Abstract
Invasion-promoting MT1-MMP is directly linked to tumorigenesis and metastasis. Our studies led us to identify those genes, the expression of which is universally linked to MT1-MMP in multiple tumor types. Genome-wide expression profiling of MT1-MMP–overexpressing versus MT1-MMP–silenced cancer cells and a further data mining analysis of the preexisting expression database of 190 human tumors of 14 cancer types led us to identify 11 genes, the expression of which correlated firmly and universally with that of MT1-MMP (P < 0.00001). These genes included regulators of energy metabolism (NNT), trafficking and membrane fusion (SLCO2A1 and ANXA7), signaling and transcription (NR3C1, JAG1, PI3Kδ, and CK2α), chromatin rearrangement (SMARCA1), cell division (STK38/NDR1), apoptosis (DAPK1), and mRNA splicing (SNRPB2). Our subsequent extensive analysis of cultured cells, tumor xenografts, and cancer patient biopsies supported our data mining. Our results suggest that transcriptional reprogramming of the specific downstream genes, which themselves are associated with tumorigenesis, represents a distinctive “molecular signature” of the proteolytically active MT1-MMP. We suggest that the transactivation activity of MT1-MMP contributes to the promigratory cell phenotype, which is induced by this tumorigenic proteinase. The activated downstream gene network then begins functioning in unison with MT1-MMP to rework the signaling, transport, cell division, energy metabolism, and other critical cell functions and to commit the cell to migration, invasion, and, consequently, tumorigenesis.
Introduction
Matrix metalloproteinases (MMP) comprise a family of zinc enzymes. A transmembrane domain and a cytoplasmic tail distinguish MT1-MMP from the soluble MMPs. MT1-MMP is directly involved in cell locomotion and matrix degradation and in the activation pathway of the soluble MMPs. The presence of invasion-promoting MT1-MMP, either in tumor cells or in the surrounding stroma, is linked to tumorigenesis and metastasis (1).
MT1-MMP is synthesized as a zymogen, which requires proteolytic activation to remove the NH2 terminal, inhibitory prodomain. The release of the prodomain exposes the active site of the enzyme and creates the respective enzyme with full functional activity. MT1-MMP is regulated by multifaceted mechanisms, which include inhibition by tissue inhibitor of metalloproteinases (TIMP), oligomerization, shedding, glycosylation, trafficking, internalization, and recycling (2-5).
Coordinated control of MT1-MMP is critically important for cell migration and tumorigenesis. Thus, the tailless MT1-MMP mutant that lacks the cytoplasmic tail functions efficiently by inducing the activation of MMP-2 and matrix remodeling. In contrast with wild-type MT1-MMP-WT, this mutant, however, is not efficiently internalized and is persistently present on the cell surface because of its recruitment to caveolin-enriched lipid rafts. These events antagonize the contribution of the proteolytically proficient tailless MT1-MMP to cell locomotion and malignant growth (6).
MT1-MMP is a broad-specificity proteinase that cleaves multiple cellular and extracellular targets. A partial list of cleavage targets of MT1-MMP includes MMP-2, MMP-13, type I collagen α1 and α2 chains, amyloid precursor protein, connective tissue growth factor, interleukin 8, leukocyte protease inhibitor, pro–tumor necrosis factor α, KiSS, CD44, EMMPRIN, gC1q-R, integrin αV, syndecan 1, semaphorin 4D, laminin 5 γ2 chain, tissue transglutaminase, lumican, and pericentrin (7-12). Transfection of only one MT1-MMP gene causes the phenotype change and increases tumorigenicity of cancer cells (13). These changes suggest that MT1-MMP proteolysis regulates the multiple cell functions and affects, either directly or indirectly, a number of signaling and transcriptional regulation pathways (14, 15). These MT1-MMP regulated genes and downstream pathways are insufficiently characterized.
We used gene expression profiling as a global assay to identify the target genes whose transcriptional activity is directly linked to MT1-MMP in cancer. Our results led us, for the first time, to identify the MT1-MMP-mediated effects on the genome-wide transcriptional regulation.
Materials and Methods
Reagents
All reagents, unless otherwise indicated, were from Sigma. Rabbit antibody against the hinge region of MT1-MMP (Ab815), murine monoclonal antibodies against the catalytic domain of MT1-MMP (clone 5D1), and CK2α (clone 1AD9), the enzyme-free cell dissociation solution, a TMB/M substrate, and a hydroxamate inhibitor of MMPs (GM6001) were from Chemicon. Rabbit antibody against CK2α was from Bethyl. Rabbit antibody against PI3Kδ was from Abcam. Murine monoclonal antibody against the catalytic subunit of PKAα (clone 5B) was from Transduction Laboratories. Matrigel and murine monoclonal antibody against NR3C1 (clone 41) was from Becton Dickinson. Rabbit antibody Ab-1 against NR3C1 was from Calbiochem. F(ab′)2 fragments of goat anti-mouse IgG, goat anti-rabbit and goat anti-mouse IgG conjugated with horseradish peroxidase (HRP), FITC-conjugated goat anti-rabbit IgG (Fc-specific), and Alexa Fluor 488–conjugated goat anti-mouse IgG (Fc-specific) were from Jackson ImmunoResearch.
Cell lines
The parental human fibrosarcoma HT1080 (HT cells), glioma U251 (U cells), and breast carcinoma MCF-7 (MCF7 cells) were from American Type Culture Collection. Cells were cultured in DMEM supplemented with 10% FCS (DMEM/FCS) and 10 Ag/mL gentamicin. The human MT1-MMP-WT (Genbank number NM_004995) was cloned into the pcDNA3-zeo and the pcDNA3-neo plasmids (Invitrogen). The catalytically inert MT1-MMP E240A mutant with the substitution of the active site Glu-240 was isolated earlier (16). To perform MT1-MMP silencing, the small interfering RNA (siRNA) expression cassette 5′-GAAGCCTGGCTGCAGCAGTAT-3′, corresponding to the 106-126 sequence of MT1-MMP, was cloned into the pSEC-puro plasmid (Ambion) under the control of the human U6 promoter (pSEC::siRNA). The control plasmid was generated by ligation of the scrambled sequence 5′-GGTCCATGCTGCAGAAAAAC-3′ into the pSEC-puro plasmid (pSEC::siRNAscr).
HT1080 cells were stably transfected with MT1-MMP-WT and MT1-MMP-siRNA using LipofectAMINE (Invitrogen) to generate HT-MT and HT-siRNA cells, respectively. U251 cells were stably transfected with MT1-MMP-WT and MT1-MMP-E240A also using LipofectAMINE to generate U-MT and U-MT-E240A cells, respectively. HT-MT, HT-siRNA and HTsiRNAscr clones resistant to 0.1 mg/mL neomycin and 2 μg/mL puromycin, respectively, and U-MT and U-MT-E240A cells resistant to 0.6–0.8 mg/mL of zeocin were selected for cell surface expression of MT1-MMP by flow cytometry on a FACStar flow cytometer (Becton Dickinson). For these purposes, cells were incubated with 5 μg/mL control rabbit IgG or MT1-MMP Ab815 antibody followed by incubation with an FITC-conjugated F(ab′)2 fragment of goat anti-rabbit IgG. To avoid clone-specific effects, transfected cell lines were generated as pools of positive cell clones (three to six clones for each cell line). Control cells transfected with the original pcDNA3-neo plasmid (HT-neo cells), the pcDNA3-zeo plasmid (U-zeo cells), and the pSEC::siRNAscr (HT-siRNAscr) were generated as a pool of neomycin-resistant, zeocin-resistant, and puromycin-resistant cells, respectively. Transfected HT-MT and H-neo cells were grown in DMEM/FCS supplemented with 0.05 mg/mL neomycin. U-MT, U-MT-E240A, and U-zeo cells were grown in DMEM/FCS with 0.2 mg/mL zeocin, whereas HT-siRNA and HT-siRNAscr cells were grown in the presence of 1 μg/mL puromycin. MCF-7 cells stably expressing MT1-MMP-WT (MCF7-MT cells) and the inert MT1-MMP-E240A mutant (MCF7-MT-E240A cells), as well as the mock cells transfected with the original pcDNA3.1-zeo plasmid (MCF7-zeo) cells, were described earlier (16).
Immunoprecipitation, Western blotting, and gelatin zymography
For Western blotting, cells were surface-biotinylated with sulfo-NHS-LC-biotin (Pierce; ref. 17). Biotin-labeled MT1-MMP was immunoprecipitated using the antibody Ab815. Precipitated samples were analyzed by Western blotting using Extravidin-HRP and a TMB/M substrate. Tumor samples were homogenized in PBS supplemented with a protease inhibitor cocktail containing 1 mmol/L phenylmethylsulfonyl fluoride and aprotinin, pepstatin, and leupeptin (1 μg/mL each). The samples were lysed in 50 mmol/L N-octyl-β-d-glucopyranoside in TBS supplemented with 1 mmol/L CaCl2, 1 mmol/L MgCl2, and a protease inhibitor cocktail. The soluble fractions (30 μg protein/lane) were used for the analysis. For gelatin zymography, cells were incubated for 8 h in DMEM. Because MCF-7 cells do not synthesize MMP-2 naturally, the MCF7 samples were supplemented with the purified pro–MMP-2 (25–50 ng/mL). Medium aliquots (10 μL) were analyzed by gelatin zymography (16).
In vitro cell invasion
Cell invasion assays were performed in wells of a 24-well, 8-μm pore size, Transwell plate (Costar). Transwell inserts were coated with 100 μL Matrigel (0.15 mg/mL) and dried. Inserts were rehydrated in 500 μL DMEM for 2 h immediately before the experiments. The bottom chamber contained 600 μL of DMEM/FCS. Cells (1×106/mL; 0.1 mL/well) were loaded in the upper chamber and allowed to invade for 12 h. The medium was then aspirated. Crystal violet (0.2%) in a 20% methanol/water solution (300 μL) was added to the bottom of each well for 30 min. The stained cells were lysed in 1% SDS (250 μL). The A570 value of the lysates was measured.
Tumorigenesis and metastasis assays and immunohistochemistry (IHC)
Athymic C57BL/6J, Foxn1nu/nu mice were purchased from Harlan Laboratories. To generate xenografts, HT1080 and U251 cells (5 × 106 in 0.1 mL PBS) were injected s.c. (8–10 animals per group). Estrogen-dependent MCF-7 cells were injected in the mammary fat pads of young female mice, which naturally produce a sufficient level of estrogen. Tumor growth was closely monitored by caliper measurements of two perpendicular diameters of xenografts (D1 and D2). Tumor volume was calculated by the formula π / 6(D1 × D2)3/2 and expressed as mean volume ± SE (mm3).
For immunohistochemical (IHC) analysis, tumors were excised and fixed overnight in 4% paraformaldehyde. The tumors were then embedded in paraffin and sectioned. For antigen retrieval, sections were soaked in xylene (30 min) followed by 100% C2H5OH (30 min) and by 70% C2H5OH (30 min). Sections were then heated at 100°C in 50 mmol/L Tris (pH 9.5) for 1 h and blocked in PBS supplemented with 1% casein, 0.05% Tween 20, and the F(ab′)2 fragments of goat anti-mouse IgG (10 μg/mL). Sections were stained for 2 h using the MT1-MMP 5D1 antibody, and the antibodies to PKAα, CK2α, PI3Kδ, and NR3C1 (2 μg/mL each in PBS supplemented with 1% casein, 0.05% Tween 20), and the F(ab′)2 fragments of goat anti-mouse IgG (10 μg/mL) followed by staining with FITC-conjugated goat anti-rabbit IgG (Fc-specific) and Alexa Fluor 488–conjugated goat anti-mouse IgG (Fc-specific) for 2 h. Images were acquired on a BX51 fluorescence microscope equipped with a MagnaFire camera (Olympus). Nuclear DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI).
To generate lung metastasis, the cells (1 × 106) were injected through the tail vein. In 25 d, mice were sacrificed. The lungs were inflated intratracheally with 2 mL of 15% Indian ink solution, fixed, and then bleached with Fekete's solution. Metastatic loci in the lungs were counted using a stereoscope.
Isolation of total RNA and DNA-chip RNA expression profiling
Cells were grown in DMEM/FCS to produce a subconfluent culture. Total RNA was extracted using an RNeasy kit (Qiagen). RNA (500 ng) was reverse-transcribed by M-MLV reverse transcriptase. The transcripts were labeled with biotin using an RNA amplification kit (Ambion). The cDNA samples were mixed with a Hyb E1 hybridization buffer containing 37.5% (w/w) formamide. The hybridization mix was dispensed on the Sentrix Human Ref-8 BeadChip (Illumina) containing 24,000 transcripts of the ∼22,000 genes represented in the consensus Reference Sequence (RefSeq) human genome database. Hybridization was performed for 18 h at 55°C. Array chips were then washed with an E1BC solution, then with 100% ethanol, and lastly with the E1BC solution again. The chips were blocked with an E1 blocking buffer followed by staining with streptavidin-Cy3, washing with the E1BC solution, and drying. Array chips were scanned using a BeadArray Reader (Illumina). The resulting images were analyzed using the BeadStudio image processing software (Illumina). Each strip on the chip contained ∼30 to 40 beads with the attached oligonucleotide DNA corresponding to an individual gene of the RefSeq database. Our experiments were performed in replicates. BeadStudio averages the values of each gene across the samples, and its algorithm automatically takes into account the statistical power of the replicates to provide a more sensitive determination of detection and differential expression. For quantifying gene expression signal levels, we performed a DirectHyb analysis, which was included in the BeadStudio software. Associated hybridization intensities were normalized by the cubic spline method. To produce output files for determining the probability that a gene signal was changed between groups of samples, we performed the BeadStudio's DirectHyb Differential Expression Analysis. The Illumina custom error model was used as a statistical model to determine the P values for differential expression.
Reverse transcription and quantitative reverse transcription–PCR
Total RNA (1.0 μg) was primed with oligo-dT and then reverse-transcribed using the QuantiTech Reverse Transcription kit (Qiagen). Quantitative reverse transcription–PCR (qRT-PCR) was performed using an Mx3000p instrument (Stratagene). The cDNA (10 ng) was used in a 25-μL reaction with the Power SYBR Green PCR Master Mix (Applied Biosystems). The sequences of the primers used in the RT-PCR reactions are shown in Supplementary Table S1. RT-PCR reactions were performed as follows: denaturing step at 95°C for 10 min, followed by 40 cycles of 30 s at 95°C, 1 min at 55°C, and 30 s at 72°C. PCR product amplification was monitored by SYBR green fluorescence and normalized relative to the Rox dye standard. A standard curve was generated with glyceraldehyde-3-phosphate dehydrogenase (GADPH) primers for each tested cDNA sample, and it proved to be linear over four orders of magnitude. This curve was used to determine the relative differences in cDNA from changes in Ct values.
Human tumor gene expression databases
Human tumor gene expression data from the Global Cancer Map (GCM) [190 specimens of 14 different types (breast, pancreas, lung, bladder, ovary, melanoma, uterus, renal, prostate, central nervous system, lymphoma, colorectal, mesothelioma, and leukemia), 16,063 genes, Affymetrix GeneChip Hu6800 and Hu35KsubA; ref. 18] were downloaded from the Website.1 Gene expression data from the normal tissues were discarded. Only the data related to cancerous tissues were further analyzed. The GeneNeighbors module of the GenePattern software was used to identify those genes, the expression of which was closely correlated with that of MT1-MMP (19). The expression level of each gene, across all tumor specimens, was normalized by setting the mean to 0 and the SD to 1. We discarded the genes with either overly low or overly high expression levels (<50 and >15,000 relative units, respectively). We also did not include, in our further analysis, the genes that had either less than a 2-fold difference or less than a 50 relative unit difference across all tumor tissues. We used the Euclidean distance as a measure of similarity in the expression pattern. This algorithm produced a numerical score that represented the calculated Euclidean distance for each gene relative to the MT1-MMP gene. The genes were then ranked so that the low score indicates the close similarity of the expression pattern of the particular gene with that of the MT1-MMP gene.
Kolmogorov-Smirnov statistics
To evaluate the significance of the coexpression pattern of genes, we used the Kolmogorov-Smirnov (KS) statistics. For our analysis, we selected the genes that are differentially expressed in HT-MT and HT-siRNA cells when compared with HT cells with at least a 2-fold difference. We next determined the positional distribution of these selected genes within the list ordered by the Euclidean distance relative to the MT1-MMP gene in the 190 tumor tissues and selected the target gene set of 11 genes. These genes are both coexpressed with the MT1-MMP gene in the tumor specimens and induced by MT1-MMP in HT1080 cells. We next calculated the KS score for these 11 genes using the score module of the GenePattern platform (19). The higher the KS score, the more the expression pattern of the particular gene set is analogous across all tumors. We next performed the same KS analysis for 11 randomly selected genes using 100,000 permutations. The frequency of events when the KS score of the randomly chosen gene set was equal to or exceeded that of the target gene set was taken as a P value.
Transient expression assay of the transcriptional activity of the NR3C1 promoter
The 2.7-kb NR3C1 promoter was recloned from the phGR plasmid (20) into the pGL4.10 (luc2) reporter vector (Promega). The resulting pGL4.10::NR3C1 plasmid that expressed an NR3C1 promoter-firefly luciferase chimera was used in the cotransfection assay with the pGL4.74 control vector that carried the Renilla luc gene under the control of the thymidine kinase promoter. Transient transfection of HT, HT-neo, HT-MT, HT-siRNA, and HT-siRNAscr cells was accomplished using Fugene HD reagent (Roche). The transcriptional activity of the NR3C1 promoter was measured using the Dual-Glo luciferase assay system (Promega).
Phosphorylated antibody screening
HT-neo, HT-MT, and HT-siRNA cells grown in DMEM/FCS were lysed in 20 mmol/L MOPS (pH 7.0), containing 2 mmol/L EGTA, 5 mmol/L EDTA, 30 mmol/L Na-fluoride, 40 mmol/L β-glycerophosphate, 10 mmol/L Na-PPi, 2 mmol/L Naorthovanadate, 1 mmol/L phenylmethylsulfonyl fluoride, 3 mmol/L benzamidine, 5 μmol/L pepstatin, 10 μmol/L leupeptin, and 0.5% Triton X-100. Insoluble material was removed by ultracentrifugation (100,000 × g; 30 min). The supernatant aliquots (500 μg protein each) were analyzed using the phosphorylated site screening KPSS-1.3 and the total kinase protein screening KPKS-1.2 (Kinexus) followed by the quantification of the images by Bio-Rad Quantity One software.
Patient specimens, tissue microarrays, and IHC
The tissue microarrays (TMA), each containing 60 to 140 specimens, represented multiple types of human epithelial malignancies, including breast, gastric, colorectal, ovarian, and cervical carcinomas with adjacent normal tissues, and several additional cancer types. The staining of PKAα, the expression of which does not correlate with MT1-MMP, was used as a control. After dewaxing, TMAs were stained with the rabbit polyclonal MT1-MMP Ab815 antibody, the murine monoclonal NR3C1 antibody (clone 41), and the murine monoclonal antibody 5B against the catalytic subunit of PKAα. The use of the secondary species-specific antibodies, a diaminobenzidine-based detection method using Envision Plus HRP system (DakoCytomation), and an automated Dako Universal Staining System immunostainer followed (21).
In the double-labeling experiments, TMAs were first incubated with murine monoclonal antibodies to PKAα and NR3C1, followed by the use of the Envision Mouse-HRP detection system (DakoCytomation) with a diaminobenzidine chromagen (brown color). Next, the slides were incubated with rabbit antibody Ab815 against MT1-MMP followed by the use of Envision Rabbit-HRP detection system (DakoCytomation) with an SG chromagen (Vector Laboratories) that yields gray-black color (22). Nuclear red was used for nuclear counterstaining. The slides were scanned on a Scanscope CM-1 scanner. The images were processed using ImageScope software (Aperio Technology) with color deconvolution and separation algorithms (23).
Results
Expression and activity of cell surface–associated MT1-MMP
We specifically selected HT1080 cells (HT cells) for our studies because these cells express MT1-MMP naturally. To increase the MT1-MMP expression, we transfected HT cells with MT1-MMP-WT and isolated a pool of the stably transfected HT-MT cells. As a control, we used HT-neo cells transfected with the original plasmid. To silence MT1-MMP, we transfected HT cells with the siRNA construct and isolated HT-siRNA cells. As a second control, we isolated HT-siRNAscr cells which were selected after stable transfection of HT cells with the scrambled siRNA. Immunoprecipitation of cell surface–associated, biotin-labeled MT1-MMP showed the near-complete silencing of the protease in HT-siRNA cells. In HT-MT cells, MT1-MMP was degraded and predominantly represented by the 40-kDa to 45-kDa degradation, membrane-tethered, inactive species (3), as well as by the full-length protease. GM6001 (a hydroxamate inhibitor of MMPs) inhibited both the activity and self-proteolysis of MT1-MMP in HT-MT cells and helped to determine the increased MT1-MMP expression in HT-MT cells compared with HT cells. Similar amounts of the full-length MT1-MMP were detected in the control HT, HT-neo, and HT-siRNAscr cells (Supplementary Fig. S1A). These data correlated with the inactivation of cell surface–associated MT1-MMP by TIMP-2 in HT1080 cells and with the our earlier and other's studies. Therefore, the MT1-MMP enzyme was incapable of self-proteolysis in HT, HT-neo, and HT-siRNAscr cells, but it was proteolyzed in the overexpressing HT-MT cells (24).
The expression of cell surface MT1-MMP was insignificant in U-zeo cells (Supplementary Fig. S1A). Transfection of U cells with either MT1-MMP-WT (U-MT cells) or the active site mutant MT1-MMP-E240A (U-MT-E240A cells) increased the levels of both full-length and degraded forms of the protease. Endogenous MT1-MMP contributed to the degradation of the inert mutant MT1-MMP-E240A in U-MT-E240A cells. Immunoprecipitation also detected significant amounts of MT1-MMP in MCF7-MT and MCF7-MT-E240A cells, whereas MCF7-zeo cells were MT1-MMP–negative. MT1-MMP was largely self-degraded in MCF7-MT cells. Because the inactive E240A mutant is incapable of self-proteolysis, the individual full-length MT1-MMP form has been found in MCF7-MT-E240A cells.
Consistent with the presence of active MT1-MMP on cell surfaces, HT-MT cells readily activated MMP-2 and converted its 68-kDa proenzyme into the 64-kDa intermediate and the 62-kDa mature enzyme. Because of the silencing of MT1-MMP, the activation of MMP-2 was blocked in HT-siRNA cells. The levels of MMP-2 activation were insignificant in the control HT, HT-neo, and HT-siRNAscr cells (Supplementary Fig. S1B). As expected, U-zeo, U-MT-E240A, MCF7-zeo, and MCF7-MT-E240A cells were unable to activate MMP-2, whereas U-MT and MCF7-MT cells efficiently activated MMP-2 (Supplementary Fig. S1B).
To confirm the silencing of MT1-MMP by siRNA, we assessed the relative levels of the MT1-MMP mRNA in HT-MT, HT-neo, and HT-siRNA cells by RT-PCR with the primers complementary to the 3′ untranslated region (Fig. 1C). Because the 3′ untranslated region is missing in the recombinant MT1-MMP gene expressed by the plasmid vector, the primers detected only the message encoded by the chromosomal MT1-MMP gene. The transcriptional efficiency of the endogenous chromosomal MT1-MMP gene was similar both in HT-MT and HT-neo cells (not shown). In turn, and consistent with the results of the Western blotting and gelatin zymography analyses, the transcription activity of the chromosomal MT1-MMP gene was nearly completely silenced in HT-siRNA cells (Supplementary Fig. S1C).
Figure 1.
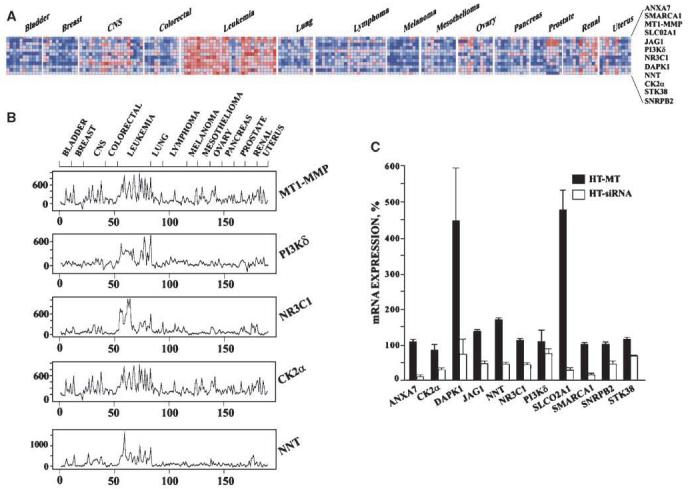
MT1-MMP target genes. A, clustergram of MT1-MMP and its target genes. The colored clustergram shows the expression pattern of the 11 MT1-MMP target genes and MT1-MMP itself in the 190 human tumors of the 14 most common cancer types which are included in the GCM database. Each colored square represents the normalized expression level of an individual gene in a single tumor sample using red and blue to denote high and low levels of expression, respectively. B, expression level correlation. The normalized expression level (Y axes) of MT1-MMP and the four MT1-MMP target genes (PI3Kδ, NR3C1, CK2α, and NNT) in each of 190 tumor samples (X axes) was plotted. Tumor types are shown above the plot. C, cellular mRNA levels of the identified target genes. mRNA levels were measured by qRT-PCR using the RNA samples isolated from HT, HT-MT, and HT-siRNA cells. The expression values were normalized relative to GAPDH. The levels of mRNA in HT-MT and HT-siRNA cells are shown in percentage relative to HT cells (100%).
Invasion-promoting, tumorigenic function of MT1-MMP
Consistent with other multiple MT1-MMP–transfected cell types, HT-MT cells acquired the ability to invade Matrigel more efficiently when compared with HT-neo and HT-siRNAscr cells. There was a statistically significant difference between the invasion efficiency of HT-MT and HT-siRNA cells (Supplementary Fig. S1D). The effects of MT1-MMP we observed in the HT1080 cell system correlate with the earlier results by others (25).
To determine the tumorigenic and prometastatic roles of MT1-MMP, we used in vivo tumor xenograft and lung metastasis assays, respectively, in immunodeficient mice (Supplementary Fig. S1E and F). HT-neo, HT-MT, HT-siRNA, U-zeo, U-MT, and U-MT-E240A cells were each injected in mice. The HT-MT xenografts grew at a rate that exceeded that of the HT-neo and HT-siRNA tumors. At day 21 postimplantation, the HT-MT tumors were ∼2-fold and 8-fold larger than the HT-neo and HT-siRNA tumors, respectively, thus providing solid evidence of the tumorigenic function of MT1-MMP. These data are in agreement with the cytoplasmic tail signaling of the inert MT1-MMP-E240 mutant we recently identified (14).
Similar results were obtained with U251 cells. U-MT cells transfected with MT1-MMP-WT were strongly tumorigenic in comparison with mock-transfected U-zeo cells. The tumorigenicity of U-MT-E240A cells transfected with the catalytically inert protease exceeded that of U-zeo cells only slightly. Similar results were obtained when MCF-7 cells (MCF7-zeo, MCF7-MT, and MCF7-MT-E240A) were used in the xenograft studies.
To determine the role of MT1-MMP in metastasis, HT-neo, HT-MT, and HT-siRNA cells were each injected i.v. into nude mice. In 25 days, the mice were sacrificed. Metastatic loci were counted in the lungs (Supplementary Fig. S1F). The number of metastases was higher by 30% and 90% in mice that received HT-MT cells when compared with HT-neo and HT-siRNA cells, respectively, thus supporting the prometastatic function of MT1-MMP. Overall, our results show the prometastatic, tumorigenic function of both the naturally expressed and overexpressed MT1-MMP.
Gene expression profiling
We used gene expression profiling to determine the genome-wide effect of MT1-MMP on the transcriptional response in HT1080 cells. RNA isolated from HT, HT-neo, HT-MT, HT-siRNAscr, and HT-siRNA cells was reverse-transcribed and amplified in the PCR reactions, and the transcripts were hybridized on a Sentrix HumanRef-8 BeadChip containing 24,000 transcripts from the 22,000 genes represented in the RefSeq database. Gene expression profiles of HT-MT and HT-siRNA cells were then compared with those of HT cells (Supplementary Table S2). We discarded the genes, the expression levels of which were affected at least 2-fold in HT-neo cells and HT-siRNAscr cells when compared with HT cells. The subsequent analysis of the gene array results using the BeadStudio DirectHyb detection software identified 312 genes (P < 0.01) which were differentially expressed in HT-MT and HT-siRNA cells when compared with HT cells and the expression of which differed at least 2-fold, either directly or inversely, in HT-MT cells relative to that in HT-siRNA cells (Supplementary Table S3).
The most obvious feature of MT1-MMP was its effect on the genes, the expression of which is either directly or indirectly linked to angiogenesis and the integrity of extracellular matrix (ECM; collagens type IV, V, and XVIII, fibronectin, laminin, procollagen C-endopeptidase enhancer, hyaluronan synthase, HIF-1α, osteoprotegerin, and plexin; Table 1).
Table 1.
Genes linked to the ECM maintenance and angiogenesis: HT-MT/HT-siRNA, the ratio of gene expression in HT-MT cells versus HT-siRNA cells (P < 0.01)
| Accession | Gene | HT-MT/HT-siRNA | |
|---|---|---|---|
| 1 | NM_022475 | Hedgehog interacting protein (HHIP) | 7.4 |
| 2 | NM_005328 | Hyaluronan synthase 2 (HAS2) | 7.1 |
| 3 | NM_012098 | Angiopoietin-like 2 (ANGPTL2) | 7.1 |
| 4 | NM_001845 | Collagen, type IV, alpha 1 (COL4A1) | 7.0 |
| 5 | NM_000362 | TIMP metallopeptidase inhibitor 3 (TIMP3) | 6.5 |
| 6 | NM_004004 | Gap junction protein, beta 2, 26kDa (GJB2) | 6.3 |
| 7 | NM_000093 | Collagen, type V, alpha 1 (COL5A1) | 3.9 |
| 8 | NM_014391 | Ankyrin repeat domain 1 (cardiac muscle) (ANKRD1) | 3.8 |
| 9 | NM_182943 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (PLOD2) | 3.5 |
| 10 | NM_033140 | Caldesmon 1 (CALD1) | 3.5 |
| 11 | NM_004105 | EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1) | 3.4 |
| 12 | NM_002391 | Midkine (neurite growth-promoting factor 2) (MDK) | 3.4 |
| 13 | NM_032048 | Elastin microfibril interfacer 2 (EMILIN2) | 3.4 |
| 16 | NM_002290 | Laminin, alpha 4 (LAMA4) | 3.4 |
| 15 | NM_032603 | Lysyl oxidase-like 3 (LOXL3) | 3.3 |
| 16 | NM_002571 |
Progestagen-associated endometrial protein (placental protein 14, pregnancy-associated endometrial alpha-2-globulin, alpha uterine protein) (PAEP) |
3.3 |
| 17 | NM_130445 | Collagen, type XVIII, alpha 1 (COL18A1) | 3.1 |
| 18 | NM_054034 | Fibronectin 1 (FN1) | 3.0 |
| 19 | NM_002291 | Laminin, beta 1 (LAMB1) | 2.7 |
| 20 | NM_018837 | Sulfatase 2 (SULF2) | 2.7 |
| 21 | NM_001530 |
Hypoxia-inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) (HIF-1α) |
2.7 |
| 22 | NM_080591 |
Prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (PTGS1) |
2.6 |
| 23 | NM_001235 |
Serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) (SERPINH1) |
2.3 |
| 24 | NM_022356 | Leucine proline-enriched proteoglycan (leprecan) 1 (LEPRE1) | 2.3 |
| 25 | NM_030583 | Matrilin 2 (MATN2) | 2.3 |
| 26 | NM_015103 | Plexin D1 (PLXND1) | 2.3 |
| 27 | NM_014918 | Carbohydrate (chondroitin) synthase 1 (CHSY1) | 2.2 |
| 28 | NM_001849 | Collagen, type VI, alpha 2 (COL6A2) | 2.2 |
| 29 | NM_014584 | ERO1-like (S. cerevisiae) (ERO1L) | 2.2 |
| 30 | NM_002546 |
Tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) (TNFRSF11B) |
2.2 |
| 31 | NM_000176 |
Nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) (NR3C1) |
2.1 |
| 32 | NM_001848. | Collagen, type VI, alpha 1 (COL6A1) | 2.0 |
| 33 | NM_001846 | Collagen, type IV, alpha 2 (COL4A2) | 2.0 |
Data mining and validation
We next determined how significantly the expression of the putative MT1-MMP target genes we identified in HT1080 cells (Supplementary Table S3) correlated with the levels of MT1-MMP in human tumors. For these purposes, we performed data mining using the GCM database. This database includes the expression profiling data of ∼16,000 genes of 190 individual tumors of the 14 human cancer types. Using the GeneNeighbors module of the GenePattern platform for the analysis of GCM database, we ranked 15,191 genes according to their level of coexpression with MT1-MMP (Supplementary Table S4). The Euclidean distance was used as an unbiased measure of the expression pattern similarity of the target gene with the expression pattern of MT1-MMP. Using the data of Supplementary Table S4, we then selected the 11 individual genes, the expression of which most uniformly correlated with that of MT1-MMP both in HT1080 cells and in 190 human tumors (Table 2 and Fig. 1A).
Table 2.
MT1-MMP target genes
| Accession | Gene | Ranking | HT-MT/HT-siRNA | |
|---|---|---|---|---|
| 1 | NM_004938 | DAPK1 | 113 | 2.6 |
| 2 | NM_182977 | NNT | 257 | 2.1 |
| 3 | NM_005630 | SLCO2A1 | 297 | 2.2 |
| 4 | NM_000214 | JAG1 | 444 | 2.0 |
| 5 | NM_005026 | PI3Kδ | 585 | 2.3 |
| 6 | NM_000176 | NR3C1 | 640 | 2.1 |
| 7 | NM_177560 | CK2α | 677 | 2.0 |
| 8 | NM_139035 | SMARCA1 | 684 | 3.7 |
| 9 | NM_007271 | STK38 | 810 | 2.0 |
| 10 | NM_198220 | SNRPB2 | 912 | 2.0 |
| 11 | NM_001156 | ANXA7 | 944 | 3.8 |
NOTE: Ranking of the genes corresponds to the Euclidean distance from the MT1-MMP reference gene, as shown in Supplementary Table S4 that includes approximately 16,000 genes. HT-MT/HT-siRNA, the ratio of gene expression levels in HT-MT cells versus HT-siRNA cells.
Abbreviations: ANXA7, annexin 7; CK2α, protein kinase 2 alpha; DAPK1, death-associated protein kinase 1; JAG1, Jagged 1; NR3C1, nuclear glucocorticoid receptor; NNT, nicotinamide nucleotide transhydrogenase; PI3Kδ, phosphoinositide-3-kinase delta; SLCO2A1, solute carrier organic anion transporter; SMARCA1, actin dependent regulator of chromatin; SNRPB2, small nuclear ribonucleoprotein polypeptide B'; STK38/NDR1, serine-threonine kinase 38/N-myc downstream regulated gene 1.
To confirm that the coexpression of these 11 genes with MT1-MMP is statistically significant we used the KS statistics. One hundred thousand trials with a randomly selected set of 11 genes showed the high statistical significance of the 11 identified genes (P < 0.00001). The appearance of several target genes early in the ordered list and how this appearance contributes to a high KS statistics score is evident from a graphical representation that shows the expression pattern of MT1-MMP, PI3Kδ, NR3C1, CK2α, and NNT in the 190 tumor samples (Fig. 1B). The expression pattern of the other seven genes was similar (not shown).
To corroborate our results, we used qRT-PCR to determine the mRNA levels of the 11 identified genes in HT, HT-MT, and HTsiRNA cells (Fig. 1C). According to our results, the expression levels of these genes were the lowest in HT-siRNA cells. HT-MT and HT cells exhibited similar mRNA levels of these genes, except DAPK1 and SLCO2A1.
To show that the MT1-MMP–mediated regulation is not unique to HT1080 cells, we stained both the HT1080 and the U251 tumor xenografts for MT1-MMP, NR3C1, and PI3Kδ. Staining of the cells for PKAα and α-tubulin was used as controls (Fig. 2). The immunoreactivity of NR3C1, PI3Kδ, and CK2α, but not of PKAα and α-tubulin, was up-regulated by the WT protease, but not by the E240A inert mutant. The studies of other genes we identified are exceedingly complicated because the availability of reliable antibodies is limited.
Figure 2.
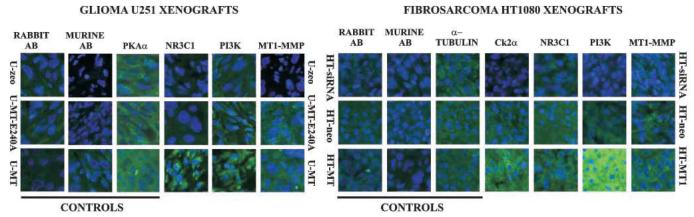
Immunostaining of MT1-MMP, NR3C1, CK2α, and PI3Kδ in tumor xenografts. Paraffin-embedded tumor xenograft sections were stained using the antibodies to MT1-MMP, NR3C1, CK2α, and PI3Kδ. As a control, tumors were stained for α-tubulin and PKAα. FITC-labeled murine and rabbit-negative isotype control IgG were used as additional controls which were clearly negative. Nuclei were stained with DAPI. Original magnification, 400×.
Western blotting of HT, HT-siRNA, and HT-MT xenograft extracts confirmed the increased levels of PI3Kδ in the latter (Fig. 3A). An additional Western blotting analysis of MCF7 xenografts also confirmed the elevated expression of NR3C1 and CK2α in MCF7-MT cells relative to the MCF7-MT-E240A cells. We conclude that the expression of these genes is regulated through different mechanisms: the expression of CK2α and NR3C1 was largely affected by the MT1-MMP gene silencing, whereas the expression of PI3Kδ was affected by an overexpression of the exogenous MT1-MMP.
Figure 3.
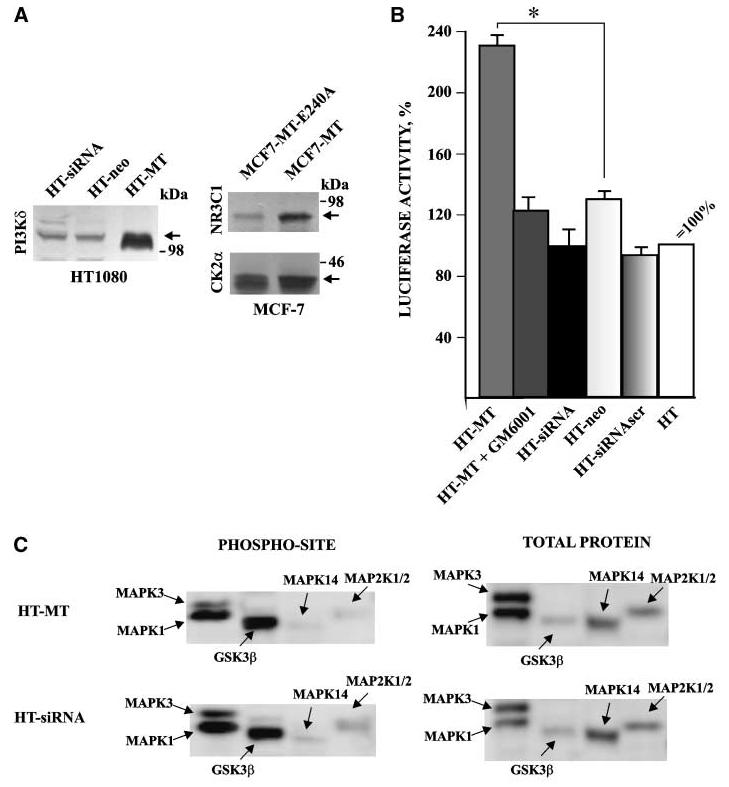
The transactivation function of MT1-MMP. A, Western blotting of HT1080 (left) and MCF-7 cells (right) with the PI3Kδ,NR3C1, and CK2α antibodies. The incubation with the primary antibody was followed by the species-specific HRP-conjugated secondary antibody and a TMB/M substrate. Note the up-regulation of the target genes in cells expressing MT1-MMP-WT. Arrows point to the PI3Kδ,NR3C1, and CK2α bands. B, transient expression assay of the transcriptional activity of the NR3C1 promoter-luciferase chimera. HT, HT-neo, HT-MT, HT-siRNA, and HT-siRNAscr cells were each transiently transfected with both the NR3C1 promoter–firefly luciferase chimera and the pGL4.74 control vector that carried the Renilla luc gene under the control of the thymidine kinase promoter. The mean of four measurements was taken to obtain the firefly luciferase activity that was expressed as relative units defined by the ratio of firefly luciferase to Renilla luciferase activity. Where indicated, cells were coincubated for 14 h with GM6001 (50 μmol/L) before activity measurement. The activity measured in the parental HT cells was taken as 100%. Columns, mean of at least three independent transfection experiments; bars, SE. The data are statistically significant (P < 0.05). C, phosphorylated site and total protein screening. HT-MT and HT-siRNA cell lysate samples were analyzed using phosphorylated site and total kinase protein screening. A portion of the Western blot image that included pMAPK1, pMAPK3, pMAPK14, pMAP2K1/2, and GSKβ (a control). Normalized cpm of pMAPK1 (T185/Y187), pMAPK3 (T202/Y204), pMAPK14 (T180/Y182), pMAP2K1/2 (S217/S221), and GSKβ(Y216) in HT-MT/HT-siRNA cells is 2040/3940, 620/1250, 80/230, 130/430, and 2900/2600, respectively. Normalized cpm of MAPK1, MAPK3, MAPK14, MAP2K1/2, and GSKβ total protein in HM-MT/HT-siRNA cells is 2290/960, 2190/1240, 1410/1370, 720/760, and 420/390, respectively. Normalized cpm is the trace quantity of the band normalized to a 60-s scan time. Values represent the average of the duplicate determinations performed by Kinexus.
Transcription assay of the NR3C1 promoter-luciferase chimera
To directly confirm transactivation activity of MT1-MMP, we used the chimeric construct, in which the sequence of the NR3C1 promoter was linked to the firefly luciferase. The construct was transiently transfected in HT, HT-neo, HT-MT, HT-siRNA, and HT-siRNAscr cells, and the luciferase activity was measured. To control the transfection efficiency the cells were also transfected with the Renilla luciferase. The luciferase activity was expressed as relative units defined by the ratio of firefly luciferase to Renilla luciferase activity. There was a statistically significant increase of the firefly luciferase activity in HT-MT cells when compared with HT-neo cells (Fig. 3B). A potent GM6001 hydroxamate inhibitor inhibited proportionally the transcriptional activity of the chimera in HT-MT cells. Because naturally expressed MT1-MMP is inhibited by TIMP-2 in the original HT cells (24), both siRNA and siRNAscr were without any silencing effect in these cells.
Phosphorylated site screening
NR3C1 is known to repress the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) signaling cascade (26). To confirm this repression and to further characterize the MT1-MMP–linked signal pathway alterations, we determined the levels of phosphorylation of 37 phosphorylation sites in cellular kinases and phospho-proteins. The samples of HT-neo, HT-MT, and HT-siRNA cell lysates were analyzed by phosphorylated site screening KPSS-1.3 (Kinexus). In parallel, the samples were analyzed by total kinase protein screening KPKS-1.2. In a consistent manner, there was a 2-fold decrease in phosphorylation of the four MAPKs: MAPK3 (p44 MAPK), MAPK1 (p42 MAPK), MAP2K1/2 (MEK1/2), and MAPK14 (p38α MAPK) in HT-MT when compared with HT-siRNA cells (Fig. 3C). In contrast, total kinase protein, especially of MAPK1 and MAPK3, was high in HT-MT cells. These results are consistent with the up-regulation of NR3C1 in HT-MT cells. As expected, GSK3β (control) was unaffected by MT1-MMP.
Direct correlations of MT1-MMP with NR3C1
To additionally confirm a link of MT1-MMP with NR3C1, we used an IHC approach. IHC was used to determine the expression of MT1-MMP and NR3C1 in tumor specimens arranged in the TMAs. Because PKAα is not a target of MT1-MMP transactivation, PKAα was used as a control marker. As expected, PKAα immunoreactivity did not correlate with MT1-MMP in tumor specimens, and therefore, MT1-MMP–positive/PKAα–positive, MT1-MMP–negative/PKAα-positive, MT1-MMP–positive/PKAα-negative, and MT1-MMP–negative/PKAα-negative biopsies were readily identified (Fig. 4). In contrast, MT1-MMP staining directly correlated with NR3C1 immunoreactivity. As a result, only the MT1-MMP–positive/NR3C1-positive and MT1-MMP–negative/NR3C1-negative specimens were identified in gastric, breast, lung small cell, ovarian, and endometrial carcinomas. The staining of the TMAs using the isotype control antibodies was clearly negative (not shown). Based on our immunostaining data and the subsequent extensive color separation and deconvolution analyses, we concluded that the expression of MT1-MMP is coupled to an up-regulation of NR3C1 in carcinomas.
Figure 4.
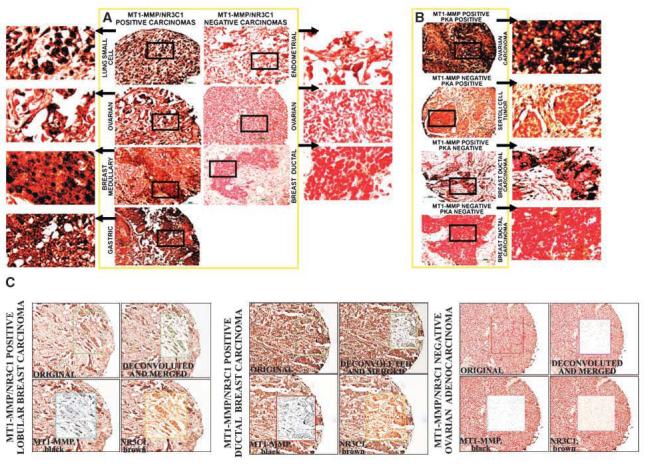
Representative immunostaining of MT1-MMP, NR3C1, and PKAα in human cancer specimens arranged in TMAs. A, MT1-MMP and NR3C1 double staining. B, MT1-MMP and PKAα double staining. TMAs were stained with the antibodies to PKAα and NR3C1 (diaminobenzidine, brown) and with the antibody Ab815 to MT1-MMP (gray black) and counterstained with Nuclear red. The areas marked with squares were enlarged to facilitate the visual analysis of the colors. Bar, either 50 or 100 μm. C, analysis of the images. The selected regions (boxed) were subjected to the image analysis system using Scanscope-HT (Aperio Technology). The black and brown colors of the original image were separated using a color deconvolution algorithm (“deconvoluted and merged” panels). The separated colors are shown in the “MT1-MMP, black” and the “NR3C1, brown” panels. There is an obvious colocalization of MT1-MMP and NR3C1 in the breast carcinoma biopsies, whereas the ovarian carcinoma sample was negative in both MT1-MMP and NR3C1 markers. Original magnification, 300×.
Discussion
MT1-MMP is a key player in stimulating cell locomotion, tumorigenesis, and metastasis of multiple cancer types (1, 27). MT1-MMP functions as an oncogene, usurps control of tumors, and leads to invasion-promoting cell changes (28). These changes suggest alterations of signal transduction and transcription pathways, which MT1-MMP affects either directly or indirectly. To identify these alterations and to determine the identity of the universal genes, the expression of which is affected by MT1-MMP, we up-regulated the expression of the protease in HT1080 cells. We also silenced MT1-MMP in HT1080 cells and performed gene expression profiling of these cells relative to the original HT1080 cells which express MT1-MMP naturally. The intriguing feature of the 312 identified genes whose expression was affected by MT1-MMP was the positive regulation of multiple genes linked to angiogenesis and ECM maintenance. Consistent with our results, the enzymatic activity of MT1-MMP is required for the induction of vascular endothelial growth factor (VEGF; ref. 29). Reciprocally, the MT1-MMP gene promoter exhibits the HIF-1α response element. In agreement, both hypoxia conditions and the addition of VEGF are known to up-regulate MT1-MMP in various cell types (30). It is likely that the MT1-MMP–mediated up-regulation of VEGF and HIF-1α represents a positive feedback to the induction of angiogenesis, which in turn stimulates the expression of MT1-MMP. In agreement with our data, the overexpression of MT1-MMP in human 184B5 mammary fibroblasts and U251 glioma cells resulted in a 6-fold and a 15-fold increase of the fibronectin gene expression, respectively (13), also suggesting a link between MT1-MMP and the synthesis of the ECM components, including fibronectin. We believe tumor cells balance the functional activity of MT1-MMP with the enhanced synthesis of the ECM components to replace the digested ECM with the newly synthesized matrix.
To provide unbiased evidence for the involvement of MT1-MMP in genome-wide transcriptional regulation, regardless of the cell type, we performed a data mining analysis using the GCM database of 190 human tumors of 14 different types. The purpose of this data mining effort was to determine which individual genes from the 312 putative MT1-MMP target genes we identified in HT1080 cells most closely correlated with the expression of MT1-MMP in 190 tumor samples. This unbiased analysis determined the identity of 11 MT1-MMP universal target genes: proapoptotic DAPK1, mitochondrial NNT, organic anion transporter SLCO2A1, WNT-dependent Notch signaling activator JAG1, kinase PI3Kδ, glucocorticoid receptor NR3C1, kinase CK2α, regulator of chromatin SMARCA1, kinase STK38/NDR1, ribonucleoprotein SNRPB2, and synexin/annexin 7 ANXA7. The expression of these genes is tightly correlated with that of MT1-MMP (P < 0.00001). The results of our data mining and our subsequent statistical analyses were validated by using qRT-PCR, Western blotting, and immunostaining of HT1080, U251, and MCF-7 tumor xenografts. Our unbiased IHC analysis of the archival paraffin-embedded tumor specimens confirmed that MT1-MMP immunoreactivity is coupled to that of NR3C1 in carcinomas of different tissue origin. In contrast, there was no correlation between MT1-MMP and PKA (a control marker) in the tumor specimens we evaluated. Based on these results, we suggest that proteolytically active MT1-MMP stimulates multiple transcriptional changes and affects several cellular pathways, including energy metabolism (NNT), trafficking and membrane fusion (SLCO2A1 and ANXA7), signaling and transcriptional regulation (NR3C1, JAG1, PI3Kδ and CK2α), chromatin organization (SMARCA1), cell division (STK38/NDR1), apoptosis (DAPK1), and mRNA splicing (SNRPB2).
Many of these genes are directly associated with cancer. A nuclear glucocorticoid receptor NR3C1 exhibits low transcriptional activity in nontumor fibroblasts but is highly active in fibrosarcoma cells (31). The functional activity of NR3C1 leads to the repression of the stress-activated protein kinase (SAPK)/c-Jun-NH2 kinase (JNK) and MAPK/ERK signaling cascades (26). In agreement, phosporylation/activation of all MAPKs detected in the phosphorylated site screening was repressed in HT-MT cells when compared with HT-siRNA cells. NR3C1 is a known positive regulator of fibronectin (32). Consistently, our data suggest that the MT1-MMP/NR3C1 axis is directly involved in the transcriptional regulation of fibronectin.
A nuclear matrix–associated serine/threonine kinase CK2 is activated by stress and plays an important role in tumor cell survival and proliferation. CK2 is overexpressed in breast (33), prostate (34), head and neck (35) and lung tumors (36), and leukemia (37), suggesting its broad-range tumorigenic function (38). Similar to NR3C1, CK2 negatively regulates SAPK/JNK and MAPK/ERK signaling (39). PI3K the activation, of which results in the generation of phosphatidylinositol (3,4,5)-trisphosphate is directly linked to tumorigenesis and metastasis (40). Both CK2 and PI3Kδ are involved in the activation of the nuclear factor-κB pathway, which is essential to the inflammatory response and, consequently, to cancer cell proliferation. PI3K is also known to up-regulate both MT1-MMP synthesis and activity (41). The expression of ANXA7 is enhanced in metastatic breast cancer and is associated with poor patient survival (42). DAPK1 is overexpressed in breast tumors, suggesting its role in tumorigenesis (43). SNRPB2 is an essential component of the mRNA splicing machinery (44). In agreement with our results, ANXA7, SMARCA1, NR3C1, NNT, CK2α, STK38/NDR1, and SNRPB2 exhibit the cancer-specific alternative splicing variants according to the Alternative Splicing Annotation Project (45). STK38/NDR1 is involved in centrosome duplication, suggesting its role in chromosome segregation, and it is also up-regulated in hypoxia (46). JAG1, the receptor Notch1 ligand, is overexpressed in tumors and plays a role in tumor cell survival, proliferation, and metastasis (47). SNRPB2 is up-regulated in oncogene-transformed fibroblasts and plays a role in cell proliferation (48). Mitochondrial NNT is important for balancing the energy supply to the cells (49). A link of MT1-MMP with NNT, which we detected, seems to associate MT1-MMP–dependent migration and tumor growth with energy metabolism in the cells.
We also inferred from our data mining that MT1-MMP is predominantly and consistently up-regulated in leukemias. Based on our data that MT1-MMP protects malignant cells from the immune surveillance and complement attack in the blood (17), we hypothesized here that MT1-MMP protects leukemia cells from an immune attack in the blood (17). MT1-MMP–dependent down-regulation of MAPKs is also likely to contribute to prosurvival signaling in hematopoetic cancers (50).
Our unbiased genome-wide studies deconvoluted, for the first time, the molecular signature of MT1-MMP in cancer and led us to identify the genes which are functionally, universally, and most consistently linked with MT1-MMP in multiple tumor types. The identified molecular signature of MT1-MMP is linked to the promigratory changes in cell phenotype. The induction of the MT1-MMP target genes, which themselves are directly associated with tumorigenesis, suggests alterations of the signaling, transport, energy metabolism, and other cell functions which jointly, as opposed to MT1-MMP alone, contribute to stimulating cell migration and invasion. The identification of the MT1-MMP target genes is an important step toward the ultimate understanding of the complete complex network of cellular events, which are linked to the MT1-MMP oncogene. The identification of the functional importance of these individual genes in the overall tumorigenic effect of promigratory, tumorigenic MT1-MMP requires additional, extensive studies, which are currently in progress.
Supplementary Material
Acknowledgments
Grant support: NIH grants CA83017, CA77470, and RR020843 and Susan G. Komen Breast Cancer Foundation grant BCTR123106 (A.Y. Strongin).
We thank Xianshu Huang for her professional assistance with TMAs, Dr. Vilma R. Martins (Ludwig Institute for Cancer Research) for her gift of the phGR plasmid, and Dr. Allen H. Olson (Aperio Technology) for his support in image analysis using the color deconvolution and colocalization algorithms.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–74. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 2.Remacle AG, Chekanov AV, Golubkov VS, Savinov AY, Rozanov DV, Strongin AY. O-glycosylation regulates autolysis of cellular membrane type-1 matrix metalloproteinase (MT1-MMP) J Biol Chem. 2006;281:16897–905. doi: 10.1074/jbc.M600295200. [DOI] [PubMed] [Google Scholar]
- 3.Osenkowski P, Toth M, Fridman R. Processing, shedding, and endocytosis of membrane type 1-matrix metalloproteinase (MT1-MMP) J Cell Physiol. 2004;200:2–10. doi: 10.1002/jcp.20064. [DOI] [PubMed] [Google Scholar]
- 4.Jiang A, Lehti K, Wang X, Weiss SJ, Keski-Oja J, Pei D. Regulation of membrane-type matrix metalloproteinase 1 activity by dynamin-mediated endocytosis. Proc Natl Acad Sci U S A. 2001;98:13693–8. doi: 10.1073/pnas.241293698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Ma D, Keski-Oja J, Pei D. Co-recycling of MT1-MMP and MT3-MMP through the trans-Golgi network. Identification of DKV582 as a recycling signal. J Biol Chem. 2004;279:9331–6. doi: 10.1074/jbc.M312369200. [DOI] [PubMed] [Google Scholar]
- 6.Rozanov DV, Deryugina EI, Monosov EZ, Marchenko ND, Strongin AY. Aberrant, persistent inclusion into lipid rafts limits the tumorigenic function of membrane type-1 matrix metalloproteinase in malignant cells. Exp Cell Res. 2004;293:81–95. doi: 10.1016/j.yexcr.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura H, Suenaga N, Taniwaki K, et al. Constitutive and induced CD44 shedding by ADAM-like proteases and membrane-type 1 matrix metalloproteinase. Cancer Res. 2004;64:876–82. doi: 10.1158/0008-5472.can-03-3502. [DOI] [PubMed] [Google Scholar]
- 8.Egawa N, Koshikawa N, Tomari T, Nabeshima K, Isobe T, Seiki M. Membrane type 1 matrix metalloproteinase (MT1-MMP/MMP-14) cleaves and releases a 22-kDa extracellular matrix metalloproteinase inducer (EMMPRIN) fragment from tumor cells. J Biol Chem. 2006;281:37576–85. doi: 10.1074/jbc.M606993200. [DOI] [PubMed] [Google Scholar]
- 9.Rozanov DV, Ghebrehiwet B, Postnova TI, Eichinger A, Deryugina EI, Strongin AY. The hemopexin-like C-terminal domain of membrane type 1 matrix metalloproteinase regulates proteolysis of a multifunctional protein, gC1qR. J Biol Chem. 2002;277:9318–25. doi: 10.1074/jbc.M110711200. [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Aoki T, Mori Y, et al. Cleavage of lumican by membrane-type matrix metalloproteinase-1 abrogates this proteoglycan-mediated suppression of tumor cell colony formation in soft agar. Cancer Res. 2004;64:7058–64. doi: 10.1158/0008-5472.CAN-04-1038. [DOI] [PubMed] [Google Scholar]
- 11.Golubkov VS, Boyd S, Savinov AY, et al. Membrane type-1 matrix metalloproteinase (MT1-MMP) exhibits an important intracellular cleavage function and causes chromosome instability. J Biol Chem. 2005;280:25079–86. doi: 10.1074/jbc.M502779200. [DOI] [PubMed] [Google Scholar]
- 12.Basile JR, Holmbeck K, Bugge TH, Gutkind JS. MT1-MMP controls tumor-induced angiogenesis through the release of semaphorin 4D. J Biol Chem. 2007;282:6899–905. doi: 10.1074/jbc.M609570200. [DOI] [PubMed] [Google Scholar]
- 13.Golubkov VS, Chekanov AV, Savinov AY, Rozanov DV, Golubkova NV, Strongin AY. Membrane type-1 matrix metalloproteinase confers aneuploidy and tumorigenicity on mammary epithelial cells. Cancer Res. 2006;66:10460–5. doi: 10.1158/0008-5472.CAN-06-2997. [DOI] [PubMed] [Google Scholar]
- 14.D'Alessio S, Ferrari G, Cinnante K, et al. Tissue inhibitor of metalloproteinases-2 binding to membrane-type 1 matrix metalloproteinase induces MAPK activation and cell growth by a non-proteolytic mechanism. J Biol Chem. 2008;283:87–99. doi: 10.1074/jbc.M705492200. [DOI] [PubMed] [Google Scholar]
- 15.Takino T, Watanabe Y, Matsui M, et al. Membrane-type 1 matrix metalloproteinase modulates focal adhesion stability and cell migration. Exp Cell Res. 2006;312:1381–9. doi: 10.1016/j.yexcr.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Rozanov DV, Deryugina EI, Ratnikov BI, et al. Mutation analysis of membrane type-1 matrix metalloproteinase (MT1-MMP). The role of the cytoplasmic tail Cys(574), the active site Glu(240), and furin cleavage motifs in oligomerization, processing, and self-proteolysis of MT1-MMP expressed in breast carcinoma cells. J Biol Chem. 2001;276:25705–14. doi: 10.1074/jbc.M007921200. [DOI] [PubMed] [Google Scholar]
- 17.Rozanov DV, Savinov AY, Golubkov VS, Tomlinson S, Strongin AY. Interference with the complement system by tumor cell membrane type-1 matrix metalloproteinase plays a significant role in promoting metastasis in mice. Cancer Res. 2006;66:6258–63. doi: 10.1158/0008-5472.CAN-06-0539. [DOI] [PubMed] [Google Scholar]
- 18.Ramaswamy S, Tamayo P, Rifkin R, et al. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci U S A. 2001;98:15149–54. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zong J, Ashraf J, Thompson EB. The promoter and first, untranslated exon of the human glucocorticoid receptor gene are GC rich but lack consensus glucocorticoid receptor element sites. Mol Cell Biol. 1990;10:5580–5. doi: 10.1128/mcb.10.10.5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krajewski S, Krajewska M, Ellerby LM, et al. Release of caspase-9 from mitochondria during neuronal apoptosis and cerebral ischemia. Proc Natl Acad Sci U S A. 1999;96:5752–7. doi: 10.1073/pnas.96.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krajewska M, Olson AH, Mercola D, Reed JC, Krajewski S. Claudin-1 immunohistochemistry for distinguishing malignant from benign epithelial lesions of prostate. Prostate. 2007;67:907–10. doi: 10.1002/pros.20578. [DOI] [PubMed] [Google Scholar]
- 23.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23:291–9. [PubMed] [Google Scholar]
- 24.Hernandez-Barrantes S, Toth M, Bernardo MM, et al. Binding of active (57 kDa) membrane type 1-matrix metalloproteinase (MT1-MMP) to tissue inhibitor of metalloproteinase (TIMP)-2 regulates MT1-MMP processing and pro-MMP-2 activation. J Biol Chem. 2000;275:12080–9. doi: 10.1074/jbc.275.16.12080. [DOI] [PubMed] [Google Scholar]
- 25.Takino T, Miyamori H, Watanabe Y, Yoshioka K, Seiki M, Sato H. Membrane type 1 matrix metalloproteinase regulates collagen-dependent mitogen-activated protein/extracellular signal-related kinase activation and cell migration. Cancer Res. 2004;64:1044–9. doi: 10.1158/0008-5472.can-03-1843. [DOI] [PubMed] [Google Scholar]
- 26.Kassel O, Sancono A, Kratzschmar J, Kreft B, Stassen M, Cato AC. Glucocorticoids inhibit MAP kinase via increased expression and decreased degradation of MKP-1. EMBO J. 2001;20:7108–16. doi: 10.1093/emboj/20.24.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Itoh Y. MT1-MMP: a key regulator of cell migration in tissue. IUBMB Life. 2006;58:589–96. doi: 10.1080/15216540600962818. [DOI] [PubMed] [Google Scholar]
- 28.Hotary K, Li XY, Allen E, Stevens SL, Weiss SJ. A cancer cell metalloprotease triad regulates the basement membrane transmigration program. Genes Dev. 2006;20:2673–86. doi: 10.1101/gad.1451806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deryugina EI, Soroceanu L, Strongin AY. Up-regulation of vascular endothelial growth factor by membrane-type 1 matrix metalloproteinase stimulates human glioma xenograft growth and angiogenesis. Cancer Res. 2002;62:580–8. [PubMed] [Google Scholar]
- 30.Miyoshi A, Kitajima Y, Ide T, et al. Hypoxia accelerates cancer invasion of hepatoma cells by upregulating MMP expression in an HIF-1α-independent manner. Int J Oncol. 2006;29:1533–9. doi: 10.3892/ijo.29.6.1533. [DOI] [PubMed] [Google Scholar]
- 31.Vivanco MD, Johnson R, Galante PE, Hanahan D, Yamamoto KR. A transition in transcriptional activation by the glucocorticoid and retinoic acid receptors at the tumor stage of dermal fibrosarcoma development. EMBO J. 1995;14:2217–28. doi: 10.1002/j.1460-2075.1995.tb07216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver N, Newby RF, Furcht LT, Bourgeois S. Regulation of fibronectin biosynthesis by glucocorticoids in human fibrosarcoma cells and normal fibroblasts. Cell. 1983;33:287–96. doi: 10.1016/0092-8674(83)90357-4. [DOI] [PubMed] [Google Scholar]
- 33.Munstermann U, Fritz G, Seitz G, Lu YP, Schneider HR, Issinger OG. Casein kinase II is elevated in solid human tumours and rapidly proliferating non-neoplastic tissue. Eur J Biochem. 1990;189:251–7. doi: 10.1111/j.1432-1033.1990.tb15484.x. [DOI] [PubMed] [Google Scholar]
- 34.Ahmed K. Significance of the casein kinase system in cell growth and proliferation with emphasis on studies of the androgenic regulation of the prostate. Cell Mol Biol Res. 1994;40:1–11. [PubMed] [Google Scholar]
- 35.Faust RA, Gapany M, Tristani P, Davis A, Adams GL, Ahmed K. Elevated protein kinase CK2 activity in chromatin of head and neck tumors: association with malignant transformation. Cancer Lett. 1996;101:31–5. doi: 10.1016/0304-3835(96)04110-9. [DOI] [PubMed] [Google Scholar]
- 36.Daya-Makin M, Sanghera JS, Mogentale TL, et al. Activation of a tumor-associated protein kinase (p40TAK) and casein kinase 2 in human squamous cell carcinomas and adenocarcinomas of the lung. Cancer Res. 1994;54:2262–8. [PubMed] [Google Scholar]
- 37.Guerra B, Issinger OG. Protein kinase CK2 and its role in cellular proliferation, development and pathology. Electrophoresis. 1999;20:391–408. doi: 10.1002/(SICI)1522-2683(19990201)20:2<391::AID-ELPS391>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 38.Pinna LA, Meggio F. Protein kinase CK2 (“casein kinase-2”) and its implication in cell division and proliferation. Prog Cell Cycle Res. 1997;3:77–97. doi: 10.1007/978-1-4615-5371-7_7. [DOI] [PubMed] [Google Scholar]
- 39.Heriche JK, Lebrin F, Rabilloud T, Leroy D, Chambaz EM, Goldberg Y. Regulation of protein phosphatase 2A by direct interaction with casein kinase 2α. Science. 1997;276:952–5. doi: 10.1126/science.276.5314.952. [DOI] [PubMed] [Google Scholar]
- 40.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 41.Hess AR, Seftor EA, Seftor RE, Hendrix MJ. Phosphoinositide 3-kinase regulates membrane Type 1-matrix metalloproteinase (MMP) and MMP-2 activity during melanoma cell vasculogenic mimicry. Cancer Res. 2003;63:4757–62. [PubMed] [Google Scholar]
- 42.Srivastava M, Bubendorf L, Raffeld M, et al. Prognostic impact of ANX7-GTPase in metastatic and HER2-negative breast cancer patients. Clin Cancer Res. 2004;10:2344–50. doi: 10.1158/1078-0432.ccr-03-0278. [DOI] [PubMed] [Google Scholar]
- 43.Dairkee SH, Ji Y, Ben Y, Moore DH, Meng Z, Jeffrey SS. A molecular ‘signature’ of primary breast cancer cultures; patterns resembling tumor tissue. BMC Genomics. 2004;5:47. doi: 10.1186/1471-2164-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fong YW, Zhou Q. Stimulatory effect of splicing factors on transcriptional elongation. Nature. 2001;414:929–33. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 45.Lee C, Atanelov L, Modrek B, Xing Y. ASAP: the Alternative Splicing Annotation Project. Nucleic Acids Res. 2003;31:101–5. doi: 10.1093/nar/gkg029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen B, Nelson DM, Sadovsky Y. N-myc down-regulated gene 1 modulates the response of term human trophoblasts to hypoxic injury. J Biol Chem. 2006;281:2764–72. doi: 10.1074/jbc.M507330200. [DOI] [PubMed] [Google Scholar]
- 47.Santagata S, Demichelis F, Riva A, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64:6854–7. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- 48.Laitinen J, Saris P, Holtta E, Pettersson I. U2-snRNP B″ protein gene is an early growth-inducible gene. J Cell Biochem. 1995;58:490–8. doi: 10.1002/jcb.240580412. [DOI] [PubMed] [Google Scholar]
- 49.Freeman H, Shimomura K, Horner E, Cox RD, Ashcroft FM. Nicotinamide nucleotide transhydrogenase: a key role in insulin secretion. Cell Metab. 2006;3:35–45. doi: 10.1016/j.cmet.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 50.Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


