220. [DOI] [PubMed] [Google Scholar]
- 24.Brewer BJ, Fangman WL. A replication fork barrier at the 3' end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 25.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–1919. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert S, Carr AM. Checkpoint responses to replication fork barriers. Biochimie. 2005;87:591–602. doi: 10.1016/j.biochi.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 27.Lambert S, Froget B, Carr AM. Arrested replication fork processing:Interplay between checkpoints and recombination. DNA Repair. 2007;6:1042–1061. doi: 10.1016/j.dnarep.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 28.Tourriere H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair Replication Fork Repair Processes. 2007;6:900–913. doi: 10.1016/j.dnarep.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Woynarowski J. AT Islands - Their Nature and Potential for Anticancer Strategies. Current Cancer Drug Targets. 2004;4:219–234. doi: 10.2174/1568009043481524. [DOI] [PubMed] [Google Scholar]
- 30.Lobachev KS, Rattray A, Narayanan V. Hairpin- and cruciform-mediated chromosome breakage: causes and consequences in eukaryotic cells. Front Biosci. 2007;12:4208–4220. doi: 10.2741/2381. [DOI] [PubMed] [Google Scholar]
- 31.Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Current Opinion in Structural Biology. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 32.Durkin SG, Glover TW. Chromosome Fragile Sites. Annual Review of Genetics. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 33.Lukusa T, Fryns JP. Human chromosome fragility. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2008;1779:3–16. doi: 10.1016/j.bbagrm.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 34.Casper AM, Nghiem P, Arlt MF, Glover TW. ATR Regulates Fragile Site Stability. Cell. 2002;111:779–789. doi: 10.1016/s0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 35.Durkin SG, Arlt MF, Howlett NG, Glover TW. Depletion of CHK1, but not CHK2 induces chromosomal instability and breaks at common fragile sites. 2006;25:4381–4388. doi: 10.1038/sj.onc.1209466. [DOI] [PubMed] [Google Scholar]
- 36.Ivessa AS, Zhou J-Q, Zakian VA. The Saccharomyces Pif1p DNA Helicase and the Highly Related Rrm3p Have Opposite Effects on Replication Fork Progression in Ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 37.Weitao T, Budd M, Hoopes LLM, Campbell JL. Dna2 Helicase/Nuclease Causes Replicative Fork Stalling and Double-strand Breaks in the Ribosomal DNA of Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:22513–22522. doi: 10.1074/jbc.M301610200. [DOI] [PubMed] [Google Scholar]
- 38.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective Telomere Lagging Strand Synthesis in Cells Lacking WRN Helicase Activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 39.Caburet S, Conti C, Schurra C, Lebofsky R, Edelstein SJ, Bensimon A. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 2005;15:1079–1085. doi: 10.1101/gr.3970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melcher R, von Golitschek R, Steinlein C, Schindler D, Neitzel H, Kainer K, Schmid M, Hoehn H. Spectral karyotyping of Werner syndrome fibroblast cultures. Cytogenetic and Genome Research. 2000;91:180–185. doi: 10.1159/000056841. [DOI] [PubMed] [Google Scholar]
- 41.Pirzio LM, Pichierri P, Bignami M, Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 2008;180:305–314. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Freudenreich CH. An AT-Rich Sequence in Human Common Fragile Site FRA16D Causes Fork Stalling and Chromosome Breakage in S.cerevisiae. Molecular Cell. 2007;27:367–379. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fry and M, Loeb LA. Human Werner Syndrome DNA Helicase Unwinds Tetrahelical Structures of the Fragile X Syndrome Repeat Sequence d(CGG)n. J.Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 44.Henry-Mowatt J, Jackson D, Masson JY, Johnson PA, Clements PM, Benson FE, Thompson LH, Takeda S, West SC, Caldecott KW. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Molecular Cell. 2003;11:1109–1117. doi: 10.1016/s1097-2765(03)00132-1. [DOI] [PubMed] [Google Scholar]
- 45.Johansson F, Lagerqvist A, Erixon K, Jenssen D. A method to monitor replication fork progression in mammalian cells: nucleotide excision repair enhances and homologous recombination delays elongation along damaged DNA. Nucl Acids Res. 2004;32:e157. doi: 10.1093/nar/gnh154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y. The Intra-S-Phase Checkpoint Affects both DNA Replication Initiation and Elongation: Single-Cell and -DNA Fiber Analyses. Mol. Cell. Biol. 2007;27:5806–5818. doi: 10.1128/MCB.02278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conti C, Seiler JA, Pommier Y. The mammalian DNA replication elongation checkpoint: implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle. 2007;6:2760–2767. doi: 10.4161/cc.6.22.4932. Epub 2007 Aug 2722. [DOI] [PubMed] [Google Scholar]
- 48.Lopes M, Foiani M, Sogo JM. Multiple Mechanisms Control Chromosome Integrity after Replication Fork Uncoupling and Restart at Irreparable UV Lesions. Molecular Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 49.cHiggins NP, Kato K, Strauss B. A model for replication repair in mammalian cells. Journal of Molecular Biology. 1976;101:417–425. doi: 10.1016/0022-2836(76)90156-x. [DOI] [PubMed] [Google Scholar]
- 50.Fujiwara Y, Tatsumi M. Replicative repair of UV damage to normal human cells. Mutation Research. 1976;38:335–336. doi: 10.1016/0165-1161(76)90159-x. [DOI] [PubMed] [Google Scholar]
- 51.Lopes M, Cotta-Ramusino C, Liberi G, Foiani M. Branch Migrating Sister Chromatid Junctions Form at Replication Origins through Rad51/Rad52-Independent Mechanisms. Molecular Cell. 2003;12:1499–1510. doi: 10.1016/s1097-2765(03)00473-8. [DOI] [PubMed] [Google Scholar]
- 52.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nagaraju G, Scully R. Minding the gap: The underground functions of BRCA1 and BRCA2 at stalled replication forks. DNA Repair. 2007;6:1018–1031. doi: 10.1016/j.dnarep.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao GY, Sonoda E, Barber LJ, Oka H, Murakawa Y, Yamada K, Ikura T, Wang X, Kobayashi M, Yamamoto K, Boulton SJ, Takeda S. A Critical Role for the Ubiquitin-Conjugating Enzyme Ubc13 in Initiating Homologous Recombination. Molecular Cell. 2007;25:663–675. doi: 10.1016/j.molcel.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 55.Helleday T, Lo J, van Gent DC, Engelward BP. DNA double-strand break repair: From mechanistic understanding to cancer treatment. DNA Repair. 2007;6:923–935. doi: 10.1016/j.dnarep.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 56.Gangavarapu V, Prakash S, Prakash L. Requirement of RAD52 Group Genes for Postreplication Repair of UV-Damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:7758–7764. doi: 10.1128/MCB.01331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, Kannouche PL, Green CM. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 58.Pillaire MJ, Betous R, Conti C, Czaplicki J, Pasero P, Bensimon A, Cazaux C, Hoffmann JS. Upregulation of error-prone DNA polymerases beta and kappa slows down fork progression without activating the replication checkpoint. Cell Cycle. 2007;6:471–477. doi: 10.4161/cc.6.4.3857. Epub 2007 Feb 2019. [DOI] [PubMed] [Google Scholar]
- 59.Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Dual Roles for DNA Polymerase [eta] in Homologous DNA Recombination and Translesion DNA Synthesis. Molecular Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 60.McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA Polymerase [eta] Promotes DNA Synthesis from Strand Invasion Intermediates of Homologous Recombination. Molecular Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 61.McIlwraith MJ, West SC. DNA Repair Synthesis Facilitates RAD52-Mediated Second-End Capture during DSB Repair. Molecular Cell. 2008;29:510–516. doi: 10.1016/j.molcel.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 62.Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3'-->5' exonuclease. Nature Genetics. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kamath-Loeb AS, Shen JC, Loeb LA, Fry M. Werner syndrome protein.II. Characterization of the integral 3' --> 5' DNA exonuclease. Journal of Biological Chemistry. 1998;273:34145–34150. doi: 10.1074/jbc.273.51.34145. [DOI] [PubMed] [Google Scholar]
- 64.Shen JC, Gray MD, Oshima J, Kamath-Loeb AS, Fry M, Loeb LA. Werner syndrome protein. I. DNA helicase and dna exonuclease reside on the same polypeptide. Journal of Biological Chemistry. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 65.Gray MD, Shen J-C, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki N, Shimamoto A, Imamura O, Kuromitsu J, Kitao S, Goto M, Furuichi Y. DNA helicase activity in Werner's syndrome gene product synthesized in a baculovirus system. Nucleic Acids Res. 1997;25:2973–2978. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.von Kobbe C, Thoma NH, Czyzewski BK, Pavletichc NP, Bohr VA. Werner Syndrome Protein Contains Three Structure-specific DNA Binding Domains. J. Biol. Chem. 2003;278:52997–53006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 68.Machwe A, Xiao L, Groden J, Matson SW, Orren DK. RecQ Family Members Combine Strand Pairing and Unwinding Activities to Catalyze Strand Exchange. J. Biol. Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 69.Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom Syndrome Proteins Catalyze Regression of a Model Replication Fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 70.Machwe A, Xiao L, Lloyd RG, Bolt E, Orren DK. Replication fork regression in vitro by the Werner syndrome protein (WRN): Holliday junction formation, the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucl. Acids Res. 2007;35:5729–5747. doi: 10.1093/nar/gkm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Reports. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao H-I, Bambara RA, Brosh RM., Jr WRN helicase and FEN-1 form a complex upon replication arrest and together process branch-migrating DNA structures associated with the replication fork. Mol. Biol. Cell. 2004:734–750. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Orren DK, Theodore S, Machwe A. The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-loops in vitro. Biochemistry. 2002;41:13483–13488. doi: 10.1021/bi0266986. [DOI] [PubMed] [Google Scholar]
- 74.Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proceedings of the National Academy of Sciences. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harrigan JA, Wilson DM, III, Prasad R, Opresko PL, Beck G, May A, Wilson SH, Bohr VA. The Werner syndrome protein operates in base excision repair and cooperates with DNA polymerase {beta} Nucl. Acids Res. 2006;34:745–754. doi: 10.1093/nar/gkj475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kamath-Loeb AS, Loeb LA, Johansson E, Burgers PMJ, Fry M. Interactions between the Werner Syndrome Helicase and DNA Polymerase delta Specifically Facilitate Copying of Tetraplex and Hairpin Structures of the d(CGG)n Trinucleotide Repeat Sequence. J. Biol. Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- 77.Brosh RM, Jr, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and Physical Interaction between WRN Helicase and Human Replication Protein A. J. Biol. Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 78.Shen J-C, Lao Y, Kamath-Loeb A, Wold MS, Loeb LA. The N-terminal domain of the large subunit of human replication protein A binds to Werner syndrome protein and stimulates helicase activity. Mechanisms of Ageing and Development. 2003;124:921–930. doi: 10.1016/s0047-6374(03)00164-7. [DOI] [PubMed] [Google Scholar]
- 79.Shen B, Singh P, Liu R, Qiu J, Zheng L, Finger LD, Alas S. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. BioEssays. 2005;27:717–729. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- 80.Henricksen LA, Tom S, Liu Y, Bambara RA. Inhibition of Flap Endonuclease 1 by Flap Secondary Structure and Relevance to Repeat Sequence Expansion. J. Biol. Chem. 2000;275:16420–16427. doi: 10.1074/jbc.M909635199. [DOI] [PubMed] [Google Scholar]
- 81.Brosh RM, Jr, von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. Embo J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brosh RM, Driscoll HC, Dianov GL, Sommers JA. Biochemical Characterization of the WRN-FEN-1 Functional Interaction. Biochemistry. 2002;41:12204–12216. doi: 10.1021/bi026031j. [DOI] [PubMed] [Google Scholar]
- 83.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;11:11. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 84.Sharma S, Sommers JA, Driscoll HC, Uzdilla L, Wilson TM, Brosh RM., Jr The Exonucleolytic and Endonucleolytic Cleavage Activities of Human Exonuclease 1 Are Stimulated by an Interaction with the Carboxyl-terminal Region of the Werner Syndrome Protein. J. Biol. Chem. 2003;278:23487–23496. doi: 10.1074/jbc.M212798200. [DOI] [PubMed] [Google Scholar]
- 85.Sharma S, Sommers JA, Brosh RM., Jr In vivo function of the conserved non-catalytic domain of Werner syndrome helicase in DNA replication. Hum.Mol. Genet. 2004;13:2247–2261. doi: 10.1093/hmg/ddh234. [DOI] [PubMed] [Google Scholar]
- 86.Watts FZ. Sumoylation of PCNA: Wrestling with recombination at stalled replication forks. DNA Repair. 2006;5:399–403. doi: 10.1016/j.dnarep.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 87.Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. 2008;18:162–173. doi: 10.1038/cr.2007.114. [DOI] [PubMed] [Google Scholar]
- 88.Motegi A, Sood R, Moinova H, Markowitz SD, Liu PP, sMyung K. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J. Cell Biol. 2006;175:703–708. doi: 10.1083/jcb.200606145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Blastyák A, Pintér L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 Protein Required for Postreplication Repair Has a DNA Helicase Activity Specific for Replication Fork Regression. Molecular Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pfander B, Moldovan G-L, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. advanced online publication. 2005 doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 91.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Crosstalk between SUMO and Ubiquitin on PCNA Is Mediated by Recruitment of the Helicase Srs2p. Molecular Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 92.Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 93.Kawabe Y-i, Seki M, Seki T, Wang W-S, Imamura O, Furuichi Y, Saitoh H, Enomoto T. Covalent Modification of the Werner's Syndrome Gene Product with the Ubiquitin-related Protein, SUMO-1. J. Biol. Chem. 2000;275:20963–20966. doi: 10.1074/jbc.C000273200. [DOI] [PubMed] [Google Scholar]
- 94.Woods YL, Xirodimas DP, Prescott AR, Sparks A, Lane DP, Saville MK. p14 Arf Promotes Small Ubiquitin-like Modifier Conjugation of Werners Helicase. Journal of Biological Chemistry. 2004;279:50157–50166. doi: 10.1074/jbc.M405414200. [DOI] [PubMed] [Google Scholar]
- 95.Dong YP, Seki M, Yoshimura A, Inoue E, Furukawa S, Tada S, Enomoto T. WRN functions in a RAD18-dependent damage avoidance pathway. Biol Pharm Bull. 2007;30:1080–1083. doi: 10.1248/bpb.30.1080. [DOI] [PubMed] [Google Scholar]
- 96.Kawabe Y-i, Branzei D, Hayashi T, Suzuki H, Masuko T, Onoda F, Heo S-J, Ikeda H, Shimamoto A, Furuichi Y, Seki M, Enomoto T. A Novel Protein Interacts with the Werner's Syndrome Gene Product Physically and Functionally. J. Biol. Chem. 2001;276:20364–20369. doi: 10.1074/jbc.C100035200. [DOI] [PubMed] [Google Scholar]
- 97.Hishida T, Iwasaki H, Ohno T, Morishita T, Shinagawa H. A yeast gene,MGS1, encoding a DNA-dependent AAA+ ATPase is required to maintain genome stability. Proceedings of the National Academy of Sciences. 2001;98:8283–8289. doi: 10.1073/pnas.121009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hishida T, Ohno T, Iwasaki H, Shinagawa H. Saccharomyces cerevisiae MGS1 is essential in strains deficient in the RAD6-dependent DNA damage tolerance pathway. Embo J. 2002;21:2019–2029. doi: 10.1093/emboj/21.8.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bish RA, Myers MP. Werner Helicase-interacting Protein 1 Binds Polyubiquitin via Its Zinc Finger Domain. J. Biol. Chem. 2007;282:23184–23193. doi: 10.1074/jbc.M701042200. [DOI] [PubMed] [Google Scholar]
- 100.Barbour L, Xiao W. Regulation of alternative replication bypass pathways at stalled replication forks and its effects on genome stability: a yeast model,Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. DNA Replication, the Cell Cycle and Genome Stability. 2003;532:137–155. doi: 10.1016/j.mrfmmm.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 101.Hishida T, Ohya T, Kubota Y, Kamada Y, Shinagawa H. Functional and Physical Interaction of Yeast Mgs1 with PCNA: Impact on RAD6-Dependent DNA Damage Tolerance. Mol. Cell. Biol. 2006;26:5509–5517. doi: 10.1128/MCB.00307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hand R, German J. Bloom's syndrome: DNA replication in cultured fibroblasts and lymphocytes. Hum Genet. 1977;38:297–306. doi: 10.1007/BF00402156. [DOI] [PubMed] [Google Scholar]
- 103.Hand R, German J. A retarded rate of DNA chain growth in Bloom's syndrome. Proc Natl Acad Sci U S A. 1975;72:758–762. doi: 10.1073/pnas.72.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rao VA, Conti C, Guirouilh-Barbat J, Nakamura A, Miao Z-H, Davies SL, Sacca B, Hickson ID, Bensimon A, Pommier Y. Endogenous {gamma}-H2AX-ATM-Chk2 Checkpoint Activation in Bloom's Syndrome Helicase Deficient Cells Is Related to DNA Replication Arrested Forks. Mol Cancer Res. 2007;5:713–724. doi: 10.1158/1541-7786.MCR-07-0028. [DOI] [PubMed] [Google Scholar]
- 105.Lebel M, Spillare EA, Harris CC, Leder P. The Werner Syndrome Gene Product Co-purifies with the DNA Replication Complex and Interacts with PCNA and Topoisomerase I. J. Biol. Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- 106.Laine J-P, Opresko PL, Indig FE, Harrigan JA, von Kobbe C, Bohr VA. Werner Protein Stimulates Topoisomerase I DNA Relaxation Activity. Cancer Res. 2003;63:7136–7146. [PubMed] [Google Scholar]
- 107.Forterre P, Gribaldo S, Gadelle D, Serre M-C. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 108.Mankouri HW, Hickson ID. The RecQ helicase-topoisomerase III-Rmi1 complex: a DNA structure-specific 'dissolvasome'? Trends in Biochemical Sciences. 2007;32:538–546. doi: 10.1016/j.tibs.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 109.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 112.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proceedings of the National Academy of Sciences. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. Epub 2007 Jun 2024. [DOI] [PubMed] [Google Scholar]
- 114.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 115.Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner's Syndrome Protein Is Required for Correct Recovery after Replication Arrest and DNA Damage Induced in S-Phase of Cell Cycle. Mol. Biol. Cell. 2001;12:2412–2421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hoehn H, Bryant EM, Au K, Norwood TH, Boman H, Martin GM. Variegated translocation mosaicism in human skin fibroblast cultures. Cytogenet Cell Genet. 1975;15:282–298. doi: 10.1159/000130526. [DOI] [PubMed] [Google Scholar]
- 117.Franchitto A, Pichierri P. Werner syndrome protein and the MRE11 complex are involved in a common pathway of replication fork recovery. Cell Cycle. 2004;3:1331–1339. doi: 10.4161/cc.3.10.1185. Epub 2004 Oct 1318. [DOI] [PubMed] [Google Scholar]
- 118.Poot M, Jin X, Hill JP, Gollahon KA, Rabinovitch PS. Distinct functions for WRN and TP53 in a shared pathway of cellular response to 1-[beta]--arabinofuranosylcytosine and bleomycin. Experimental Cell Research. 2004;296:327–336. doi: 10.1016/j.yexcr.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 119.Suzuki N, Shiratori M, Goto M, Furuichi Y. Werner syndrome helicase contains a 5'-->3' exonuclease activity that digests DNA and RNA strands in DNA/DNA and RNA/DNA duplexes dependent on unwinding. Nucl. Acids Res. 1999;27:2361–2368. doi: 10.1093/nar/27.11.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]