Abstract
Understanding how steroid hormones regulate physiological functions has been significantly advanced by structural biology approaches. However, progress has been hampered by significant misfolding of the ligand binding domains in heterologous expression systems and by conformational flexibility that interferes with crystallization. Here, we show that protein folding problems common to steroid hormone receptors are circumvented by a mutations that stabilize well-characterized conformations of the receptor. We use this approach to present the first structure of an apo steroid receptor, revealing a ligand-accessible channel, allowing soaking of preformed crystals. Furthermore, crystallization of different pharmacological classes of compounds allowed us to define the structural basis of NFκB selective signaling through ER, revealing a unique conformation of the receptor that allows selective suppression of inflammatory gene expression. The ability to crystallize many receptor-ligand complexes with distinct pharmacophores allows one to define the structural features of signaling specificity that would not be apparent in a single structure.
Introduction
The Nuclear receptor (NR) superfamily of transcription factors are effective drug targets because their function is regulated by small molecule ligands, including steroids, lipid metabolites, and synthetic compounds1. The estrogen receptor (ER) exists as two subtypes, ERα and ERβ whicḥ are derived from distinct genes but display a high degree of structural conservation in their DNA and ligand binding domains. In addition to the Selective Estrogen Receptor Modulators (SERMs) used to treat breast cancer or osteoporosis, ER ligands are sought for treatment of a variety of anti-inflammatory and neurological conditions, through either targeting ERβ,. or via ER ligands that selectively suppress the NFκB inflammatory transcriptional cascade 2,3.
While structural biology approaches have provided insights into the development of improved therapeutics, there is still little understanding of the how subtle changes in small molecule chemistry can affect widely different physiological outcomes through the NRs. The ligand-binding domain (LBD) of NRs represents a structurally conserved protein fold, comprised of three layers of α-helices, which contain a buried ligand binding pocket and a solvent exposed coregulator-binding site. The most C-terminal helix, helix 12, acts as a ligand-regulated molecular switch that forms part of the coregulator-binding site. The mechanism of gene activation by nuclear receptors involves the recruitment of transcriptional coactivator proteins to the coregulator binding site, termed AF2, which is formed by helices 3–5 and helix 12 4,5.
Steroid receptor LBDs have proven especially difficult to crystallize due to misfolding in heterologous expression systems. This problem has traditionally required co-fermentation and dedicated purification with a specific ligand to obtain ligand-receptor complexes suitable for X-ray analysis. A second hurdle is that many of the pharmacologically interesting classes of compounds do not fully stabilize the receptor, hindering crystallization. For example, partial agonists, long sought for anti-inflammatory therapies through the glucocorticoid 6, and estrogen receptors 2,3, do not efficiently stabilize the coactivator-binding site, presumably through a destabilization of helix 12. These problems have limited the field of NR crystallography in general to a few structures per year, and have also limited structural analysis to examining individual structures rather than structures from families of ligands representing different potencies and pharmacological classes.
We present a novel approach to crystallizing the LBD through mutations in helix 12 that stabilize well-characterized conformations of the receptor. Specifically, by adding a hydrogen bond to the surface of the protein, helix 12 can be stabilized in the active conformation seen with agonist ligands, or the well characterized inactive conformation seen with a variety of antagonists. We show here that these mutations solve the protein misfolding problem common to the steroid receptor LBDs, allowing ligands to be added in parallel to the purified, concentrated receptor, or soaked into preformed crystals of the apo receptor. This novel technique was used to examine the structures of ER bound to classes of compounds, allowing the discernment of small structural differences. This approach defined the structural basis of partial agonist/NFκB selective signaling through ER. Our findings demonstrate that helix 12 stabilizing mutations provide a tool of broad applicability for rapid structural analysis, thereby more effectively revealing the relationships between biostructural features of ligand-receptor complexes and ligand bioactivity.
Results
Structurally stabilizing mutations
At the base of helix 12 in the ERα LBD lies Tyr-537, a residue implicated in receptor activation. For example, mutation of this residue to serine is sufficient to direct constitutive, ligand-independent activity of the receptor in cell-based assays 7. However, this mutant is still antagonized by tamoxifen (SI Table 1, 1), suggesting that both ligand binding and the associated shift in the localization of helix 12 are intact within the Tyr-537-Ser mutant. Further, this mutant receptor displays high affinity binding to estradiol (2), constitutive association with coactivators, and resistance to proteolysis and urea-induced unfolding 7–12. Therefore, the Tyr-537-Ser mutant mimics the ligand-occupied ERα and this is associated with structural stabilization of the receptor.
To gain insight into mechanisms directing ligand binding and dissociation, the Tyr-537-Ser mutant ERα LBD (amino acids 298–554, Fig. 1a) was expressed in E. coli and purified in the absence of added ligand using immobilized nickel affinity and ion exchange chromatography. The Tyr-537-Ser mutation caused a significant increase in the percent of properly folded protein, as shown by saturated binding of tritiated estradiol (Fig. 1b), thus allowing us to circumvent the protein-misfolding problem common amongst steroid receptors produced in heterologous expression systems. As a result, we were able to obtain diffraction-quality crystals with the receptor co-crystallized with an NR box II peptide from the coactivator GRIP1 5. This peptide helps stabilize the agonist conformation of the structure. The structure was solved by molecular replacement and refined to 2.1 Å (SI Table 2), revealing the canonical agonist conformation (Fig. 1c), with helix 12 folded across helix 3 and helix 11, which stabilizes the coactivator-binding pocket. We also obtained an apo structure in the absence of the Grip1 peptide, but at less than 3 Å resolution (not shown). The mutant Tyr-537-Ser structure was compared to the wild type ERα bound to the full agonist diethylstilbesterol (3, Fig. 1d) to compare their overall secondary structures. The two structures could be superimposed with an R.M.S. deviation of 0.68 Å for the backbone atoms, establishing a high degree of similarity.
Fig. 1. Overall Structure of the Tyr-537-Ser ERα/Grip1 complex.
(a) Cartoon of ER, showing domain boundaries and names, and abbreviations.
(b) The Tyr-537-Ser ERα mutant or the Wt ERα LBD were incubated with excess tritiated estradiol, bound to glass beads, and the unbound ligand washed away. Picomoles of bound ligand were calculated, and compared with the numbers of picomoles of the receptor, to determine % of ligand bound. Shown is mean + SEM.
(c) The secondary structure is shown as ribbon diagram, with the GRIP1 peptide colored red, the helices colored blue, and the beta sheet colored yellow.
(d) The alpha carbon trace of the mutant ER (orange) was superimposed on the DES/ERα structure (blue) using all the backbone atoms. The green boxes denote regions of significant difference in secondary structure.
The initial electron density maps of the mutant Tyr-537-Ser ERα/GRIP1 peptide structure suggested the presence of a small molecule in the ligand-binding pocket, which remained during refinement, but could not be identified using several mass spectrometry approaches (not shown). The apparent electron density does not interact with the other end of the pocket, including helix 11 (SI Fig. 1), strongly suggesting that the significant differences in helix 11 as compared to liganded structures are not artifactual.
To add to the structural toolbox, we also undertook studies to identify mutations that would stabilize the inactive, or antagonist conformation of the receptor. Starting from the tamoxifen-bound ERα structure, we examined the interface between helix 12 and helices 3–5 to identify possible sites for mutagenesis. Our general approach was to find a polar or charged amino acid at the interface that is close to a non-polar residue, which we could then mutate. Fig. 2 shows how the mutation of a leucine in helix 12 to serine allows formation of a hydrogen bond with a glutamic acid in helix 5 that was predicted using this molecular modeling approach.
Fig 2. Identification of a mutation that stabilizes the antagonist conformation of ERα.
(a) Shown is the structure of tamoxifen-bound ERα (PDB:3ERT) as a ribbon diagram. Helices 3–5 are colored pink, and helix 12 is colored red.
(b) Molecular modeling suggests that the mutation Leu-536-Ser of ERα promotes a stabilizing interaction between helix 12 and Glu-380 in helix 3.
ERα Leu-536-Ser was purified, concentrated, and then incubated overnight with tamoxifen (1), raloxifene (4), or the full antagonist ICI 184,780 (5) before setting up crystallization trials. From this, we obtained a 1.8 Å data set for the raloxifene-bound LBD, demonstrating that our technique is not specific to a single mutation, or to the conformation of helix 12. The ligand could be clearly and unambiguously docked into the electron density (SI Fig 1), which interacted with the receptor identically as with the published wild type raloxifene-bound ERα structure. Superpositioning of the wild type and mutant structures demonstrates a 0.32 Å R.M.S. deviation for amino acids within 4.2 Å of the ligand. Though we do not show the function of the mutant in cells, this structure demonstrates that mutations allow the stabilization of the antagonist conformation and proper folding of the receptor in bacteria.
A Ligand-Regulated Solvent-Accessible Channel
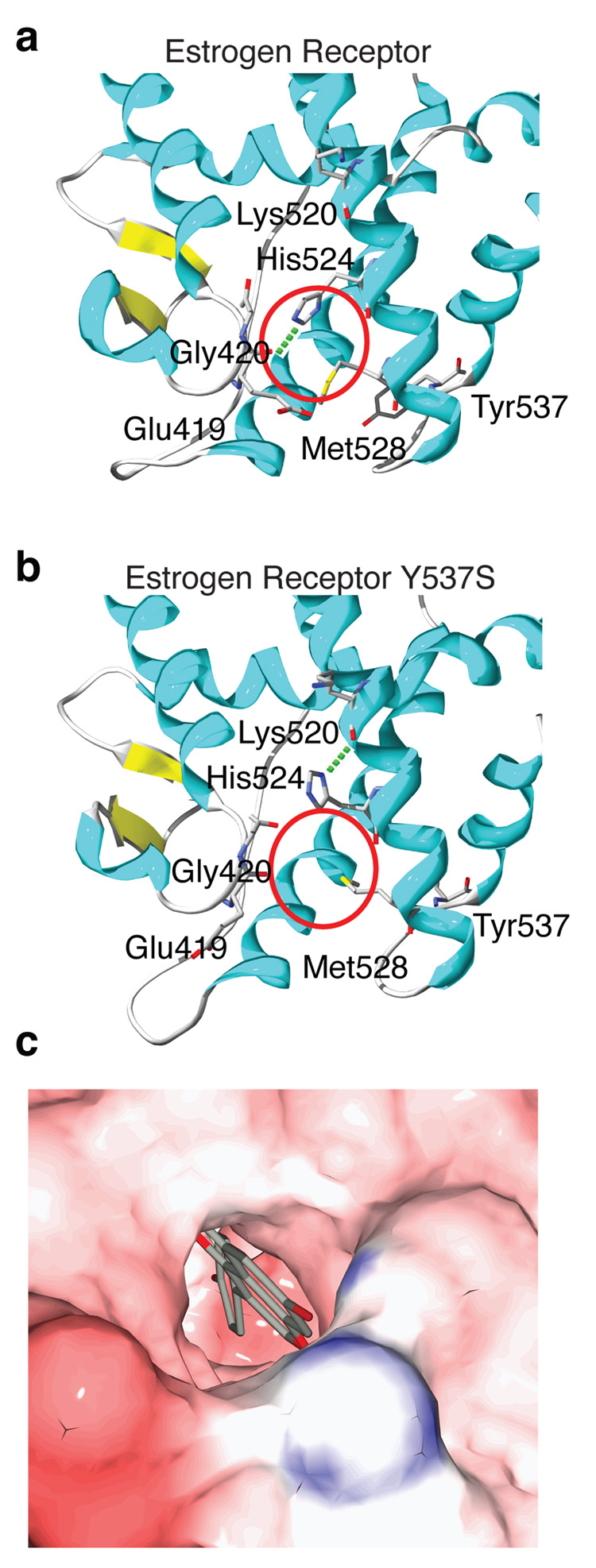
The Tyr-537-Ser mutant ERα LBD structure reveals a cavity connecting the ligand-binding pocket to the solvent, between helix 11 and the loop between helices 7–8 (L7-8) (Fig. 3). In the liganded wild type structures, such as genistein (6)-bound ERα (Fig. 3a), the ligand interacts with His-524 in helix 11, which in turn forms an electrostatic interaction with the backbone of the L7-8. By contrast, in the apo mutant structure, His-524 is flipped out of the ligand binding pocket, H-bonding with the backbone of helix 11 rather than the L7-8 (Fig. 3b). This switch in the position of His-524 opens a channel between the ligand binding cavity and the solvent (red circle in Fig. 3b vs. 3a). The mutant structure was superimposed onto the genistein/ERα structure, allowing a graphical representation of the access of the ligand-binding pocket to the solvent (Fig. 3c). Thus, the mutant Tyr-537-Ser ERα structure reveals a novel solvent channel that connects the ligand-binding cavity to the surface. A notable feature of this channel is that it contains a ligand-mediated switch through His-524, which closes the channel following ligand binding.
Fig 3. A solvent-accessible channel in the Tyr-537-Ser ERα LBD.
(a) The structure of genistein-bound ERα Tyr-537-Ser is depicted as ribbon diagram, showing only a portion of the molecule that interacts with the ligand. The closed interface between helix 11 and L7-8 is shown by a red circle.
(b) The mutant ERα structure with no added ligand is rendered as in panel A, with the red circle highlighting the altered conformation of His-524, and the solvent accessible channel.
(c) The apo and genistein-bound ERα Tyr-537-Ser structures were superimposed over the backbone residues. A surface rendering of the apo structure was colored by electrostatic potential, revealing the solvent accessible channel and the superimposed genistein molecule as a stick rendering.
Other members of the steroid receptor family also demonstrate the structural features consistent with a ligand-mediated switch in helix 11. Specifically, with the glucocorticoid, mineralocorticoid, progesterone, and androgen receptors, an aromatic amino acid is similarly positioned in helix 11 to interact with the ligand. With each of these receptors, molecular modeling was used to examine the most common rotamers for the switch residue. In each case, one of the common rotamers reveals a solvent channel similar to that seen with the ERα̣. For example, for the progesterone receptor (PR, SI Fig. 3a–c), Tyr-890 interacts directly with the progestin ligand and residues in the N-terminus of helix 8 (SI Fig. 3a). A search of the most common rotamers identified an energetically favorable conformation of Tyr-890 that has no clashes with neighboring residues (SI Fig. 3b), and that has a channel to the surface (SI Fig. 3a–c). These observations suggest that the channel is conserved among the steroid receptors. Other ligand channels have been observed, including in the antagonist conformation of ER, and PPARγ, for example, but this novel channel in the apo-ER structure is the first example of a ligand accessible channel into the agonist conformation ER. As discussed below, this channel allowed us to soak apo crystals with a variety of agonist and partial agonist ligands.
Parallel Ligand-Receptor Crystallization
The proper folding of of the Tyr-537-Ser mutant, and the existence of a channel into the ligand binding pocket suggest that the mutant might allow the parallel soaking of ligands into the purified, concentrated receptor. With the wild-type receptor, properly folded protein is commonly obtained by adding ligand during the bacterial fermentation and subsequent purification, making it labor intensive to crystallize multiple ligand-receptor complexes.
To test the hypothesis that the Tyr-537-Ser mutant receptor was competent to bind ligand, a diverse set of 40 ligands (1–41) 13–24 for the ER were incubated with the purified mutant protein overnight, and then used for crystallization trials, resulting in a total of ten solved crystal structures in a few weeks time, including several that have been described elsewhere 13,14,21,22. The ligands could be clearly placed in the electron density of each of the ten structures (SI Fig. 4). These structures were refined with a range of resolutions, from 1.85 Å to 2.7 Å. Details of six of these structures are presented in SI Table 2.
Among the wild-type structures for which structure factors have been deposited, we examined the electron density maps for this region and found that Leu-536 is solvent exposed in approximately 80% of the molecules, and is buried in the remainder (Fig. 4a). With all of the Tyr-537-Ser structures, Leu-536 was uniformly, and unambiguously, rotated into the buried position (Fig. 4b). Given the well-described role of helix 12 as an obligate step in receptor activation, this suggests that a remodeling of the helix 11–12 loop contributes to the stabilization of helix 12 with the mutant receptor. A second observed difference in the ligand-bound Tyr-537-Ser mutant structure is the presence of an altered hydrogen bond pattern in helix 3. In the wild type receptor, Tyr-537 H-bonds to Asn-348. The resultant conformation of Tyr-537 stabilizes the backbone of the helix 11–12 loop, thus fixing Leu-536 into the solvent-exposed position. In the mutant structure, however, a novel hydrogen bond links Ser-537 to Asp-351, which allow burial of Leu-536 in a solvent inaccessible conformation. Thus, both the novel hydrogen bond and altered conformation of the helix 11–12 loop could contribute to constitutive activity of Tyr-537-Ser ERα.
Fig 4. Comparison of ERα wild type and Tyr-537-Ser crystal structures.
(a) The wild type Tyr-537 ERα is shown from the structure of genistein-bound ERα (PDB code:1X7R). Helices 3, 11, and 12 are shown as ribbons, while the loop between helices 11–12 is rendered as a stick figure. This illustrates the hydrogen bond formed with Asn-348, and the location of Leu-536 in a solvent exposed position.
(b) The mutant Tyr-537-Ser ER bound to genistein forms a hydrogen bond with Asp-351, allowing the rotation of Leu-536 out of the solvent.
(c) The amino acids lining the pocket are shown for the mutant and wild-type ERα (PDB code:1X7R) bound to genistein. The two structures were superimposed using the backbone atoms of amino acids within 4.2 Å of the ligands. The Tyr-537-Ser mutant ERα with genistein is colored gray, with the corresponding amino acids colored green. The wild-type ERα structure is colored orange. A structurally conserved water molecule is also shown.
A critical feature of the Tyr-537-Ser mutant ERα receptor is that the surface mutation appears to have no impact on the structure of the ligand-binding pocket. This is clearly demonstrated by a comparison of the Tyr-537-Ser mutant and wild-type ERα bound to genistein (Fig. 4c), which shows that the amino acids within a 4.2 Å radius of the ligand are highly superimposable, with a backbone R.M.S. deviation of 0.2 Å. Thus, by stabilizing the agonist conformation, the Tyr-537-Ser mutation limits the interpretation of helix 12 structural dynamics, but allows for the rapid analysis of receptor-ligand interactions.
It is noteworthy that concentrated receptor was also able to efficiently bind low affinity ligands, such as the oxabicyclic compound (17, SI Fig. 3f), which has a relative binding affinity of 0.02% relative to estradiol 20, corresponding to an IC50 of 1 µM. Therefore, this parallel crystallization approach can be applied to chemistry projects in early stages of development, where ligand affinities are likely to be low, and it allows the application of structural information to optimizing ligand potency.
Ligand Soaking into Apo ERα Tyr-537-Ser Crystals
The crystallization of the apo ERα receptor, the existence of a solvent accessible channel, and our success with co-crystallizing ligands with the purified mutant receptor suggested that we might be able to soak ligands into preformed apo crystals. With ERα Tyr-537-Ser, we soaked apo crystals of ERα Tyr-537-Ser with compounds 38, 41–2, 44–45. Importantly, compound 38 gave identical results to our previously published structure with this compound 14, highlighting that the soaking did not alter ligand interactions. From compounds 41–42, 44–45, three crystals gave data sets to 1.7–2.3 Å resolution, and showed unambiguous electron density for the ligands (SI Fig. 5). Several of these compounds act as selective inhibitors of NFκB through ERα, allowing us to define the structural basis for this signaling specificity, as discussed below.
Structural Analysis of NFκB selective ER ligands
While it is known that the ER and other NRs are involved in the inflammatory pathway via their connection with the NF-κB family of transcription factors 2,25,26, it is not clear how ER ligands specifically signal to this pathway. To further investigate the basis of this selectivity, we desired a set of compounds that show limited activation of estrogen response element (ERE) dependant transcription, but strongly suppress NF-κB dependant transcriptional activity. PhIP (45) and the indazolyl phenol (42) displayed less than 50% activity, while pyrazolopyrimidine (20) and a diethyl oxabicyclic compound (17) have only approximately 10% efficacy in an ERE-luciferase assay relative to estradiol (Fig. 5a). In contrast, these compounds display robust suppression of an NFκB dependant luciferase reporter in heterologous systems (Fig. 5b) as well as of three native NFκB responsive genes in the MCF-7 cell line (Fig 5c), demonstrating strong suppression of TNFκ induced inflammatory gene expression, equivalent to the effects of estradiol.
Fig 5. Transcriptional activity of NF-κB selective ER ligands.
(a) MCF-7 cells were transfected with a 3×ERE-luciferase reporter. The next day, cells treated for 24 hrs with the indicated ligands, and processed for luciferase activity. Shown is mean + SEM from 4–8 wells for each dose.
(b) MCF-7 cells were transfected with a 5×NFκB-luciferase reporter. The next day, cells were treated for 6 hrs with TNFα and the indicated ligands, and then processed for luciferase activity.
(c) MCF-7 cells were treated for 2 hrs with TNFα and the indicated ligands, and then processed for RT-qPCR. The mRNA for IL-6, IL-8, or MCP-1 was normalized to 18S mRNA. Shown is the mean + SEM for duplicate measurements.
We then obtained crystal structures of the most selective compounds: the protein with both the pyrazolopyrimidine and diethyl oxabicyclic shows a dramatic shift in the positioning of helix 11 (Fig. 6a–b, shown in comparison to a mutant structure bound to a full agonist (39), colored green). In the agonist conformation, helix 12 docks against helix 11 to form one side of the coactivator binding site. In the presence of the ligands, the dramatic repositioning of helix 11 His-524 eliminates a key hydrogen bond between His-524 and the loop between helices 7–8. Thus this loss of stabilizing contacts between secondary structural elements likely renders helix 11 more dynamic in solution, interfering with the stabilization of helix 12 in the agonist conformation.
Fig 6. Crystal structures of ER bound to NFκB selective compounds.
(a) The structure of the oxabicyclic diarlyethylene compound/ERα LBD is shown as a ribbon diagram, with the ligand and selected residues in the ligand binding pocket shown as sticks.
(b) The structures of ERα Tyr-537-Ser bound to the indicated ligands were each superimposed with the structure bound to the full agonist, ether estradiol compound (38), using all of the main chain atoms. Shown are selected residues in the pocket, and alpha carbon traces for helices 11 and 12. The ether estradiol bound structure is colored green (ligand not shown), and the NFκB selective structures, and compounds, are colored gray. The red arrows denote the shifts in helix 11 induced by the NFκB selective compounds.
The two compounds with intermediate ERE-dependant transcriptional activity also displayed different conformations of helix 11, but less dramatic changes than seen with the more selective compounds. Fig. 6c shows that PhIP does stabilize His-524 similarly to a full agonist, but induces a shift in the last three turns of helix 11, out of the pocket, and towards helix 12. This suboptimal positioning of helix 11 also likely renders helix 12 more dynamic in solution. The indazolyl phenol is positioned in the pocket so as to draw Leu-525 into an alternate conformation, away from helix 12, resulting in a loss of stabilizing contacts between helices 11 and 12 (Fig 6d). Thus a total of 4 crystal structures of ER bound to compounds with partial agonist/NFκB selective activity provides strong support for the idea that helix 11 conformation is a critical regulator of transcriptional signaling specificity.
To further test this hypothesis, we chemically modified one of the ligands in order to alter the interaction with helix 11. We noted that one of the ethyl groups of the diethyl oxabicyclic directly contacts and repositions His 524 in helix 11, and reasoned that a smaller substitution would allow His 524 to adopt the conformation seen with full agonist compounds. The dimethyl oxabicyclic (43) was synthesized as previously described 20, and tested for cellular activity. Like the diethyl oxabicyclic, the dimethyl compound displayed strong suppression of NFκB responsive luciferase activity (Fig. 7a). However, the dimethyl substituted compound activated the ERE-luciferase reporter to 50–60% efficacy (Fig 7b), in contrast to the very low activity seen with the oxabicyclic diarylethylene (Fig. 7a). We also obtained a crystal structure of the dimethyl oxabicyclic bound to ER, which clearly demonstrates that the change of a single methyl group allows helix 11 to shift closer to the agonist conformation (Fig. 7c), as seen with the other two intermediate agonists. Thus the combination of chemistry, biology, and structural approaches allows us to clearly define how the ligand induced positioning of helix 11 contributes to selectivity for NF-κB signaling, versus activation of estrogen response element dependant transcription.
Fig 7. Structural and Biological Characterization of an Intermediate Agonist.
(a–b) Luciferase activity was assayed as described in Fig 5.
(c) The structure ERα Tyr-537-Ser bound to the oxabicycic diarylmethylene was superimposed with the full agonist, ether estradiol structure, as described in Fig 6.
Discussion
The conserved role of helix 12 in NR function suggests that allosteric mutations may be used to stabilize a variety of receptor conformations. Indeed, we have identified mutations that stabilize naturally occurring conformations of the LBDs of steroid receptors. These strategically designed changes markedly augment NR crystallization and structure determination, and they amplify the power of X-ray crystallography as a tool for relating NR biostructure to ligand biological activity. Among the dozens of published ER structures, there is currently only one in which the ligand caused a complete dislocation of helix 12, which was unstructured 27. This suggests that our approach will be widely applicable to studying most types of ligands. It is important that the mutations are found on the surface, and have no impact in the interaction of the ligand with the receptor. This is directly visualized by comparing our structure of genistein-bound ER with the previously published structure, which showed an identical binding mode for the ligand. Similarly the raloxifene bound mutant ER identically to the published with type structure.
Further, the apo Tyr-537-Ser ERα revealed a novel solvent channel, allowing the soaking of preformed crystals with ligands of interest. Because ligand crystallization trials can thus be set up in parallel with unliganded Tyr-537-Ser ERα, this allows X-ray crystallographic analyses of multiple ER ligands in a high throughput manner. This technique worked even for low affinity compounds, which allows structure to guide chemistry in improving the affinity and selectivity of several of the compounds 21,22. This approach may be particularly advantageous in crystallizing partial agonist compounds, which are expected to render helix 12 more dynamic, a conformational heterogeneity that likely inhibits crystallization. We have since extended this technique to other systems, and preliminary studies with thyroid hormone bound to the thyroid hormone receptor, which is also prone to misfolding, confirm the generality of the approach.
Little is known about how ligands associate or dissociate from nuclear receptors. Structures of the steroid receptors bound to agonist ligands show that the receptor completely encloses the ligand in the agonist conformation. Our crystallization of the Tyr-537-Ser ERα ligand-binding domain in the absence of added ligand has revealed a ligand-mediated switch that controls a channel between the solvent and ligand-binding pocket. The direct interaction of ligand with His-524, a residue known to be important for binding to estradiol and ER’s transcriptional response 28, stabilizes the closed conformation. The conservation of these structural features among the steroid hormone receptors suggests that this channel may be used for rapid crystallization of other steroid receptors.
Another route for ligand binding or dissociation may involve helix 12 dynamics. When Helix 12 is not in the agonist position, most NRs show a solvent accessible channel to the ligand, between helices 3 and 11. This is consistent with the observation that the Tyr-537-Ser mutation slows both the off and on rate of ligand exchange 9. However, other nuclear receptors display additional channels to the ligand. For example, we recently crystallized a series of ligands with the wild-type PPARγ LBD using soaking of apo crystals 29. PPARγ displays a large channel near the beta sheet, which presumably allows ligand binding. Studies of the apo-PPARγ LBD with NMR 30 and hydrogen/deuterium exchange mass spectrometry 29 demonstrate that the apo ligand-binding pocket retains secondary structure, but exists in an ensemble of rapidly exchanging conformations. Thus there may exist other transient openings for ligand access.
In addition to markedly improving the rate of obtaining crystal structures, this parallel crystallization approach also allows one to simultaneously crystallize whole classes of compounds and thereby to identify subtle structural features that would not be apparent with individual structures. As proof-in-principle of this approach, we characterized a series of compounds that strongly suppress NFκB dependant transcription, but show reduced activation of ERE dependent transcriptional activity. Two compounds that showed little to no activation of ERE-luciferase activity demonstrated a remodeling of helix 11 His-524 into the conformation seen with the apo ER, disrupting a canonical hydrogen bond that stabilizes helix 11 against the loop between helices 7–8. Compounds with intermediated ERE dependant transactivation showed the conformation of His-524 seen with full agonist ligands, but showed other disruptions in helix 11, including altered interaction between Leu-525 and Helix 12, and shifts in the last three turns of helix 11. Following up these observations with targeted chemistry of the oxabicyclic ligand allowed us to generate targeted transactivation of the ERE dependant activity, while maintaining strong suppression of NFκB dependant gene expression. The crystal structures we were able to obtain with this set of oxabicyclic compounds provides the first structural demonstration that modulation of helix 11 conformation directly accounts for altered transactivation of ERE dependant transcriptional activity, and separation of pathway selective signaling.
Materials and Methods
Protein Purification and Crystallization
The ERα LBD (amino acids 298–554) was mutated (Tyr-537-Ser or Leu-536-Ser) with the Stratagene Quickchange Mutagenesis kit, and cloned into a modified PET vector with a ligation independent cloning site, 6× His tag, and TEV protease site 31. The protein was induced in BL21 (DE3) cells, and purified with immobilized nickel affinity chromatography. The eluted protein was mixed with a 1:30 ratio (by weight) of his-tagged TEV protease and dialyzed overnight in 20 mM Tris pH 8, 50 mM NaCl, 50 mM β-mercaptoethanol, and 10% glycerol. The next day, the solution was passed through nickel-NTA beads (Qiagen) to remove uncut ERα, the cut tags, and the TEV protease. The flow through was diluted 2× in H2O and subjected to ion exchange chromatography with a Q-FF column (GEHealth). The protein was eluted in 175–185 mM NaCl, 20 mM Tris pH 8.0, 50 mM β-mercaptoethanol, and 10% glycerol, and then concentrated to 0.3 mM (10 mg/ml). The concentrated protein was aliquoted and mixed with 1mM ligands and 1–2 mM GRIP peptide, and incubated overnight. The next day, the protein-ligand slurries were centrifuged at maximum speed for 15 minutes in a 4 °C microcentrifuge, and the supernatant used to set up crystal trials. For each receptor-ligand complex, both the Emerald Biosciences Wizard I&II and Hampton Research Index I&II screens were probed. Initial hits were optimized using a grid screen around pH, precipitant concentration, and protein concentration. The crystals were cryoprotected in either glycerol, PEG, or sucrose added to the mother liquor.
Data was collected at the Structural Biology Center, Biocars, and SER-CAT beamlines at the Advanced Photon Source, Argonne, Illinois, as well as SSRL BL1-11 and scaled with HKL2000. The structures were solved with molecular replacement using Molrep/CCP4. Refinement and rebuilding were performed with CCP4, CNX, Coot, and XtalView. Coordinates and structure factors have been deposited in the protein data bank with the following accession codes: 2QR9, 2QA6, 2QSE, 2QXM, 2QGT, 2QGW, 2QAB, 2QH6, 2QA8, 2B23
Structural Superimposition
The SwissPDBviewer software was used to superimpose backbone atoms in the respective structures. For the comparison of ER subtypes, the structures were superimposed onto the 3ERD structure using all the residues located within 4.2 Å of any of the ligands.
Plasmid DNA and Transient Transfections
The ERE and NFκB luciferase assays, and native gene analysis were performed in MCF-7 cells as previously described 14. MCF-7 cells were maintained in phenol red free DMEM with 10% FBS charcoal/dextran treated (Hyclone). The cells were transfected using Fugene HD (Roche) with a 3×ERE-TATA-luciferase reporter, or 5×NFκB-luciferase reporter. After 6 hours, the cells were passaged and transferred into 384 well plates using a WellMate Microplate Dispenser (Matrix). For the ERE-luc assays, ligands were added the next day and allowed to incubate overnight before processing for luciferase activity. For the NFκB-luc assay, compounds were added with 15ng/ml TNFα, and incubated for 6 hours. An equal volume of Britelite (PerkinElmer) was dispensed by a WellMate Microplate Dispenserand the luminescence was measured by ViewLux ultraHTS Microplate Image (PerkinElmer).
RNA isolation and QPCR
Total RNA was isolated from MCF7 cells using RNeasy (Qiagen), which was used to generate cDNA. PCR analysis was performed on an ABI PRISM 7900HT. Values are normalized with GAPDH content.
Supplementary Material
describe the synthesis and characterization of novel compounds used in the crystallization screens.
Acknowledgements
We are very grateful to S. Rajan for her work on refinement and model building of the structures. We would like to thank T. Tellinghiusen and J. Cleveland for comments on the manuscript. The authors also wish to thank J. Chrzas, G. Sahle, and J. Habel for data collection at APS SER-CAT and C. Smith, G. Card, and J. Habel for data collection at SSRL beamlines. Portions of data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID (or 22-BM) beamline at the Advanced Photon Source, Argonne National Laboratory. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. This work was supported by the National Institutes of Health (GLG 5R01 CA89489; JAK 5R37 DK15556; BSK 5R01 CA18119; RH R01 HL61432 and R01 CA37799), the Ludwig Fund for Cancer Research, (GLG) and DOD Grant W81XWH-04-1-0791 (GLG).
Footnotes
Accession Codes
Coordinates and structure factors were deposited in the Protein Data Bank with the following codes: 2B23 (Apo), 2QGT (ether estradiol, 39), 2QGW (ethyl indazole, 9), 2QAB (chloro indazole, 7), 2QH6 (diethyl oxabicyclic, 17), 2QA8 (genistein, 6), 2QR9 (dimethyl oxabicyclic, 43), 2QA6 (indazolyl phenol, 42), 2QSE (4OH-PhIP, 44), 2QXM (PhIP, 45).
References
- 1.Schulman IG, Heyman RA. The flip side: Identifying small molecule regulators of nuclear receptors. Chem Biol. 2004;11:639–646. doi: 10.1016/j.chembiol.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 2.Chadwick CC, et al. Identification of pathway-selective estrogen receptor ligands that inhibit NF-kappaB transcriptional activity. Proc Natl Acad Sci U S A. 2005;102:2543–2548. doi: 10.1073/pnas.0405841102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steffan RJ, et al. Synthesis and activity of substituted 4-(indazol-3-yl)phenols as pathway-selective estrogen receptor ligands useful in the treatment of rheumatoid arthritis. J Med Chem. 2004;47:6435–6438. doi: 10.1021/jm049194+. [DOI] [PubMed] [Google Scholar]
- 4.Darimont BD, et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiau AK, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 6.De Bosscher K, et al. A fully dissociated compound of plant origin for inflammatory gene repression. Proc Natl Acad Sci U S A. 2005;102:15827–15832. doi: 10.1073/pnas.0505554102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weis KE, Ekena K, Thomas JA, Lazennec G, Katzenellenbogen BS. Constitutively active human estrogen receptors containing amino acid substitutions for tyrosine 537 in the receptor protein. Mol Endocrinol. 1996;10:1388–1398. doi: 10.1210/mend.10.11.8923465. [DOI] [PubMed] [Google Scholar]
- 8.Zhong L, Skafar DF. Mutations of tyrosine 537 in the human estrogen receptor-alpha selectively alter the receptor's affinity for estradiol and the kinetics of the interaction. Biochemistry. 2002;41:4209–4217. doi: 10.1021/bi0121095. [DOI] [PubMed] [Google Scholar]
- 9.Carlson KE, Choi I, Gee A, Katzenellenbogen BS, Katzenellenbogen JA. Altered ligand binding properties and enhanced stability of a constitutively active estrogen receptor: evidence that an open pocket conformation is required for ligand interaction. Biochemistry. 1997;36:14897–14905. doi: 10.1021/bi971746l. [DOI] [PubMed] [Google Scholar]
- 10.Tremblay GB, Tremblay A, Labrie F, Giguere V. Ligand-independent activation of the estrogen receptors alpha and beta by mutations of a conserved tyrosine can be abolished by antiestrogens. Cancer Res. 1998;58:877–881. [PubMed] [Google Scholar]
- 11.Yudt MR, et al. Function of estrogen receptor tyrosine 537 in hormone binding, DNA binding, and transactivation. Biochemistry. 1999;38:14146–14156. doi: 10.1021/bi9911132. [DOI] [PubMed] [Google Scholar]
- 12.Lazennec G, Ediger TR, Petz LN, Nardulli AM, Katzenellenbogen BS. Mechanistic aspects of estrogen receptor activation probed with constitutively active estrogen receptors: correlations with DNA and coregulator interactions and receptor conformational changes. Mol Endocrinol. 1997;11:1375–1386. doi: 10.1210/mend.11.9.9983. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh RW, et al. Identification of ligands with bicyclic scaffolds provides insights into mechanisms of estrogen receptor subtype selectivity. J Biol Chem. 2006;281:17909–17919. doi: 10.1074/jbc.M513684200. [DOI] [PubMed] [Google Scholar]
- 14.Nettles KW, et al. Structural plasticity in the oestrogen receptor ligand-binding domain. EMBO Rep. 2007 doi: 10.1038/sj.embor.7400963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Compton DR, et al. Pyrazolo[1,5-a]pyrimidines: estrogen receptor ligands possessing estrogen receptor beta antagonist activity. J Med Chem. 2004;47:5872–5893. doi: 10.1021/jm049631k. [DOI] [PubMed] [Google Scholar]
- 16.De Angelis M, Stossi F, Carlson KA, Katzenellenbogen BS, Katzenellenbogen JA. Indazole estrogens: highly selective ligands for the estrogen receptor beta. J Med Chem. 2005;48:1132–1144. doi: 10.1021/jm049223g. [DOI] [PubMed] [Google Scholar]
- 17.De Angelis M, Stossi F, Waibel M, Katzenellenbogen BS, Katzenellenbogen JA. Isocoumarins as estrogen receptor beta selective ligands: Isomers of isoflavone phytoestrogens and their metabolites. Bioorg Med Chem. 2005;13:6529–6542. doi: 10.1016/j.bmc.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 18.Kim SH, Katzenellenbogen JA. Hormone-PAMAM dendrimer conjugates: polymer dynamics and tether structure affect ligand access to receptors. Angew Chem Int Ed Engl. 2006;45:7243–7248. doi: 10.1002/anie.200601923. [DOI] [PubMed] [Google Scholar]
- 19.Muthyala RS, Sheng S, Carlson KE, Katzenellenbogen BS, Katzenellenbogen JA. Bridged bicyclic cores containing a 1,1-diarylethylene motif are high-affinity subtype-selective ligands for the estrogen receptor. J Med Chem. 2003;46:1589–1602. doi: 10.1021/jm0204800. [DOI] [PubMed] [Google Scholar]
- 20.Zhou HB, Comninos JS, Stossi F, Katzenellenbogen BS, Katzenellenbogen JA. Synthesis and evaluation of estrogen receptor ligands with bridged oxabicyclic cores containing a diarylethylene motif: estrogen antagonists of unusual structure. J Med Chem. 2005;48:7261–7274. doi: 10.1021/jm0506773. [DOI] [PubMed] [Google Scholar]
- 21.Zhou HB, et al. Elemental Isomerism: A Boron-Nitrogen Surrogate for a Carbon-Carbon Double Bond Increases the Chemical Diversity of Estrogen Receptor Ligands. Chem Biol. 2007;14:659–669. doi: 10.1016/j.chembiol.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Zhou HB, et al. Structure-guided optimization of estrogen receptor binding affinity and antagonist potency of pyrazolopyrimidines with basic side chains. J Med Chem. 2007;50:399–403. doi: 10.1021/jm061035y. [DOI] [PubMed] [Google Scholar]
- 23.Zhang JX, Labaree DC, Hochberg RB. Nonpolar and short side chain groups at C-11beta of estradiol result in antiestrogens. J Med Chem. 2005;48:1428–1447. doi: 10.1021/jm049352x. [DOI] [PubMed] [Google Scholar]
- 24.Bennion BJ, et al. PhIP carcinogenicity in breast cancer: Computational and experimental evidence for competitive interactions with human estrogen receptor. Chem Res Toxicol. 2005;18:1528–1536. doi: 10.1021/tx0501031. [DOI] [PubMed] [Google Scholar]
- 25.Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14:2314–2329. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nettles KW, et al. CBP is a dosage dependent regulator of NF{kappa}B suppression by the estrogen receptor. Mol Endocrinol. 2007 doi: 10.1210/me.2007-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pike AC, et al. Structural insights into the mode of action of a pure antiestrogen. Structure (Camb) 2001;9:145–153. doi: 10.1016/s0969-2126(01)00568-8. [DOI] [PubMed] [Google Scholar]
- 28.Ekena K, Weis KE, Katzenellenbogen JA, Katzenellenbogen BS. Identification of amino acids in the hormone binding domain of the human estrogen receptor important in estrogen binding. J Biol Chem. 1996;271:20053–20059. doi: 10.1074/jbc.271.33.20053. [DOI] [PubMed] [Google Scholar]
- 29.Bruning JB, et al. Partial agonists activate PPARgamma using a helix 12 independent mechanism. Structure. 2007;15:1258–1271. doi: 10.1016/j.str.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Johnson BA, et al. Ligand-induced stabilization of PPARgamma monitored by NMR spectroscopy: implications for nuclear receptor activation. J Mol Biol. 2000;298:187–194. doi: 10.1006/jmbi.2000.3636. [DOI] [PubMed] [Google Scholar]
- 31.Stols L, et al. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
describe the synthesis and characterization of novel compounds used in the crystallization screens.