Summary
Iron deficiency and iron overload are among the most prevalent nutritional disorders worldwide. Duodenal cytochrome b (DcytB) and divalent metal transporter 1 (DMT1) are regulators of iron absorption. Their expression is increased during high systemic requirements for iron, but the molecular mechanisms that regulate DcytB and DMT1 expression are undefined. Hypoxia inducible factor (HIF) signaling was induced in the intestine following acute iron deficiency in the duodenum, resulting in activation of DcytB and DMT1 expression and an increase in iron uptake. DcytB and DMT1 were demonstrated as direct HIF-2α target genes. Genetic disruption of HIF signaling in the intestine abolished the adaptive induction of iron absorption following iron deficiency, resulting in low systemic iron and hematological defects. These results demonstrate that HIF signaling in the intestine is a critical regulator of systemic iron homeostasis.
Introduction
Hypoxia inducible factor (HIF) is a heterodimeric nuclear transcription factor consisting of an oxygen sensitive alpha subunit (HIF-1α and HIF-2α), and a ubiquitously expressed beta subunit, designated aryl hydrocarbon nuclear translocator (ARNT), also known as HIF-1β (Semenza and Wang, 1992; Tian et al., 1997; Wang et al., 1995; Wang and Semenza, 1993). Under normoxia, HIF-1α and HIF-2α are hydroxylated by an iron-dependent prolyl hydroxylase (PHD). Following hydroxylation, HIF-1α and HIF-2α are ubiquitinated by the E3 ubiquitin ligase, von Hippel-Lindau tumor suppressor (VHL) and degraded via the proteasome pathway (Ivan et al., 2001; Jaakkola et al., 2001). The importance of the ubiquitin pathway for HIF-α subunit degradation is underscored by the robust HIF activation observed in mouse models containing a conditional disruption of VHL (Haase, 2005; Kapitsinou and Haase, 2008). HIF signaling is critical in the adaptive response to low oxygen levels by activating genes involved in metabolism, angiogenesis, cell survival and iron metabolism (Hu et al., 2003; Lee and Andersen, 2006; Peyssonnaux et al., 2008; Pugh and Ratcliffe, 2003; Semenza, 2003).
Iron levels are maintained through dietary absorption by duodenal enterocytes. Dietary ferric iron (Fe3+) is reduced to ferrous iron (Fe2+) by the apical ferric reductase duodenal cytochrome b (DcytB) (Latunde-Dada et al., 2008; Mackenzie and Garrick, 2005; McKie et al., 2001). Following the reduction of iron to the ferrous form, iron crosses into the cytoplasm via an apical iron transporter divalent metal transporter-1 (DMT1, also known as Nramp2, SLC11a2 and DCT1) (Fleming et al., 1997; Gunshin et al., 1997; Mackenzie and Garrick, 2005). Both DMT1 and DcytB are localized to the absorptive microvilli structure termed brush border. Iron is either stored or transported out of the enterocyte into the circulation by the sole basolateral transporter ferroportin (FPN, also known as SLC40A1) (Abboud and Haile, 2000; Donovan et al., 2000; Donovan et al., 2005; McKie et al., 2000). When systemic iron requirements are increased, such as in iron deficiency, the expression of DcytB and DMT1 mRNA and protein in the duodenum are induced and more iron is absorbed (Gunshin et al., 1997; McKie et al., 2001). Furthermore, the liver plays a central role in regulating iron absorption by an iron regulatory hormone, hepcidin (Nicolas et al., 2001; Nicolas et al., 2002a). Hepcidin, a small antimicrobial peptide expressed in the liver and secreted into circulation, acts as an inhibitor of iron absorption. To inhibit iron absorption, hepcidin binds to basolateral FPN causing its internalization and degradation, therefore abolishing iron transport (Nemeth et al., 2004). The production of hepatic hepcidin is decreased by iron deficiency and increased during periods of excess iron (Nicolas et al., 2002b; Pigeon et al., 2001). The mechanism by which the apical absorptive proteins DMT1 and DcytB are regulated by iron status is not as clear, however a strong inverse relationship exist between the brush border enzymes and hepcidin, suggesting a role for hepcidin in regulating their expression (Frazer et al., 2002; Wessling-Resnick, 2002).
Recently, HIF was shown to regulate hepcidin transcription by directly binding to and repressing its promoter (Peyssonnaux et al., 2007). Conditional disruption of VHL in hepatocytes resulted in a significant decrease in hepcidin levels through a HIF-dependent mechanism, thus establishing a role for HIF in hepcidin regulation in vivo (Peyssonnaux et al., 2007). Furthermore, systemic hypoxia in rodents and the early period of exposure to high altitudes, both resulted in higher iron absorption (Mendel, 1961; Reynafarje and Ramos, 1961). Given the central role for HIF and oxygen signaling in iron homeostasis, it is still unclear whether HIF signaling is critical for the repression of hepcidin and reciprocal increase in iron absorption following iron deficiency.
In this study, HIF signaling was modulated in enterocytes or hepatocytes using the cre/loxP strategy where the cre transgene is under control of the murine villin and albumin promoters, respectively. A hepatocyte specific conditional knockout of Hif-1α or Arnt (inactivates all HIF-α isoforms) had marginal effects on liver and intestinal responses to low iron diets. Surprisingly, intestinal HIF-2 signaling was shown to be critical in regulating DMT1 and DcytB expression following iron deprivation. These findings indicate a previously unknown and essential role for intestinal HIF-2 in response to nutritional iron deficiency and provide further molecular evidence for a critical link between iron levels and iron absorption.
Results
Generation and characterization of hepatocyte specific ablation of HIF-1α and ARNT via Cre-loxP–mediated recombination in mice
To specifically study the role of HIF signaling in hepatocytes, mice were generated that either lack HIF-1α or the heterodimer partner ARNT, thus inhibiting both HIF-1α and HIF-2α isoforms. HIF-1αF/F or ArntF/F mice were crossed with albumin-cre transgenic mice to generate the HIF-1αΔLIV or ArntΔLIV mouse lines. The albumin promoter confers adult hepatocyte-specific expression; promoter activity increased gradually after birth to reach maximal levels at two-weeks of age (Postic et al., 1999). Hepatocyte-specific disruption of Arnt and Hif-1α genes were confirmed by Southern blotting analysis (Supplemental Figure 1A and B) and mRNA levels by quantitative RT-PCR (qPCR) analysis (Supplemental Figure 1C and D). Both Hif-1αΔLiv and ArntΔLiv mice did not exhibit any obvious deleterious phenotype under normal breeding conditions. To assess the functional effect of disrupting HIF-1α and ARNT, global gene expression profiles in the liver of Hif-1αΔLiv or ArntΔLiv mice were assessed (Supplemental Table 1 and Supplemental Table 2). These results suggest that HIF signaling is limited in the liver with no significant alterations of basal gene expression noted under normal physiological conditions. To assess the role of hepatic HIF signaling in iron homeostasis, Hif-1αΔLiv or ArntΔLiv mice were fed an iron-deficient diet for 2-weeks. ArntΔLiv mice expressed 5 fold more hepcidin compared to wild-type littermate controls on a low-iron diet, however disruption of ARNT could not fully reverse hepcidin repression following iron starvation (Figure 1A). No significant change was observed in Hif-1αΔLiv compared to wild-type littermate controls on a low-iron diet for 2-weeks (Figure 1B). Consistent with the robust decrease in hepcidin mRNA transcripts seen in Hif-1αΔLiv and ArntΔLiv mice, a marked increase in apical iron absorption genes were observed (Figure 1A and B). Both the iron-responsive isoform of Dmt1 (Dmt1 ire) and the ferric reductase Dcytb were increased to the same extent in Hif-1αΔLiv and ArntΔLiv mice compared to their littermate controls following 2-weeks of low-iron diet. Furthermore no significant changes were observed in serum iron levels (Figure 1C and D) or red blood cell parameters as assessed by hematocrit, hemoglobin content and mean cell volume (MCV) in Hif-1αΔLiv and ArntΔLiv mice compared to their littermate controls following 2-weeks of low-iron diet (Supplemental Figure 2). These results demonstrate that even in the presence of higher hepcidin levels in the ArntΔLiv mice treated with low-iron diet for 2-weeks, the HIF-independent decrease of hepcidin can fully mediate the compensatory response to acute iron-deficiency.
Figure 1. Disruption of HIF-1α or ARNT in hepatocytes does not affect iron homeostasis following iron deprivation.
qPCR analysis measuring hepcidin expression or DcytB and DMT1 IRE expression in (A) ArntΔLIV or ArntF/F or (B) Hif-1αΔLIV or Hif-1αF/F mice receiving a low-iron diet for 2-weeks. Expression was normalized to β-actin. Serum iron levels were measured in (C) Hif-1αΔIE or Hif-1αF/F mice or (D) ArntΔIE or ArntF/F mice receiving low-iron diet for 2-weeks. Each bar represents the mean value ± S.D. (*)= P<.05.
HIF signaling is activated in the small intestine of iron-deficient mice
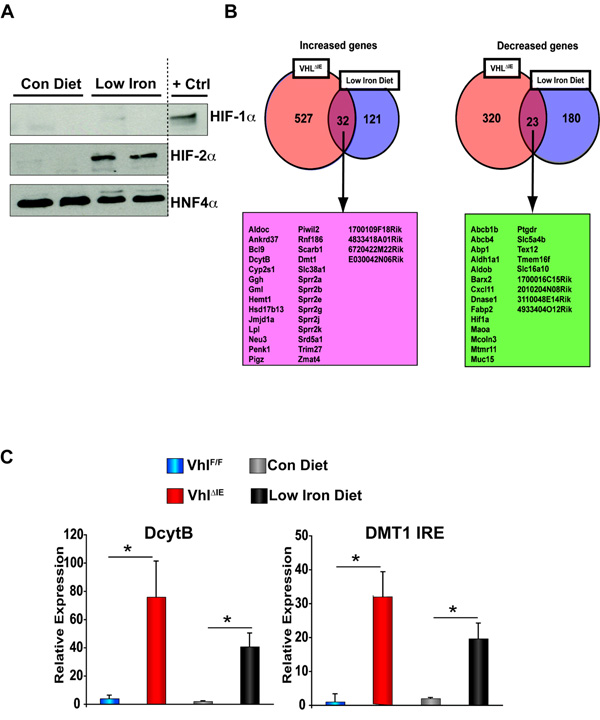
Iron status has been shown to regulate hepatic HIF signaling as mice on iron-deficient diet demonstrated a significant increase in HIF-1α protein expression (Peyssonnaux et al., 2007). However, due to the limited role of hepatic HIF signaling in acute iron homeostasis and given the link between hypoxia and iron absorption (Mendel, 1961; Reynafarje and Ramos, 1961), the potential associations between iron availability and intestinal HIF signaling were examined. Wild-type mice fed a low-iron diet for 2-weeks demonstrated a specific increase in HIF-2α protein expression (Figure 2A), whereas no increase in HIF-1α was observed (Figure 2A). Previous studies revealed that mice with an intestine specific inactivation of VHL (VhlΔIE) demonstrated nearly 100% disruption of the Vhl allele in the colon and throughout the small intestine, had markedly increased HIF-2α protein expression, whereas no HIF-1α protein was detected (Shah et al., 2008). Similar to what was observed in the colon, VhlΔIE mice exhibited a specific increase in HIF-2α protein expression in the small intestine with a concomitant increase in HIF target genes (Supplemental Figure 3A and B). Therefore, to investigate the role of intestinal HIF-2α in iron homeostasis, duodenal gene expression profile from VhlΔIE mice were compared to wild-type mice on low-iron diet. Interestingly, 32 out of 153 genes induced in the small intestine following low-iron diet were also increased in the duodenum of VhlΔIE mice, and 23 out of 203 genes inhibited following low-iron diet were also decreased in the duodenum of VhlΔIE mice (Figure 2B and Supplemental Table 3). The two major apical brush border proteins responsible for the initial absorption of iron into the enterocytes were increased in the small intestine of VhlΔIE mice as confirmed by qPCR measurement of mRNAs in independent samples. Ferric reductase Dcytb demonstrated nearly an 80 fold induction in the duodenum, and over a 30 fold increase was seen with the iron-responsive Dmt1 ire, similar to that observed in mice following 2-weeks of low-iron diet (Figure 2C). These results suggest an important role for HIF-2α in intestinal iron absorption.
Figure 2. Hif-2α modulates iron-regulated genes in the small intestine.
(A) Western blot analysis duodenal epithelial cells in wild-type C57BL/6 mice receiving a low-iron diet for 2-weeks. Expression was normalized to HNF4a protein expression. Nuclear lysate from HCT116 cells treated with 1% O2 for 24-hours were used as a positive control (+ Ctrl) (B) Global gene expression profiling was assessed in duodenal RNAs isolated from VhlF/F and VhlΔIE mice or wild-type mice receiving a low-iron diet for 2-weeks. (C) qPCR analysis of selected genes in duodenal epithelial cells from VhlF/F and VhlΔIE mice or wild-type C57BL/6 mice receiving a low-iron diet for 2-weeks. Expression was normalized to β-actin and each bar represents the mean value ± S.D. (*)= P<.05.
Disruption of VHL in the small intestine affects whole-body iron homeostasis
To confirm the gene expression data, Western blot analysis was performed to quantify DcytB and DMT1 from duodenal extracts isolated from VhlF/F and VhlΔIE mice. Both DMT1 and DcytB were increased in VhlΔIE mice as compared to the littermate controls (Figure 3A). Furthermore, a strong induction in apical staining along the duodenal brush border was evident for both DMT1 and DcytB in VhlΔIE mice as compared to littermate controls (Supplemental Figure 4A and B). Consistent with the increase in DcytB mRNA and protein, ferric reductase activity was increased as measured by nitro blue tetrazolium staining of freshly isolated duodenal samples from VhlF/F and VhlΔIE mice (Figure 3B). Next, histochemical staining for non-heme iron was performed. VhlF/F mice revealing no stainable non-heme iron in duodenal enterocytes (Figure 3C). In contrast, VhlΔIE mice had abundant enterocyte iron as visualized by blue staining along the brush border microvilli (Figure 3C), suggesting an increase in iron uptake from the diet. Furthermore an increase in serum iron levels were observed in VhlΔIE mice compared to littermate controls (Figure 3D), correlating with a significant overload of iron in the livers of VhlΔIE mice beginning as early as 2-months of age. (Figure 3E) Iron overload was also observed in the spleen, which is atypical of mouse models of human hemochromatosis (Fleming et al., 2002; Nicolas et al., 2001; Zhou et al., 1998), therefore suggesting a hepcidin-independent mechanism. Hepcidin mRNA levels were assessed by qPCR analysis and no significant change was observed from livers of VhlF/F and VhlΔIE mice. In addition, no change was observed in the expression of genes encoding proteins critical for basolateral iron export, suggesting that the robust increase in iron absorptive genes can mediate the increase in serum iron and tissue iron overload.
Figure 3. HIF signaling activates iron absorption genes in the duodenum.
(A) Western blot analysis in duodenal epithelial cells. Expression was normalized to GAPDH protein expression. (B) Ferric reductase assays in freshly isolated duodenum from VhlF/F and VhlΔIE mice. The purple stain represents increased ferric reductase activity. (C) Prussian blue iron staining in paraffin-embedded duodenal section from VhlF/F and VhlΔIE mice. The blue stain represents accumulation of non-heme iron. (D) Serum iron analysis from VhlF/F and VhlΔIE mice. Each bar represents the mean value ± S.D. (*) = P<.05. (E) Prussian blue iron staining in paraffin-embedded liver and spleen sections from VhlF/F and VhlΔIE mice. (F) qPCR analysis of selected genes in liver or duodenal epithelial cells from VhlF/F and VhlΔIE mice. Expression was normalized to β-actin and each bar represents the mean value ± S.D.
Double disruption of HIF-1α/VHL or ARNT/VHL in the intestine revealed a major role for HIF-2α in regulation of iron absorption genes
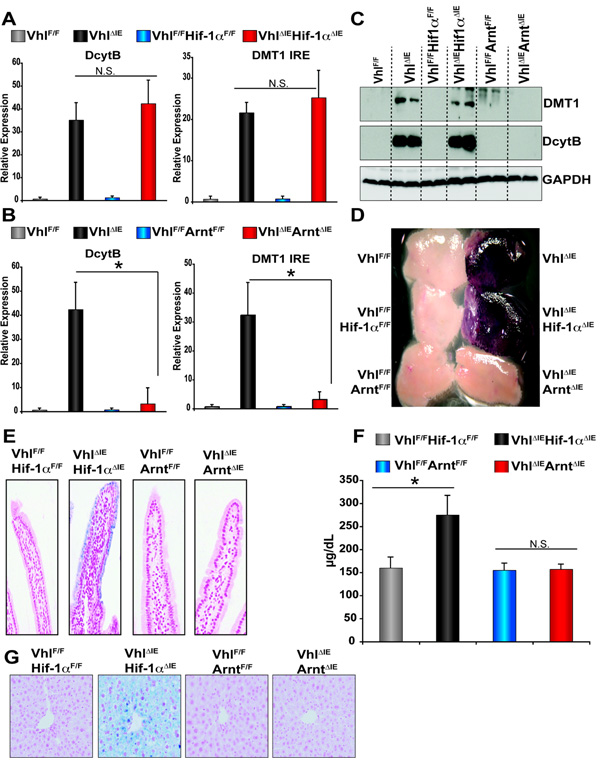
To assess the influence of the HIF-dependent pathways on iron absorption genes and iron uptake in the small intestine of VhlΔIE mice, mice with a double disruption of VHL and HIF-1α or VHL and ARNT were generated. Disruption of VHL and HIF-1α inhibits HIF-1 heterocomplexes, but not HIF-2 heterocomplexes, whereas inactivation of VHL and ARNT prevents the formation of functional HIF-1 and HIF-2. Thus, these mouse models allow delineation of HIF-1 versus HIF-2 signaling pathways and HIF-independent functions of VHL. To use littermate control mice VhlF/FHIF-1αF/+ or VhlF/FArntF/+ mice hemizygous for villin-cre were mated and expression of Dcytb and Dmt1 ire mRNAs were assessed. Disruption of both VHL and HIF-1α (VhlΔIEHif-1αΔIE) had no effect on the induction of Dcytb and Dmt1 ire observed in the VHL single disrupted mice (VhlΔIE) (Figure 4A), whereas double disruption of VHL and ARNT (VhlΔIEArntΔIE) completely abolished the induction of these genes observed in VhlΔIE single knockout mice (Figure 4B). Next, Western analysis was performed on duodenal extracts for DcytB and DMT1. The double mutant VhlΔIEHif-1αΔIE mice had no effect on the induction of DcytB and DMT IRE protein expression observed in VhlΔIE mice, whereas the induction observed in the VhlΔIE mice was completely eliminated in the VhlΔIE ArntΔIE mice (Fig 4C). Induction of ferric reductase activity in the duodenum of VhlΔIE was ameliorated in VhlΔIEArntΔIE, whereas no effect was observed in the VhlΔIEHif-1αΔIE mice (Figure 4D). An increase in non-heme iron was evident in the duodenum VhlΔIEHif-1αΔIE mice and abolished in VhlΔIEArntΔIE mice (Figure 4E). Consistent with these data, an increase in serum iron and hepatic and splenic iron were observed in VhlΔIEHif-1αΔIE mice and not detected in VhlΔIEArntΔIE mice (Figure 4F and G, and data not shown). Together this data demonstrate that the HIF-2 complex is a critical regulator of duodenal iron transport.
Figure 4. Intestine-specific disruption of VHL increases iron absorption through a HIF-dependent mechanism.
(A and B) qPCR analysis measuring Dmt1 and Dcytb expression in RNA from duodenal epithelium. Expression was normalized to β-actin and each bar represents the mean value ± S.D. (*)= P<.05. (C) Western blot analysis measuring DMT1 and DcytB expression in duodenal epithelial cells. Expression was normalized to GAPDH protein expression. (D) Ferric reductase assay in freshly isolated duodenum. (E) Prussian blue iron staining in paraffin-embedded duodenal sections. (F) Serum iron analysis and each bar represents the mean value ± S.D. (*) = P<.05. (G) Prussian blue iron staining in paraffin-embedded liver.
HIF regulates DMT1 and DcytB in the duodenum by directly binding to the regulatory regions in vivo
Dmt1 mRNA levels are increased in the duodenum of iron-deficient animals. This regulation has been observed for Dmt1 mRNA containing a 3’ UTR iron-responsive element, Dmt ire, but not for the isoform lacking the IRE, Dmt1 non-ire (Eisenstein and Ross, 2003; Gunshin et al., 2001). To assess if Dmt1 non-ire is also induced in the small intestine of VhlΔIE mice, qPCR was performed. Only a small increase in Dmt1 non-ire mRNA was observed (Supplemental Figure 3C), demonstrating that the Dmt1 ire is the predominant isoform induced by HIF-2. In addition to the 3’ UTR variants of DMT1, this gene also contains two 5’ processing variants, DMT1A and DMT1B transcribed from two distinct regulatory regions (Hubert and Hentze, 2002). To assess which regulatory region was controlled by HIF, primers were designed to specifically amplify Dmt1a ire or Dmt1b ire isoforms. Using semiquantitative RT-PCR, an increase in the Dmt1a isoform was shown, whereas no regulation of the Dmt1b ire isoform was observed in the duodenum of VhlΔIE mice, similar to the regulation observed in 2-week low-iron treated mice (Supplemental Figure 3D). The upstream sequence from the transcription initiation site of DMT1A contained 2 putative HIF response elements (HRE) (Figure 5A). The upstream sequence of DMT1A encompassing both HREs was analyzed by transient transfection using a luciferase reporter construct. When the DMT1A promoter luciferase construct was transfected into intestine-derived HCT116 cells, hypoxia (1% O2) induced luciferase expression (Figure 5B). Co-transfection with a mammalian expression plasmid for HIF-1α did not significantly increase luciferase expression, while co-transfection with HIF-2α expression plasmid strongly increased the luciferase expression. The HIF-2α increase in luciferase expression was further potentiated in cells incubated in 1% O2, thus demonstrating HIF-2α as the major isoform regulating the DMT1A, and the 5’ deletion analysis suggest that both HREs are functional. Chromatin immunoprecipitation (ChIP) assays were performed in crosslinked soluble chromatin isolated in duodenal samples from VhlF/F or VhlΔIE mice. Using primers flanking both HREs specifically amplified DNA sequence immunoprecipitated by HIF-2α antibody in VhlΔIE mice, demonstrating that HIF-2α is able to bind the DMT1A regulatory region in vivo (Figure 5C). Similar to DMT1A regulatory region the DcytB promoter sequence revealed the presence of two HREs (Figure 5D), and the promoter region of the mouse Dcytb encompassing both HREs was analyzed by transient transfection using a luciferase reporter construct. When the DcytB promoter luciferase construct was transfected into HCT116 cells, hypoxia and co-transfection of HIF-2α induced luciferase expression, whereas co-transfection with a mammalian expression plasmid for HIF-1α did not significantly increase luciferase expression. The HIF-2α increase in luciferase expression was further potentiated in cells incubated in 1% O2, thus demonstrating HIF-2α as the major isoform regulating the DcytB promoter, and the 5’ deletion analysis reveal a functional role for both HREs is DcytB regulation (Figure 5E). Next, ChIP assays were performed using crosslinked soluble chromatin isolated from duodenal samples from VhlF/F or VhlΔIE mice. Primers flanking HRE1 or HRE2 specifically amplified DNA sequence immunoprecipitated by HIF-2α antibody in VhlΔIE mice, thus demonstrating that HIF-2α is able to bind the DcytB regulatory region at both the HREs in vivo (Figure 5F).
Figure 5. HIF-2α directly binds to the endogenous promoters of DMT1A and DcytB.
Schematic diagram of the (A) DMT1A and (C) DcytB promoter illustrating the HREs in the regulatory region, the upstream regions are numbered in relation to the transcription initiation site, which is designated +1. Luciferase-reporter constructs under the control of the regulatory region of the mouse (B) DMT1A and (D) DcytB gene. HCT116 cells transiently transfected with the luciferase construct, and co-transfected with empty vector, HIF-1α or HIF-2α expression plasmids. Standard dual luciferase assays were performed on cell extracts as described in the Materials and Methods. Each bar represents the mean value ± S.D. (*) = P<.05 compared to control incubated cells. (†) = P<.05 compared to Hif-2α transfected cells. In vivo ChIP assays on duodenal extracts from VhlF/F and VhlΔIE mice using primers amplifying both HREs of (C) DMT1A or primers flanking HRE1 and HRE2 of (F) DcytB.
HIF-2 signaling is required for iron absorption following iron deficiency
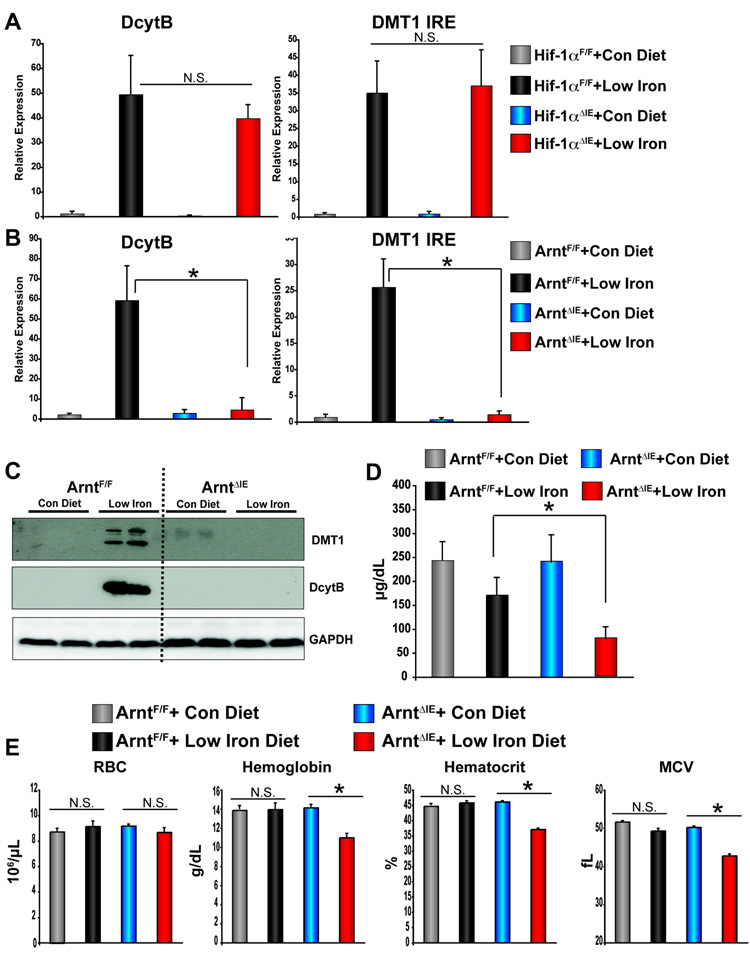
The data thus far have shown that low-iron diet induced HIF-2 signaling, and overexpression of HIF-2α in the duodenum regulated iron absorption genes and increased iron uptake in the duodenum. However, it is unclear whether HIF signaling is required for the adaptive induction of DMT1 and DcytB following iron starvation. To address this issue, mice with an intestine specific disruption of HIF-1α (Hif-1αΔIE) or ARNT (ArntΔIE) were placed on a low-iron diet for 2-weeks and the expression of Dcytb and Dmt1 ire mRNAs were assessed. Intestinal disruption of HIF-1α (Hif-1αΔIE) had no effect on induction of DcytB and DMT IRE following 2-weeks of low-iron diet (Figure 6A), whereas intestinal disruption of Arnt (ArntΔIE) completely inhibited the induction observed in littermate control mice (ArntF/F) (Figure 6B). In duodenal extracts for DcytB and DMT1, a robust induction of DcytB and DMT1 protein expression was observed in ArntF/F mice following 2-weeks of low-iron diet, whereas intestine-specific disruption of the Arnt gene eliminated the induction of DcytB and DMT1 protein expression following 2-weeks of low-iron diet (Figure 6C). The Hif-1αΔIE and ArntΔIE mice demonstrated a dramatic repression of hepcidin expression, indicating that the abrogated intestinal response to low-iron diet in ArntΔIE mice was not due to dysregulation of hepatic hepcidin expression (Supplemental Figure 5). A significant decrease in serum iron levels were observed in ArntΔIE mice following low-iron diet compared to iron-starved littermate controls (Figure 6D). The dramatic drop in serum iron levels corresponded to decreased hematocrit levels, red blood cell size (MCV) and hemoglobin content in ArntΔIE mice compared to littermate controls on low-iron diet (Figure 6E). No change in serum iron levels or hematological parameters were seen in HIF-1αΔIE mice compared to littermate control mice on a low-iron diet (data not shown). ARNT is also an obligate heterodimer for the aryl hydrocarbon receptor (AHR). To discriminate the influences of the HIF-dependent pathways from AHR pathways regulated by ARNT, Ahr-null mice were given low-iron diet for 2-weeks. Disruption of AHR had no effect on the induction of Dcytb and Dmt ire mRNA expression following 2-weeks on low-iron diet (Supplemental Figure 6), and did not demonstrate changes in serum iron levels or red blood cell parameters compared to their littermate controls (data not shown). Together these data demonstrates that HIF-2 signaling in the intestine is critical for the proper adaptive response to iron deficiency.
Figure 6. Inactivation of ARNT abolishes the intestinal response to low-iron diet.
(A) qPCR analysis measuring Dcytb and Dmt1 ire mRNA expression in duodenal epithelial cells in (A) Hif-1αΔIE or Hif-1αF/F mice or (B) ArntIE or ArntF/F mice receiving low-iron diet for 2-weeks. (C) Western blot analysis measuring DcytB and DMT1 expression from duodenal epithelial cells in ArntIE or ArntF/F mice receiving low-iron diet for 2-weeks. (D) Serum iron analysis from ArntIE or ArntF/F mice receiving low-iron diet for 2-weeks. (E) Red blood cell analysis from ArntIE or ArntF/F mice receiving low-iron diet for 2-weeks. Each bar represents the mean value ± S.D. (*) = P<.05
Discussion
The present results reveal a role for HIF-2α as a transcription factor that regulates the expression of genes involved in iron uptake and as a critical transcription factor in the compensatory increase in iron absorption following iron deficiency. These conclusions are based on the following results: (a) Iron starvation strikingly induced HIF-2α protein expression in the small intestine (b) Overexpression of HIF-2α in the small intestine increased the expression of the DMT1 and DcytB, two major apical membrane proteins involved in the mobilization of iron into enterocytes (c) Disruption of intestinal ARNT inhibited the induction of iron transport genes following iron starvation in a HIF-2α dependent manner (d) ArntΔIE mice on low-iron diet demonstrated a dramatic decrease in serum iron levels, RBC size, hemoglobin, and hematocrit content, suggestive of microcytic anemia. These data established a working model for iron absorption in which low iron levels decreased the expression hepatic hepcidin and stabilized the expression of FPN, thereby mobilizing iron into circulation, and leaving the enterocyte in an iron-deficient state. The decreased local iron concentration inhibited PHD activity and increased HIF-2α expression. The activation of HIF-2 signaling induced the expression of DMT1 and DcytB and upregulated transport of iron into the small intestine (Figure 7).
Figure 7. Intestinal HIF-2 activates iron uptake into the small intestine.
(A) Under iron-replete conditions basal iron absorption and export maintain iron homeostasis. Iron export is inhibited through high basal expression of hepcidin and hepcidin-mediated internalization and degradation of FPN. Iron uptake is limited due to decreased expression of absorptive genes. (B) Under iron-deficient conditions, decreased hepcidin expression stabilizes FPN expression resulting in increased iron export. HIF-2α activates iron absorptive genes leading to increased iron uptake into the intestine.
Targeted disruption of the DcytB gene suggests that additional ferric reductase genes exist (Gunshin et al., 2005b), however DcytB remains the only known iron-regulated reductase in the duodenum. Reduction of iron is required for transport through DMT1, due to the selectivity for ferrous iron. Previous studies have demonstrated that DcytB increases iron uptake into intestinal cell lines (Latunde-Dada et al., 2008). Studies in humans have demonstrated duodenal ferric reductase activity as an important factor in the regulation of intestinal iron absorption in healthy individuals and those with altered iron metabolism (Atanasova et al., 2005). The mechanism by which DcytB was induced following iron deficiency was hitherto unknown. The results presented herein demonstrate that a decrease in nutritional iron regulated DcytB expression via a HIF-2-dependent pathway by direct binding of HIF-2α to two consensus HRE elements in the regulatory region of the DcytB promoter.
Following reduction of iron to the ferrous form, DMT1 transports iron into the enterocyte, and its critical role in iron homeostasis is evident by the severe microcytic anemia observed in DMT1 mutant Belgrade rats (Fleming et al., 1998) and microcytic (mk) mice (Fleming et al., 1997). In addition, conditional disruption of Dmt1 in the intestine led to severe anemia and demonstrated DMT1 as a major regulator for iron entry into the small intestine (Gunshin et al., 2005a). The Dmt1 ire isoform was dramatically induced upon iron starvation. IREs are found in either the 5' or 3’ untranslated region (UTR) of transcripts, and are highly conserved binding sites for iron regulatory proteins (IRP). IRPs regulate protein expression post transcriptionally by either altering mRNA stability or protein translation (Pantopoulos, 2004; Rouault, 2006). Mice lacking both IRP1 and IRP2 in the intestine demonstrated an inhibition of mRNA encoded by the IRE-containing isoform of Dmt1 (Galy et al., 2008). These results suggest that IRPs regulate the basal expression of DMT1, however the precise molecular mechanisms by which the IRPs control DMT1 expression upon iron deficiency remained to be determined. In cell lines, iron regulation of DMT1 expression was shown to be through a transcriptional mechanism, and the IRE for DMT1 did not alter mRNA stability when placed at the 3’ end of a constitutively expressed mRNA (Tchernitchko et al., 2002; Zoller et al., 2002). The present study demonstrates HIF-2α as a critical transcriptional regulator of DMT1 via direct binding to the regulatory region of Dmt1. In addition, the induction of the DMT1 IRE isoform by iron depletion was completely dependent on HIF-2, suggesting molecular crosstalk between HIF-2 and IRP signaling, where IRP1 and/or IRP2 are critical for basal gene expression of Dmt1 ire isoform and HIF-2 is required for transcriptional response to low-iron.
Disruption of HIF-1 and HIF-2 signaling in hepatocytes did not influence the susceptibility to iron deficiency. However, it should be noted that a previous study demonstrated that overexpression of HIF-1α and HIF-2α directly suppressed hepcidin levels. This study revealed that feeding a low-iron diet to mice lacking expression of hepatic HIF-1α resulted in significantly higher hepcidin than in wild-type littermates (Peyssonnaux et al., 2008). However, the present study did not reveal differences in hepcidin levels compared to littermate controls in the Hif-1αΔLIV mice following low-iron diet treatment; the slight differences in genetic background of the mouse lines used may explain this discrepancy. However, low-iron treatment of ArntΔLIV mice expressed 5-fold more hepcidin than wild-type littermates, indicating that disruption of HIF-1 and/or HIF-2 regulates hepcidin repression during iron deficiency, consistent with a previous study (Peyssonnaux et al., 2007). Similar to that observed with HIF-1α liver-deficient mice, disruption of ARNT in hepatocytes was insufficient to reverse the repression mediated by an iron-deficient diet. Furthermore, in Hif-1αΔLIV and ArntΔLIV mice, no significant changes were observed in the expression of apical iron absorption genes, serum iron levels or hematological parameters following 2-weeks of low-iron diet compared to similarly treated wild-type littermates. Due to the central role of hepcidin in iron homeostasis, it is not surprising that other transcription factors may contribute or have redundant roles in the regulation hepcidin following iron deficiency. Recently, several factors were shown to regulate hepcidin, such as BMP/SMAD, STAT, and CEBP/α (Babitt et al., 2006; Babitt et al., 2007; Courselaud et al., 2002; Verga Falzacappa et al., 2007; Wang et al., 2005; Xia et al., 2008); whether these factors interact to regulate hepcidin transcription requires additional work to ascertain.
Intestinal disruption of VHL via a HIF-dependent mechanism led to an increase in serum iron and hepatic and splenic iron overload at 2-months of age. Tissue iron deposition was observed only in the liver and spleen, however older mice will be assessed in the future. The present data suggest that the iron overload was independent of the hepcidin-ferroportin axis, and therefore due to an increase in expression of iron absorptive genes. A recent study demonstrated that improper degradation of DMT1 led to overexpression of DMT1 and iron overload (Foot et al., 2008). Furthermore, the importance of iron absorption in iron overload was shown in HFE−/− mice. HFE−/− mice demonstrate a systemic increase in iron, however crossing the HFE−/− mice to mice carrying a mutation in DMT1, significantly decreased iron accumulation (Levy et al., 2000). Hereditary hemochromatosis (HH) is a human iron overload disease most commonly due to a mutation in the HFE gene. The mutation in HFE gene is the most prevalent genetic disorder. Interestingly, HH penetrance is low, indicating modifying genes critical for the pathogenesis of HH (Beutler, 2003). The pattern of iron accumulation in VhlΔIE mice differ from mouse models of HH, however any mechanism that increases iron absorption genes could potentially be involved in the pathogenesis of HH.
In the VhlΔIE mice, HIF-2α acts as the predominant transcription factor regulating the HIF pathways, which is consistent with several other studies demonstrating that loss of VHL preferentially activates HIF-2 signaling (Boutin et al., 2008; Carroll and Ashcroft, 2006; Rankin et al., 2007). Using the VhlΔIE mice and other genetic HIF-1α and ARNT mouse models, the present study demonstrates that HIF-2α dominates in the regulation of intestinal iron absorptive genes through preferential association of the HIF-2 heterocomplex to endogenous HREs. The present study provides a mechanistic basis for a novel therapeutic approach for the treatment of iron related diseases through regulating HIF signaling in the intestine.
Experimental Procedures
Luciferase assay
The mouse DcytB and DMT1A luciferase reporter plasmid was constructed by cloning the upstream regions using primers listed in Supplementary Table 4. The PCR fragments were cloned into the NheI and KpnI restriction sites in the pGL3-basic vector (Promega, Madison WI). These luciferase reporters were transfected into HCT116 cells with Fugene 6 (Invitrogen). For hypoxia treatment, HCT116 cells were incubated for 24 hours in a water-jacketed CO2 incubator containing 1% O2, 5% CO2 and 94% nitrogen gas. Standard dual luciferase assay was used and normalized to a co-transfected control reporter (Promega).
Animals and diets
VhlF/F (Haase et al., 2001), VhlΔIE (Shah et al., 2008), Hif-1αF/F (Tomita et al., 2003), Hif-1αΔIE (Ito et al., 2007), ArntF/F (Tomita et al., 2000), ArntΔIE (Ito et al., 2007), Ahr wild-type and Ahr-null mice (Fernandez-Salguero et al., 1995) were previously described. The VhlF/F and VhlΔIE are in a mixed Sv129 and C57BL/6 background. The Hif-1αF/F, Hif-1αΔIE, ArntF/F and ArntΔIE mice were backcrossed to C57BL/6 five generations. For hepatocyte disruption of HIF-1α and ARNT, Hif-1αF/F and ArntF/F mice were crossed with mice harboring the Cre recombinase under control of the albumin promoter and backcrossed to C57BL/6 5 generations. The mice were used between the ages of 6–8 weeks old for all experiments. The mice were housed in temperature and light controlled rooms, and were given water and pelleted NIH-31 chow ad libitum. In the low-iron studies, mice were given iron-replete AIN93G diet containing (250 ppm iron) or iron-deficient AIN93G diet (5 ppm iron) for 2-weeks (Dyets, Bethlehem, PA). All animal studies were carried out in accordance with Institute of Laboratory Animal Resources guidelines and approved by the National Cancer Institute Animal Care and Use Committee.
Southern blot analysis
Southern blot analysis and probes were previously described (Tomita et al., 2000; Tomita et al., 2003).
RNA analysis
qPCR was performed using primer sequences listed in Supplementary Table 4. For semi-quantitative PCR, cDNA was amplified using primers listed in Supplementary Table 4, electrophoresed on a 1% agarose gel and visualized by ethidium staining.
Western blot analysis
Small intestine epithelium was lysed using RIPA buffer for whole cell extract or lysed using the NE-PER nuclear extraction kit (Pierce, Rockford, IL) for nuclear lysate. For detection of DcytB, proteins extracts were mixed with 2X sample buffer and incubated at 65 C°, for all other antibodies used the samples were boiled for 5 min and then separated and transferred to nitrocellulose membranes using standard Western blotting techniques. The membranes were incubated with an antibody against HIF-1α, HIF-2α (Novus Biologicals, Littleton, CO), DMT1, DcytB (Alpha Diagnostic International, San Antonio, TX); the signals obtained were normalized to GAPDH (Millipore, Temecula, CA) for whole cell extract and HNF4α (Santa Cruz Biotechnology Inc, Santa Cruz, CA) for nuclear extract.
cDNA microarray analysis
Dye-coupled cDNAs were purified with a MiniElute PCR purification kit (Qiagen) and hybridized to an Agilent 44 K mouse 60-mer oligo microarray (Agilent Technologies, Santa Clara, CA). The procedures were repeated for replicate experiments with independent hybridization and processing. The data was processed and analyzed by a Genespring GX software package (Agilent Technologies).
Reductase assay
The duodenum was removed, opened lengthwise, and rinsed with 150 mM NaCl. One mm duodenal sections were incubated at 37°C in 200 µl of 1 mM nitrotetrazolium blue chloride (Sigma, St. Louis, MO) in incubation buffer (125 mM NaCl, 3.5 mM KCl, and 16 mM Hepes/NaOH (pH 7.4)) for 5 min. After incubation, tissues were rinsed twice with 150 mM NaCl and photographed with a dissecting microscope.
Immunohistochemistry
Five µm sections were cut, deparaffinized with xylene, and hydrated in an ethanol gradient. Immunohistochemical analysis was performed with DMT1 and DcytB antibodies (Alpha Diagnostic International) by a streptavidin-biotin immunoperoxidase method. The signal was visualized by diaminobenzidine staining (DAKO, Carpinteria, CA) and counterstained with hematoxylin. Tissue iron detection was performed in formalin fixed paraffin embedded sections stained with Perls Prussian blue and nuclear fast red counter stain.
Serum iron and hematological analysis
Routine erythrocyte analysis was performed by The National Institutes of Health Clinical Center and serum iron was analyzed by the QuantiChrom Iron Assay Kit (Bioassay Systems, Hayward CA) using the manufacturer protocol.
ChIP assay
Duodenal epithelium scrapings were crosslinked in 1% formaldehyde in 1X PBS at 37 C for 20 min. Nuclei were isolated using the NE-PER nuclear extraction kit (Pierce) and lysed in a SDS lysis buffer (50 mM Tris-HCl pH 8.1, 10 mM, EDTA, 1% SDS, and protease inhibitors). Chromatin was sheared in a chilled Bioruptor (Diagenode, Liege, Belgium) and the nuclei lysate was cleared by centrifugation at 16,000 g for 15 min. The soluble chromatin was immunoprecipitated with primary antibody for HIF-2α (Novus Biologicals). The decrosslinked samples were incubated with RNase A and proteinase K. DNA was purified using phenol/chloroform/isoamyl alcohol extraction, and 2 uL of sample was used for PCR using primers listed in Supplementary Table 4.
Data analysis
Results are expressed as mean ± S.D. P values were calculated by Independent t-test. p < 0.05 was considered significant.
Supplementary Material
Acknowledgements
We thank Volker H. Haase for providing the VhlF/F mice, and Richard Bruick and Oliver Hankinson for providing plasmids. We also appreciate technical assistance from John Buckley. This study was supported by the NCI Intramural Research Program and Y.M.S was supported by a postdoctoral fellowship from the American Cancer Society PF-06-014-01-CNE.
Footnotes
Competing interest statement The authors declare that they have no competing financial interest.
References
- Abboud S, Haile DJ. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J Biol Chem. 2000;275:19906–19912. doi: 10.1074/jbc.M000713200. [DOI] [PubMed] [Google Scholar]
- Atanasova BD, Li AC, Bjarnason I, Tzatchev KN, Simpson RJ. Duodenal ascorbate and ferric reductase in human iron deficiency. Am J Clin Nutr. 2005;81:130–133. doi: 10.1093/ajcn/81.1.130. [DOI] [PubMed] [Google Scholar]
- Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest. 2007;117:1933–1939. doi: 10.1172/JCI31342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler E. The HFE Cys282Tyr mutation as a necessary but not sufficient cause of clinical hereditary hemochromatosis. Blood. 2003;101:3347–3350. doi: 10.1182/blood-2002-06-1747. [DOI] [PubMed] [Google Scholar]
- Boutin AT, Weidemann A, Fu Z, Mesropian L, Gradin K, Jamora C, Wiesener M, Eckardt KU, Koch CJ, Ellies LG, Haddad G, Haase VH, Simon MC, Poellinger L, Powell FL, Johnson RS. Epidermal sensing of oxygen is essential for systemic hypoxic response. Cell. 2008;133:223–234. doi: 10.1016/j.cell.2008.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll VA, Ashcroft M. Role of hypoxia-inducible factor (HIF)-1alpha versus HIF-2alpha in the regulation of HIF target genes in response to hypoxia, insulin-like growth factor-I, or loss of von Hippel-Lindau function: implications for targeting the HIF pathway. Cancer Res. 2006;66:6264–6270. doi: 10.1158/0008-5472.CAN-05-2519. [DOI] [PubMed] [Google Scholar]
- Courselaud B, Pigeon C, Inoue Y, Inoue J, Gonzalez FJ, Leroyer P, Gilot D, Boudjema K, Guguen-Guillouzo C, Brissot P, Loreal O, Ilyin G. C/EBPalpha regulates hepatic transcription of hepcidin, an antimicrobial peptide and regulator of iron metabolism. Cross-talk between C/EBP pathway and iron metabolism. J Biol Chem. 2002;277:41163–41170. doi: 10.1074/jbc.M202653200. [DOI] [PubMed] [Google Scholar]
- Donovan A, Brownlie A, Zhou Y, Shepard J, Pratt SJ, Moynihan J, Paw BH, Drejer A, Barut B, Zapata A, Law TC, Brugnara C, Lux SE, Pinkus GS, Pinkus JL, Kingsley PD, Palis J, Fleming MD, Andrews NC, Zon LI. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 2000;403:776–781. doi: 10.1038/35001596. [DOI] [PubMed] [Google Scholar]
- Donovan A, Lima CA, Pinkus JL, Pinkus GS, Zon LI, Robine S, Andrews NC. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1:191–200. doi: 10.1016/j.cmet.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Eisenstein RS, Ross KL. Novel roles for iron regulatory proteins in the adaptive response to iron deficiency. J Nutr. 2003;133:1510S–1516S. doi: 10.1093/jn/133.5.1510S. [DOI] [PubMed] [Google Scholar]
- Fernandez-Salguero P, Pineau T, Hilbert DM, McPhail T, Lee SS, Kimura S, Nebert DW, Rudikoff S, Ward JM, Gonzalez FJ. Immune system impairment and hepatic fibrosis in mice lacking the dioxin-binding Ah receptor. Science. 1995;268:722–726. doi: 10.1126/science.7732381. [DOI] [PubMed] [Google Scholar]
- Fleming MD, Romano MA, Su MA, Garrick LM, Garrick MD, Andrews NC. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc Natl Acad Sci U S A. 1998;95:1148–1153. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming MD, Trenor CC, 3rd, Su MA, Foernzler D, Beier DR, Dietrich WF, Andrews NC. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet. 1997;16:383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- Fleming RE, Ahmann JR, Migas MC, Waheed A, Koeffler HP, Kawabata H, Britton RS, Bacon BR, Sly WS. Targeted mutagenesis of the murine transferrin receptor-2 gene produces hemochromatosis. Proc Natl Acad Sci U S A. 2002;99:10653–10658. doi: 10.1073/pnas.162360699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foot NJ, Dalton HE, Shearwin-Whyatt LM, Dorstyn L, Tan SS, Yang B, Kumar S. Regulation of the divalent metal ion transporter DMT1 and iron homeostasis by a ubiquitin-dependent mechanism involving Ndfips and WWP2. Blood. 2008 doi: 10.1182/blood-2008-04-150953. [DOI] [PubMed] [Google Scholar]
- Frazer DM, Wilkins SJ, Becker EM, Vulpe CD, McKie AT, Trinder D, Anderson GJ. Hepcidin expression inversely correlates with the expression of duodenal iron transporters and iron absorption in rats. Gastroenterology. 2002;123:835–844. doi: 10.1053/gast.2002.35353. [DOI] [PubMed] [Google Scholar]
- Galy B, Ferring-Appel D, Kaden S, Grone HJ, Hentze MW. Iron regulatory proteins are essential for intestinal function and control key iron absorption molecules in the duodenum. Cell Metab. 2008;7:79–85. doi: 10.1016/j.cmet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Gunshin H, Allerson CR, Polycarpou-Schwarz M, Rofts A, Rogers JT, Kishi F, Hentze MW, Rouault TA, Andrews NC, Hediger MA. Iron-dependent regulation of the divalent metal ion transporter. FEBS Lett. 2001;509:309–316. doi: 10.1016/s0014-5793(01)03189-1. [DOI] [PubMed] [Google Scholar]
- Gunshin H, Fujiwara Y, Custodio AO, Direnzo C, Robine S, Andrews NC. Slc11a2 is required for intestinal iron absorption and erythropoiesis but dispensable in placenta and liver. J Clin Invest. 2005a;115:1258–1266. doi: 10.1172/JCI24356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- Gunshin H, Starr CN, Direnzo C, Fleming MD, Jin J, Greer EL, Sellers VM, Galica SM, Andrews NC. Cybrd1 (duodenal cytochrome b) is not necessary for dietary iron absorption in mice. Blood. 2005b;106:2879–2883. doi: 10.1182/blood-2005-02-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase VH. The VHL tumor suppressor in development and disease: functional studies in mice by conditional gene targeting. Semin Cell Dev Biol. 2005;16:564–574. doi: 10.1016/j.semcdb.2005.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase VH, Glickman JN, Socolovsky M, Jaenisch R. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc Natl Acad Sci U S A. 2001;98:1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert N, Hentze MW. Previously uncharacterized isoforms of divalent metal transporter (DMT)-1: implications for regulation and cellular function. Proc Natl Acad Sci U S A. 2002;99:12345–12350. doi: 10.1073/pnas.192423399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Chen C, Satoh J, Yim S, Gonzalez FJ. Dietary phytochemicals regulate whole-body CYP1A1 expression through an arylhydrocarbon receptor nuclear translocator-dependent system in gut. J Clin Invest. 2007;117:1940–1950. doi: 10.1172/JCI31647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, Maxwell PH, Pugh CW, Ratcliffe PJ. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Kapitsinou PP, Haase VH. The VHL tumor suppressor and HIF: insights from genetic studies in mice. Cell Death Differ. 2008;15:650–659. doi: 10.1038/sj.cdd.4402313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latunde-Dada GO, Simpson RJ, McKie AT. Duodenal cytochrome B expression stimulates iron uptake by human intestinal epithelial cells. J Nutr. 2008;138:991–995. doi: 10.1093/jn/138.6.991. [DOI] [PubMed] [Google Scholar]
- Lee DW, Andersen JK. Role of HIF-1 in iron regulation: potential therapeutic strategy for neurodegenerative disorders. Curr Mol Med. 2006;6:883–893. doi: 10.2174/156652406779010849. [DOI] [PubMed] [Google Scholar]
- Levy JE, Montross LK, Andrews NC. Genes that modify the hemochromatosis phenotype in mice. J Clin Invest. 2000;105:1209–1216. doi: 10.1172/JCI9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie B, Garrick MD. Iron Imports. II. Iron uptake at the apical membrane in the intestine. Am J Physiol Gastrointest Liver Physiol. 2005;289:G981–G986. doi: 10.1152/ajpgi.00363.2005. [DOI] [PubMed] [Google Scholar]
- McKie AT, Barrow D, Latunde-Dada GO, Rolfs A, Sager G, Mudaly E, Mudaly M, Richardson C, Barlow D, Bomford A, Peters TJ, Raja KB, Shirali S, Hediger MA, Farzaneh F, Simpson RJ. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science. 2001;291:1755–1759. doi: 10.1126/science.1057206. [DOI] [PubMed] [Google Scholar]
- McKie AT, Marciani P, Rolfs A, Brennan K, Wehr K, Barrow D, Miret S, Bomford A, Peters TJ, Farzaneh F, Hediger MA, Hentze MW, Simpson RJ. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell. 2000;5:299–309. doi: 10.1016/s1097-2765(00)80425-6. [DOI] [PubMed] [Google Scholar]
- Mendel GA. Studies on iron absorption. I. The relationships between the rate of erythropoiesis, hypoxia and iron absorption. Blood. 1961;18:727–736. [PubMed] [Google Scholar]
- Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- Nicolas G, Bennoun M, Devaux I, Beaumont C, Grandchamp B, Kahn A, Vaulont S. Lack of hepcidin gene expression and severe tissue iron overload in upstream stimulatory factor 2 (USF2) knockout mice. Proc Natl Acad Sci U S A. 2001;98:8780–8785. doi: 10.1073/pnas.151179498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas G, Bennoun M, Porteu A, Mativet S, Beaumont C, Grandchamp B, Sirito M, Sawadogo M, Kahn A, Vaulont S. Severe iron deficiency anemia in transgenic mice expressing liver hepcidin. Proc Natl Acad Sci U S A. 2002a;99:4596–4601. doi: 10.1073/pnas.072632499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas G, Chauvet C, Viatte L, Danan JL, Bigard X, Devaux I, Beaumont C, Kahn A, Vaulont S. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest. 2002b;110:1037–1044. doi: 10.1172/JCI15686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantopoulos K. Iron metabolism and the IRE/IRP regulatory system: an update. Ann N Y Acad Sci. 2004;1012:1–13. doi: 10.1196/annals.1306.001. [DOI] [PubMed] [Google Scholar]
- Peyssonnaux C, Nizet V, Johnson RS. Role of the hypoxia inducible factors HIF in iron metabolism. Cell Cycle. 2008;7:28–32. doi: 10.4161/cc.7.1.5145. [DOI] [PubMed] [Google Scholar]
- Peyssonnaux C, Zinkernagel AS, Schuepbach RA, Rankin E, Vaulont S, Haase VH, Nizet V, Johnson RS. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J Clin Invest. 2007;117:1926–1932. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loreal O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811–7819. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, Shelton KD, Lindner J, Cherrington AD, Magnuson MA. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-pecific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9:677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- Rankin EB, Biju MP, Liu Q, Unger TL, Rha J, Johnson RS, Simon MC, Keith B, Haase VH. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest. 2007;117:1068–1077. doi: 10.1172/JCI30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynafarje C, Ramos J. Influence of altitude changes on intestinal iron absorption. J Lab Clin Med. 1961;57:848–855. doi: 10.21236/ad0268226. [DOI] [PubMed] [Google Scholar]
- Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah YM, Ito S, Morimura K, Chen C, Yim SH, Haase VH, Gonzalez FJ. Hypoxia-inducible factor augments experimental colitis through an MIF-dependent inflammatory signaling cascade. Gastroenterology. 2008;134:2036–2048. doi: 10.1053/j.gastro.2008.03.009. 2048 e2031–e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchernitchko D, Bourgeois M, Martin ME, Beaumont C. Expression of the two mRNA isoforms of the iron transporter Nramp2/DMTI in mice and function of the iron responsive element. Biochem J. 2002;363:449–455. doi: 10.1042/0264-6021:3630449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- Tomita S, Sinal CJ, Yim SH, Gonzalez FJ. Conditional disruption of the aryl hydrocarbon receptor nuclear translocator (Arnt) gene leads to loss of target gene induction by the aryl hydrocarbon receptor and hypoxia-inducible factor 1alpha. Mol Endocrinol. 2000;14:1674–1681. doi: 10.1210/mend.14.10.0533. [DOI] [PubMed] [Google Scholar]
- Tomita S, Ueno M, Sakamoto M, Kitahama Y, Ueki M, Maekawa N, Sakamoto H, Gassmann M, Kageyama R, Ueda N, Gonzalez FJ, Takahama Y. Defective brain development in mice lacking the Hif-1alpha gene in neural cells. Mol Cell Biol. 2003;23:6739–6749. doi: 10.1128/MCB.23.19.6739-6749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109:353–358. doi: 10.1182/blood-2006-07-033969. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J Biol Chem. 1993;268:21513–21518. [PubMed] [Google Scholar]
- Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, Cooperman S, Eckhaus M, Rouault T, Mishra L, Deng CX. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Wessling-Resnick M. A possible link between hepcidin and regulation of dietary iron absorption. Nutr Rev. 2002;60:371–374. doi: 10.1301/00296640260385900. [DOI] [PubMed] [Google Scholar]
- Xia Y, Babitt JL, Sidis Y, Chung RT, Lin HY. Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood. 2008;111:5195–5204. doi: 10.1182/blood-2007-09-111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XY, Tomatsu S, Fleming RE, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt EM, Ruddy DA, Prass CE, Schatzman RC, O'Neill R, Britton RS, Bacon BR, Sly WS. HFE gene knockout produces mouse model of hereditary hemochromatosis. Proc Natl Acad Sci U S A. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller H, Theurl I, Koch R, Kaser A, Weiss G. Mechanisms of iron mediated regulation of the duodenal iron transporters divalent metal transporter 1 and ferroportin 1. Blood Cells Mol Dis. 2002;29:488–497. doi: 10.1006/bcmd.2002.0587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.