Abstract
The interactions of proteins at surfaces are of great importance to biomineralizaton processes and to the development and function of biomaterials. Amelogenin is a unique biomineralization protein because it self-assembles to form supramolecular structures called “nanospheres”, spherical aggregates of monomers that are 20−60 nm in diameter. Although the nanosphere quaternary structure has been observed in solution, the quaternary structure of amelogenin adsorbed onto surfaces is also of great interest because the surface structure is critical to its function. We report studies of the adsorption of the amelogenin onto self-assembled monolayers (SAMs) with COOH and CH3 end group functionality and single crystal fluoroapatite (FAP). Dynamic light scattering (DLS) experiments showed that the solutions contained nanospheres and aggregates of nanospheres. Protein adsorption onto the various substrates was evidenced by null ellipsometry, X-ray photoelectron spectroscopy (XPS), and external reflectance Fourier transform infrared spectroscopy (ERFTIR). Although only nanospheres were observed in solution, ellipsometry and atomic force microscopy (AFM) indicated that the protein adsorbates were much smaller structures than the original nanospheres, from monomers to small oligomers in size. Monomer adsorption was promoted onto the CH3 surfaces, and small oligomer adsorption was promoted onto the COOH and FAP substrates. In some cases, remnants of the original nanospheres adsorbed as multilayers on top of the underlying subnanosphere layers. Although the small structures may be present in solution even though they are not detected by DLS, we also propose that amelogenin may adsorb by the “shedding” or disassembling of substructures from the nanospheres onto the substrates. This work suggests that amelogenin may have a range of possible quaternary structures that interact with surfaces.
Introduction
The behavior of proteins at interfaces is of great importance to a number of research areas and applications including biomineralization processes, the development of implants and medical devices, tissue engineering, biofouling, biosensors, and protein microarrays.1-3 Proteins interact at biomineral interfaces to control the nucleation and growth of hard tissues such as shell, tooth, and bone.4 Protein adsorption onto man-made surfaces has an impact on the biocompatibility of implants and the function of biomedical devices.5 Protein physisorption is controlled by electrostatic, hydrophobic, and/or hydrogen bonding interactions between surface sites and the protein.1 These interactions may lead to time dependent conformational changes in the protein, leading to changes in the tertiary or secondary structure.6 Adsorption behavior is often described by Langmuir and Freudlich isotherms with adsorption increasing with concentration until a saturation limit is obtained.7
Although most studies of proteins onto surfaces involve globular proteins that exist as monomers in solution, there is less research on the adsorption of proteins that form oligomers or other self-assembled quaternary structures in solution. An example of such a protein is amelogenin, a biomineralization protein that is involved in the formation of the high aspect ratio hydroxyapatite crystals found in tooth enamel.8 Amelogenin is highly hydrophobic for a mineralization protein, with a large hydrophobic central region and charged amino acids located primarily in the C-terminal domain. It is believed that the hydrophobic region promotes the assembly of protein monomers into supramolecular structures called “nanospheres”.9 Monomers may assemble into dimers, trimers, and hexamers which in turn assemble into the larger nanospheres.10 The nanospheres are typically 20−60 nm in diameter, with sizes and distributions in vitro depending on the solution pH, temperature, protein concentration, and buffer type.9,11 Smaller structures such as dimers have also been observed but only in acidic solutions and less polar solvents such as 60% acetonitrile in water.10,11 Nanosphere-like structures have also been observed in vivo, within growing enamel.12,13
The function of amelogenin is not completely understood, but the protein may have roles in nucleation,14 growth,15 control of crystal size and habit,16,17 and moderating crystal–crystal interactions.18 Research has shown that amelogenin can adsorb onto hydroxyapatite surfaces, consistent with the premise that amelogenin's role involves interactions with surfaces.19,20 Nanosphere-like structures have been observed on enamel and fluoroapatite surfaces.19,21 Since the function of amelogenin depends on its interactions with surfaces, the structure of amelogenin at interfaces is of great importance. The surface quaternary structure is of special interest because of the unique nanosphere quaternary structure observed in solution.
We report studies of the adsorption of amelogenin onto self-assembling monolayers (SAMs) containing COOH and CH3 functionality, and single crystal fluoroapatite (FAP). The SAM surfaces have highly controlled structures and chemistries and make good surfaces for fundamental studies of how proteins behave at interfaces.22 The FAP surface is of interest as a model biological surface, with well-known crystal faces. Our goals were to understand the quaternary structure of the protein in solution and compare it to the quaternary structure of the protein adsorbed onto the surfaces. The quaternary structure of the protein on substrates was studied using atomic force microscopy (AFM), aided by the use of molecularly smooth SAMs on gold on mica and polished single crystal FAP. We found that although the solutions only contained nanospheres as evidenced by dynamic light scattering (DLS), much smaller structures adsorbed onto the substrates, from monomer to hexamer in thickness.
Experimental Procedures
Self-Assembled Monolayer Formation
N-Hexadecanethiol (Aldrich, 92%) was purified by vacuum distillation. 16-Mercaptohexadecanoic acid was obtained from Asemblon, Inc. (Seattle, WA, 99%) and used as received. For ellipsometry, contact angle, FTIR, and XPS experiments, gold was deposited onto (100) silicon wafers or 20 mm diameter glass coverslips using 99.999% gold in a DC magnetron sputtering chamber at 10−8 Torr base pressure. An ion gun was used to clean the substrates. The deposited gold was polycrystalline with 100 nm grain sizes and (111) orientation. Films of 200 nm gold were deposited onto a 10 nm titanium or chromium adhesion layer. For AFM experiments requiring large, atomically smooth single crystal terraces, mica substrates with deposited gold were obtained from Structure Probe Inc. (West Chester, PA). The gold was freshly deposited, hydrogen annealed, and sealed with argon in glass containers. The substrates were immersed into 1 mM solutions of the thiols in absolute ethanol for at least 24 h. The CH3 SAMs were cleaned in ethanol, and the COOH SAMs were cleaned in ethanol acidified with hydrochloric acid to pH 2 to remove multilayer adsorbates and then thoroughly rinsed in ethanol. The SAMs were characterized by contact angle goniometry and ellipsometry before use. Advancing contact angles were ∼20 and 110° for the COOH and CH3 surfaces, respectively. Thicknesses determined by null ellipsometry were 22 and 20 Å, respectively.
Fluoroapatite Single Crystals
Single crystals of fluoroapatite were obtained from Excalibur Mineral Company (Peekskill, NY). The crystals were hexagonal with well defined prismatic faces ((100) or (110)) and were diamond polished on these faces. The crystals were cleaned in chloroform, ethanol, and by UV-ozone exposure (Boekel Industries). The XPS data indicated that the crystals had fluorine substitution resulting in a molecular formula of Ca10(PO4)6F2.
Protein Adsorption
Amelogenin was recombinantly synthesized as the histidine tagged rp(H)M180 as described previously.14 The expression vector for rp(H)M180 was kindly provided by Dr. M. L. Snead. Several experiments were also done using the native mouse amelogenin, rM179, kindly provided by Dr. J. Simmer and prepared as described previously.23 The protein was prepared by dissolution in 0.01 M HCl and then dissolved in buffer solutions (50 mM NaCl) and adjusted to pH 8. Several solutions contained 25 mM Tris · HCl buffer with no difference in adsorption behavior. The concentrations of the protein solutions were determined by UV–visible spectroscopy calibrated by amino acid analysis (AAA Services Laboratory, Damascus, OR). The substrates were placed into protein solutions (2−5 mL) for time periods from several minutes to 17 h. There was no change in pH in the solutions over the time course of the experiment. The substrates were removed, rinsed with ∼12 mL of water, and then dried in a stream of nitrogen. More extensive rinsing did not significantly change the thickness of the protein adsorbates. The work in this paper represented adsorption studies on over 100 samples and performed with several batches of protein and a number of researchers. Experiments were replicated at least 3 times.
Dynamic Light Scattering
DLS measurements on the protein solutions were obtained using a Brookhaven Instruments 90 Plus equipped with a 657 nm 35 mW laser. Time dependent fluctuations in the scattered intensity were measured using a Bl-APD digital correlator. Protein solutions were analyzed in triplicate using a 90° scattering angle at 25.0 °C. The buffer solutions were run through 0.2 and 0.02 μm filters and were also analyzed by DLS. Standard NIST traceable polystyrene 22 nm ± 1.8 nm latex standards and a blank, 0.02 μm filtered DI ultrapure water (VWR) were also run as standards. The autocorrelation functions were deconvoluted to obtain size distributions using both the non-negatively constrained least-squares fit (multiple pass NNLS) and the regularized LaPlace inversion (Contin) algorithms. Since the distributions were multimodal, the intensity weighted size distributions obtained from the NNLS algorithms were presented.
Contact Angle Goniometry
Contact angle measurements of water with the SAM surfaces were made using a Rame–Hart goniometer equipped with an environmental chamber. Water was placed into the chamber to form a saturated vapor. Reported contact angles were advancing.
Single Wavelength Ellipsometry
Single wavelength ellipsometer (SWE) measurements were performed on a Rudolph Autoel II null ellipsometer using a wavelength of 632.8 nm and angle of incidence of 70°. Ellipsometric constants were determined for the bare gold substrates soon after removal from the deposition chamber, and new ellipsometric constants were determined after SAM formation. A three-phase model (ambient/SAM/gold) was used to calculate the SAM thickness using a refractive index of 1.50 + 0i for the SAM. Protein thicknesses were determined similar to methods described previously.22,24,25 Ellipsometric constants were determined for the substrates after protein adsorption, and the protein thickness was determined using a refractive index of 1.50 + 0i and a four-phase model (ambient/protein/SAM/gold). The thickness was an equivalent thickness, the true thickness of the protein times the surface coverage. The refractive index of 1.50 is commonly used for proteins since it is an average over reported ranges of 1.4 to 1.6. Using refractive indices of 1.4 and 1.6 instead of 1.5 would result in 16% larger and 11% smaller thicknesses, respectively. We were not able to obtain good refractive index data using a refractometer, which we attribute to the complex nanosphere structure of the protein.
Atomic Force Microscopy (AFM)
Atomic force microscopy was conducted in air and solution using a Digital Instruments Nanoscope IIIa Multimode system (Veeco Metrology, Santa Barbara, CA) equipped with a Quadrex module. The AFM head was placed in an enclosure lined with Sonex acoustic dampening foam (Illbruck, Minneapolis, MN) and placed on a vibration isolation table (Micro-g, TMC, Peabody, MA). Substrates and rp(H)M180 coated substrates were imaged in tapping mode using TESP single beam silicon cantilevers having a nominal spring constant of ∼50 N/m (Veeco Probes, Camarillo, CA). Topography and phase images were simultaneously obtained at scan rates ranging from 1.5 to 3.5 Hz and at amplitude setpoints ranging from 0.6 to 0.8 of the cantilever excitation free amplitude. Several experiments were also done in solution using a Nanoscope solution cell. Protein solutions were introduced into the solution cell for 1 h, flushed with buffer without protein, and images were obtained.
Images from first scans were used as much as possible because the tip tended to become contaminated by the physisorbed protein over time. The images shown in this manuscript were unmodified except for tilt removal using a second-order planefit in some cases. Graphic images were obtained using the Nanoscope software (version 5.12r5) or by importing the raw data into ImageJ (rsb.info.nih.gov/ij/). The images were processed for brightness and contrast using Adobe Photoshop. The same height scale was used in comparing 1 μm scans of the bare substrates with substrates with adsorbates (usually 0−20 nm). A height scale of 0−15 nm was used for the 300 nm scans. Images of the intact nanospheres in solution were obtained by dropping solutions onto freshly cleaved mica and “fixing” the structures in place using a Karnovsky fixative (paraformaldehyde and glutaraldehyde mixtures in 0.1 M sodium cacodylate or 0.1 M sodium phosphate) similar to methods described previously.26
External Reflectance Infrared Spectroscopy
Infrared external reflection spectra of protein adsorbates on SAMs on gold were obtained using a nitrogen purged Nicolet Fourier transform instrument with external reflectance accessory and a narrowband HgCdTe detector cooled with liquid nitrogen. The incoming beam was p-polarized using a wire-grid polarizer and was focused onto the sample at an 80° angle of incidence. Spectra were obtained at 2 cm−1 resolution, and the interferograms were computed using triangular apodization and zero filling. Spectra were obtained with reference to a SAM on gold reference by coadding 1000 scans. The spectral intensities are reported as reflectivities in absorbance units, −log(R/R0), where R0 is the reflectivity of the reference sample.
X-ray Photoelectron Spectroscopy Characterization
Protein adsorbed to apatite crystals was characterized using XPS, since the substrates were not reflective enough to use ellipsometry. XPS measurements were performed using the Physical Electronics Quantum 2000 Scanning ESCA Microprobe at PNNL. This system uses a focused monochromatic Al Kα X-ray (1486.6 eV) source and a spherical section analyzer. The X-ray beam used was a 100 W, 100 mm diameter beam that was rastered over a 1.3 mm by 0.2 mm area on the sample. Wide scan data was collected using a pass energy of 117.4 eV. For the Ag3d5/2 line, these conditions produce a fwhm of 1.6 eV. High energy photoemission spectra were collected using a pass energy of 46.95. For the Ag3d5/2 line, these conditions produced a fwhm of 0.98 eV. The binding energy (BE) scale is calibrated using the Cu2p3/2 at 932.62 ± 0.05 eV and Au 4f at 83.96 ± 0.05 eV. Low energy electrons at ∼1 eV, 20 μA and low energy Ar+ ions were used to minimize charging. The spectral plots were corrected for charging using the C1s line at 285.0 eV. Thickness estimates of the protein overlayers were made using the method of Cumpson and Seah (Supporting Information).27
Results
Solution Characterization
DLS and AFM Studies
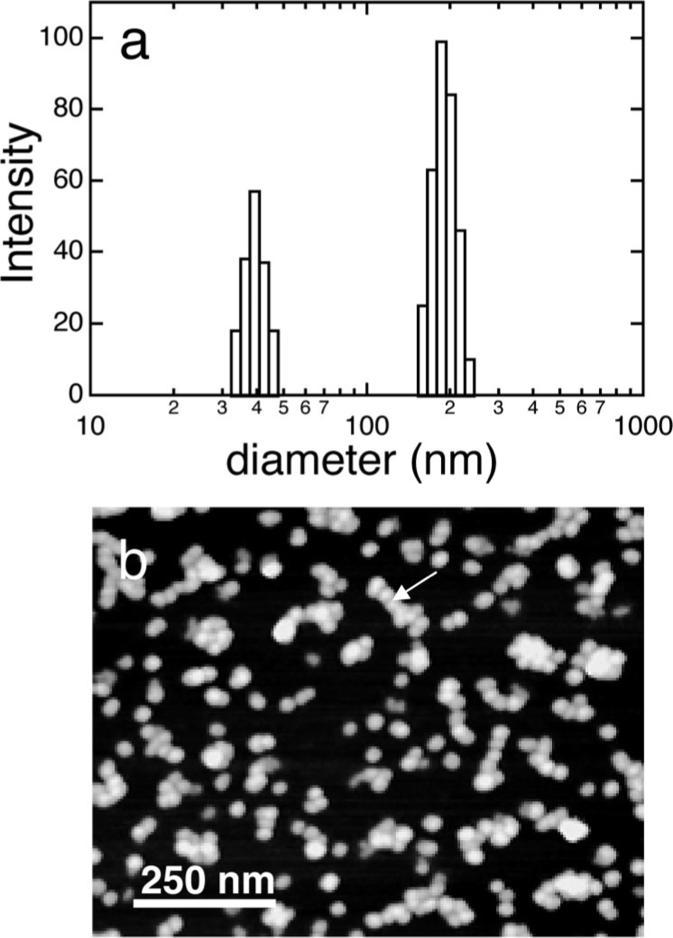
Protein solutions were prepared at a concentration of 134 μg/mL in solutions (50 mM NaCl) at pH 8. The protein solutions were characterized by dynamic light scattering and by AFM imaging of solution nanospheres immobilized onto mica substrates using a fixative. Figure 1a showed a plot of the intensity weighted size distributions of structures in the pH 8 solutions. The size distributions were bimodal, with one peak at 37.5 nm and one peak at 177 nm. The 37.5 nm peak was similar to a previously determined DLS size for rp(H)M180 that was attributed to the diameter of individual nanospheres.28 Figure 1b showed an AFM image of the solutions mixed with a fixative and dropped onto a mica substrate. We believe that the fixative method preserved the integrity of the nanospheres in solution as previous researchers have suggested.26 This image showed individual nanospheres as well as aggregates of nanospheres. More aggregates were observed in solutions that were several days old and solutions that did not contain Tris buffer. The AFM image suggested that the 177 nm peak in the DLS data represented clusters of individual nanospheres. Aggregates of nanospheres have also been seen by previous researchers.29,30 Nanosphere diameters determined by analysis of the AFM images were 24 ± 5 nm. If the 37.5 nm peak represented the size of the individual nanospheres, there were discrepancies between the DLS-determined diameter and the AFM-determined diameter. Other researchers have also observed differences between DLS-determined nanosphere sizes28 and sizes determined by AFM,26 TEM,10 and SAXS.29 We attribute the discrepancy to a small degree of clustering of nanospheres in the smaller 37.5 nm fraction. The DLS data showed that the nanospheres had a narrow size distribution with standard errors of ∼16% and that there was no evidence for the presence of smaller structures such as monomers and dimers.
Figure 1.
(a) DLS-determined size distribution of rp(H)M180 134 μg/mL solution at pH 8 showing peaks at 37.5 and 177 nm. (b) AFM image of protein solutions mixed with a fixative and dropped onto mica showing individual nanospheres and clusters of nanospheres (see arrow). The fixative method preserves the structures of the nanospheres in solution.
Adsorbate Characterization
Protein Thicknesses by Ellipsometry and XPS
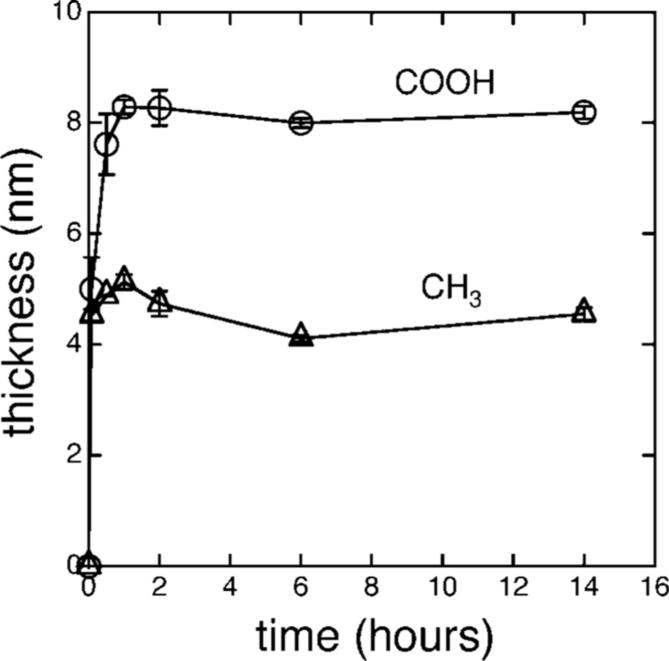
The rate of adsorption of amelogenin onto the COOH and CH3 SAMs was determined by obtaining ellipsometric thicknesses of the adsorbates as a function of time. Figure 2 showed an increase in the amount of protein adsorbed until a saturation limit was reached by 1 h of adsorption. At 14 hours, thicknesses were ∼4.5 nm for the CH3 SAM and ∼8.0 nm for the COOH SAM. Studies of protein adsorption as a function of concentration in solution showed saturation adsorption was reached at ∼20 μg/mL (Supporting Information Figure S1). XPS was used to estimate thicknesses of adsorbates onto the FAP substrates, since these substrates were not reflective enough for ellipsometry. Thickness values were obtained by the method of Cumpson and Seah27 which involved the reduction in intensity of the substrate peak, Ca2p, due to the presence of the protein overlayer. Thicknesses were ∼8.0 nm after 2 and 17 h of adsorption, indicating a saturation limit occurred over similar time periods as the SAM surfaces.
Figure 2.
Ellipsometry-determined thickness of rp(H)M180 adsorbed from 134 μg/mL solutions at pH 8 onto CH3 and COOH surfaces as a function of time.
AFM studies
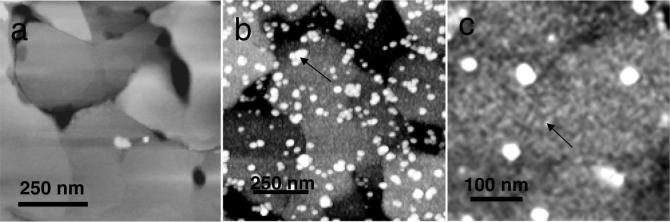
The quaternary structures of the amelogenin adsorbates were studied by obtaining AFM images of SAMs on gold on mica and molecularly polished FAP. Adsorbate structures obtained within a liquid cell were similar to the structures of the dried adsorbates. Dried state images were presented here because in some cases they were of higher resolution than the liquid state images. Figure 3a showed a scan of CH3 SAMs on gold on mica. The image showed large 500−800 nm atomically smooth gold terraces separated by 2−10 nm step edges. Amelogenin adsorbates on the CH3 SAMs (Figure 3b) consisted of large nanosphere-like structures at low coverage overlying much smaller structures at very high coverage. The smaller adsorbates were also seen more clearly in the higher magnification image shown in Figure 3c. Because the ellipsometric thicknesses of the protein on the CH3 substrates were typically 4.5−5.0 nm, we believe the small adsorbates represented the size of amelogenin monomers, thought to be 4.4 nm in diameter.10 These very small structures could not be accurately measured by AFM because it is well-known that AFM overestimates the diameter of three-dimensional structures that are smaller than the radius of curvature of the tip (10−20 nm) due to the tip broadening effect.31-33 We believe that the diameters of the larger nanospheres, however, were more accurate, because their sizes were larger than the radius of curvature of most AFM tips. Also, the height of soft structures such as proteins can be underestimated up to 6 times because of the nonlinear dynamic response of the oscillating cantilever.34-36
Figure 3.
AFM images: (a) 1 μm scan of CH3 SAMs on gold on mica showing large single crystal gold terraces; (b) 1 μm scan of rp(H)M180 adsorbates from 134 μg/mL solutions on CH3 SAMs after 14 h showing remnant nanospheres (arrow) overlying a high coverage of small adsorbates; (c) a magnified view of a similar image in part b showing a few remnant nanospheres and the monomer adsorbates at high coverage (arrow). Monomers are suggested by the ellipsometric thickness of 4.5 nm.
Although the diameters and heights of the small structures and the heights of the larger nanospheres were inaccurate, we found these measurements were useful for comparison purposes and could be calibrated with the ellipsometry and XPS measurements. A summary of the sizes of the small adsorbates obtained by AFM and ellipsometry and their suggested structures is shown in Table 1. The sizes of the larger nanosphere-like structures were 22.3 nm × 10.3 nm (diameter × height), smaller than the AFM measured sizes of the original nanosphere (24.3 nm × 12.5 nm). This indicated that the structures were remnants of the original nanospheres. The remnant nanospheres contributed about 0.34 nm to the ellipsometric thickness based on the observed area coverage of ∼1.6%. The smaller adsorbates averaged 9.3 nm × 0.8 nm. The AFM measurements, therefore, overestimated the monomer diameter by 2 times and underestimated the height by 5 times.
TABLE 1.
Sizes of Disassembled Adsorbates and Possible Structures
| surface | thicknessa (nm) | average AFM diameter (nm) | possible structure | estimated diameter of structure (nm) |
|---|---|---|---|---|
| CH3 | 4.0−5.0 | 9.3 | monomer | 4.4b |
| COOH | 7.5−8.0 | 14.1 | dimer to hexamer | 7.0−9.2c |
| FAP | 8.0 | 14.3 | dimer to hexamer | 7.0−9.2c |
From ellipsometry for COOH and CH3 and XPS for FAP.
From smallest structures observed in DLS (Du et al.).
From space filling models (Du et al.).
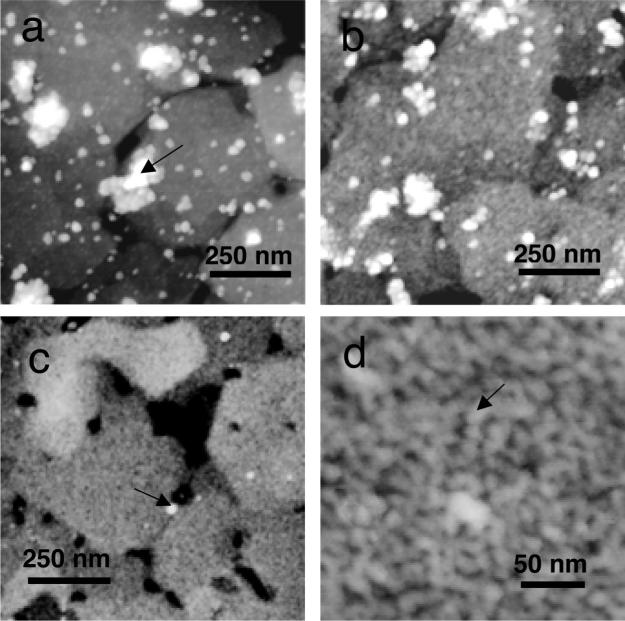
Protein adsorbate structures on COOH SAMs on gold on mica were examined as a function of adsorption time, as shown in Figure 4. Figure 4a showed adsorbates after 1 min of exposure to the protein solutions, revealing the presence of large clusters of nanosphere-like structures overlying a low coverage of smaller adsorbates. After 6 min of exposure, there was a much higher coverage of the smaller adsorbates, as shown in Figure 4b. By 17 h of adsorption, the remnant nanospheres were gone, leaving behind a layer of small adsorbates at high coverage (Figure 4c and d). The remnant nanospheres were also not observed in samples after 2 h of adsorption. The final ellipsometric thicknesses were typically 7.5−8.0 nm, suggesting that these adsorbates may be amelogenin dimers, trimers, or hexamers, structures calculated to be 7.0 to 9.2 nm in diameter10 (see Table 1). The small adsorbates were 14.1 nm in average AFM diameter, an overestimate by a factor of 2. There was a relatively wide distribution of AFM heights relative to the structures on CH3 with a significant population of structures from 1 to 1.5 nm (Supporting Information Figure S3). Although the ellipsometric thicknesses suggested that the adsorbates were small oligomers, the AFM diameter and height measurements suggested that there were some monomer-sized adsorbates mixed in with the oligomers. The monomer-sized structures, however, may also be monomer subunits of the oligomeric structures.
Figure 4.
AFM images: (a) 1 μm scan of rp(H)M180 adsorbates on COOH SAMs on gold on mica after 1 min of exposure to 134 μg/mL solutions showing large clusters of nanospheres (arrow) and individual remnant nanospheres overlying a low coverage of small adsorbates; (b) 1 μm scan of adsorbates after 6 min showing clusters of nanospheres, individual nanospheres, and a higher coverage of smaller adsorbates; (c) 1 μm scan after 17 h showing only a few remnant nanospheres (arrow) and a high coverage of oligomeric adsorbates; (d) 300 nm scan of the oligomeric adsorbates (arrow). Dimer to hexamer-sized oligomers are suggested by the ellipsometric thicknesses of 7.5−8 nm.
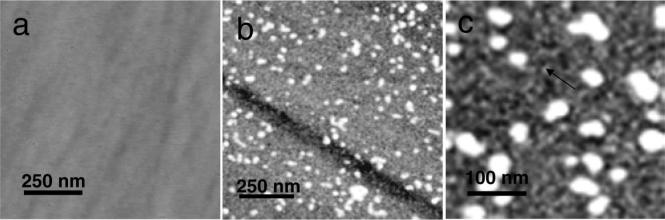
AFM imaging studies on the FAP substrates were shown in Figure 5. The FAP substrates were molecularly smooth, as shown in Figure 5a, with root-mean-square (rms) roughnesses of 0.2−0.3 nm. The adsorbates on FAP after 15 h showed the presence of a low coverage of remnant nanospheres overlying a high coverage of smaller adsorbates. The XPS-determined thicknesses were 8 nm, suggesting that the structures were small oligomers, similar to the COOH surfaces. The remnant nanospheres were 21.7 nm × 7.0 nm and would contribute about 1 nm to the XPS thickness based on the measured area coverage of 4.8% for the sample in Figure 5b and c. The AFM diameter of the small adsorbates was 14.25 nm, indicating that the AFM diameter overestimated the oligomer size by a factor of 2.
Figure 5.
AFM images: (a) 1 μm scan of bare FAP; (b) 1 μm scan of adsorbates on FAP from 134 μg/mL solutions after 14 h showing remnant nanospheres overlying dimer/trimer adsorbates (the depression is an artifact of the polishing process); (c) magnification of a similar image showing the oligomeric adsorbates (arrow). Oligomer-sized adsorbates are suggested by the XPS-determined thickness of 8 nm.
Several experiments were done using rM179, identical to rp(H)M180 but without the histidine tag. Similar adsorbate structures were found with rM179 compared to the rp(H)M180: monomer-sized adsorbates on the CH3 surfaces, and oligomer-sized adsorbates on the COOH and FAP surfaces (Supporting Information Figure S5).
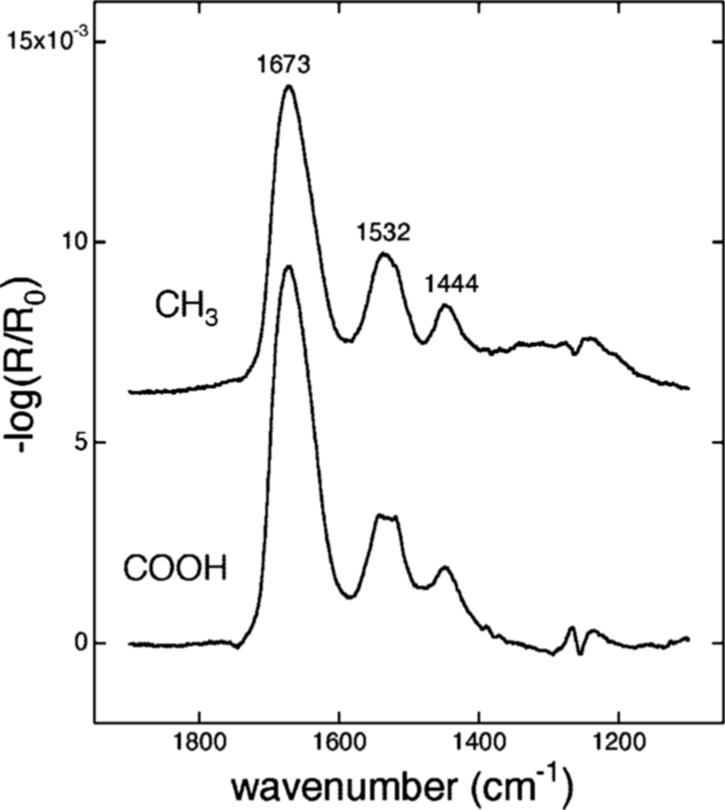
ERFTIR and XPS studies
Further evidence for protein adsorption onto the surfaces was obtained using ERFTIR and XPS. Figure 6 showed external reflectance Fourier transform infrared spectra of amelogenin adsorbates onto CH3 and COOH SAMs on gold. The peaks at 1673, 1532, 1444, and ∼1250 cm−1 were assigned to the amide I, amide II, CH2 bending, and amide III vibrational modes of the protein, respectively.37,38 Since the only significant component of the oscillating electric field at the metal surface is perpendicular to the surface, the peak intensities for each mode relate to the components of the transition dipole moment that are perpendicular to the substrate. There were differences in the relative peak intensities of the amide II and amide I peaks between the two surfaces which may reflect differences in the orientation or conformation of the protein with respect to the surface. The CH3 surface, however, had remnant nanosphere adsorbates which may complicate the interpretation. Modeling studies and further analysis of the spectra would be necessary in order to obtain orientational and conformational information.
Figure 6.
External reflectance Fourier transform infrared spectra of amelogenin adsorbates onto CH3 and COOH SAMs on gold. The peaks at 1673, 1532, and 1444 cm−1 were assigned to the amide I, amide II, and CH2 bending vibrational modes of the protein, respectively.
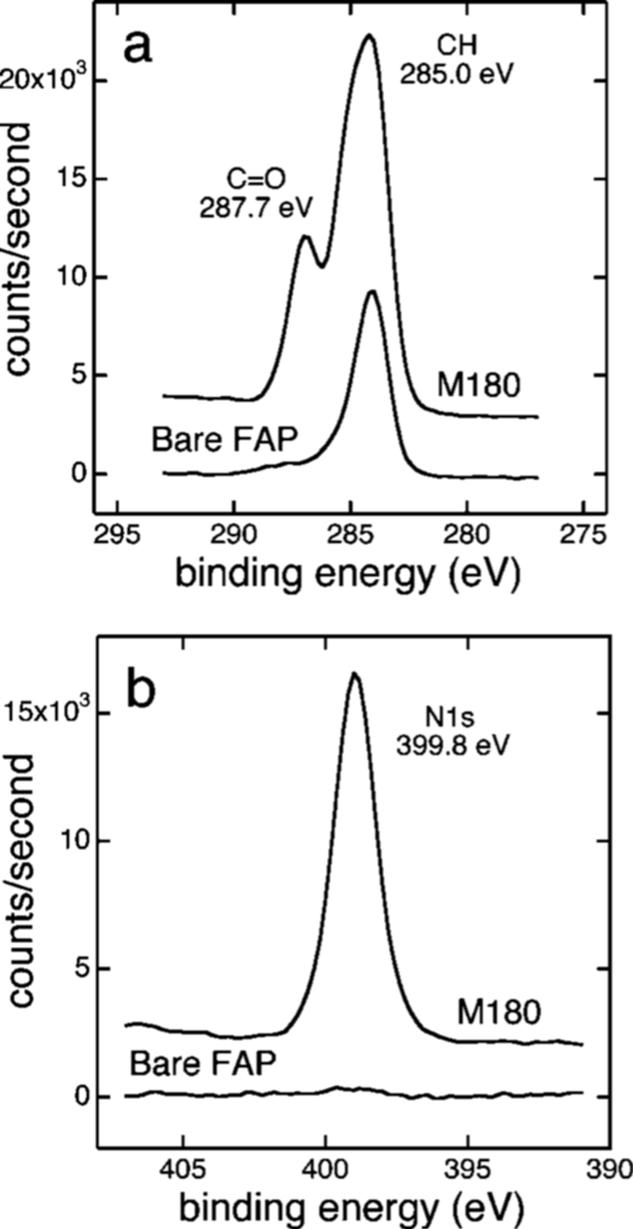
Protein adsorption onto the FAP surfaces was shown by the presence of nitrogen N1s peaks on the surfaces exposed to rp(H)M180 but not bare FAP (Figure 7a). Nitrogen was present in peptide bonds and amine groups in the side groups of a number of amino acid residues. Protein was also evidenced in Figure 7b by the presence of C=O bonds shown in the shoulder of the C1s peak at 287.7 eV. These bonds were present in peptide bonds and carboxylic acid groups in one aspartic acid and six glutamic acid residues and carbonyl groups in two asparagines and 25 glutamine residues. The Ca 2p and P 2p peaks were unique to the fluoroapatite and were attenuated upon adsorption of the protein (Supporting Information Figure S4). The relative intensities of the protein (from the N1s peak) and the substrate (from the Ca 2p3/2 peak) were used to estimate the thickness of the protein overlayer (Supporting Information).
Figure 7.
X-ray photoelectron spectroscopy of rp(H)M180 adsorbed to single crystal fluoroapatite (FAP) and bare FAP. Protein adsorption is evidenced by the N1s peak and the C=O peak from the peptide bond and a number of amino acid residues.
Discussion
Adsorption Mechanism
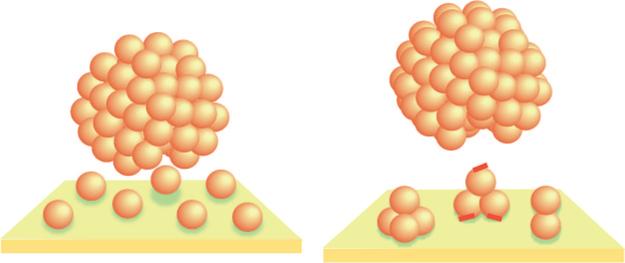
The AFM and ellipsometry studies revealed that amelogenin adsorbed onto the surfaces, not as intact nanospheres, but as much smaller adsorbates from monomer to small oligomer in size. The ERFTIR and XPS studies confirmed that the adsorbates were the amelogenin protein. Because the solutions only contained nanospheres of narrow size distribution, we think a reasonable explanation for this behavior is that the small structures “shed” or “disassemble” from the nanospheres onto the surface, as shown in the schematic in Figure 8. The presence of remnant nanospheres on the surfaces, structures smaller than the starting nanospheres in solution, also suggests that the nanospheres are losing material by disassembling onto the surfaces. If we used very small solution volumes and repeatedly exposed surfaces to the same solution, we observed smaller and smaller remnant nanospheres, indicating that the nanospheres were depleted by repeated interactions with the surfaces. Time dependent studies onto the COOH surfaces showed that nanospheres initially adsorbed onto the substrates and then disappeared over time as the underlying oligomer layer was built up. This suggests that the nanospheres were consumed by the disassembly process or that as the adsorbate layer was built up it eventually repelled the nanospheres, preventing them from adsorbing as multilayers.
Figure 8.
Schematic of nanospheres interacting with surfaces: (left) the disassembly of amelogenin monomers onto hydrophobic CH3 SAMs by the interaction of the hydrophobic domain with the hydrophobic surface; (right) the disassembly of oligomeric adsorbates onto the hydrophilic COOH and FAP surfaces by the interaction of the charged C-terminal domains on the outside of the oligomers (represented by the red tags) with the surfaces. Nanospheres are proposed to be composed of monomeric and oligomeric subunits after Du et al.
This disassembly behavior is very similar to the adsorption behavior of phospholipid vesicles onto substrates—phospholipids self-assemble into spherical vesicles in solution, but they disassemble or “fuse” onto surfaces to form planar lipid monolayers.39,40 The interactions between the phospholipids and the substrate are stronger than the interactions between the phospholipids holding together the vesicles. The phospholipid vesicles disassemble onto hydrophilic surfaces such as COOH SAMs, silica, and mica by electrostatic interactions between the polar headgroup of the lipid and the surfaces.41,42 Vesicles disassemble onto hydrophobic surfaces by interactions of the nonpolar alkyl chains of the lipid with the surface.43 Amelogenin disassembled onto both hydrophobic and hydrophilic surfaces similar to the way that vesicles can disassemble onto surfaces with a range of chemistries.
There was no evidence for the presence of small structures such as monomers to trimers in the protein solutions by DLS, so to the best of our ability to probe the quaternary structure in solution, we believe that our solutions only contained nanospheres. This is consistent with a large number of studies of amelogenin solutions at pH values in the range 6−8 using DLS,9,10,29,30,44 small-angle X-ray scattering (SAXS),29 and small angle neutron scattering (SANS).29 Small structures in the size range of monomers to dimers have been observed in solution but only in acidic solutions less than pH 429,30,45 and from less polar solvents such as 60% acetonitrile in water.10 It is possible that low concentrations of monomers and small oligomers exist in the pH 8 solutions and are not detected by DLS, SAXS, and SANS. The presence of nanospheres on the surfaces in the early stages of adsorption, adsorbed remnant nanospheres smaller than the starting solution nanospheres, and different structures on the different surfaces are pieces of evidence that are more consistent with a disassembly mechanism. Although we can't dismiss the possibility of smaller structures in the starting solutions, the experimental data we have at the present time points more strongly to a disassembly mechanism.
Effect of Surface Functionality
The amelogenin protein rp(H)M180 is a ∼21 kD protein with a large, highly hydrophobic central region and charged regions in the C-terminal domain and N-terminus. It is expected that the hydrophobic central region would promote adsorption onto the hydrophobic CH3 surfaces. It is well-known that many proteins have a high affinity for adsorption onto hydrophobic surfaces driven by hydrophobic interactions.1 A significant amount of adsorption also occurred onto the hydrophilic COOH and FAP surfaces. Both of these surfaces and the amelogenin protein would be expected to have a small net negative charge at pH 8. The COOH surface has a mixture of COOH and COO− groups (pK for ionization at ∼pH 6−7) and is net negatively charged, the FAP surface has a mixture of positive and negative sites but the overall charge at pH 8 would be expected to be neutral or slightly negative (isoelectric point at pH 4−7),46 and amelogenin has a net negative charge at pH 8 (isoelectric point at pH 6.7). Adsorption of amelogenin onto FAP and COOH suggests a role for electrostatic interactions via positively charged groups on the surface (for FAP) and/or positively charged groups on the protein. Previous researchers have suggested a role for positive sites in HAP promoting the adsorption of amelogenin.19 Advanced solid state NMR techniques have been developed to give molecular details of the orientation of the protein on hydroxyapatite surfaces by measuring the intersite spacing between 13C labeled residues on the protein and phosphate (31P) surface sites. These studies have shown that the C-terminal domain is located at the hydroxyapatite surface47 and that the domain lies down flat at the surface.48 The C-terminal domain contains a number of potential electrostatic binding sites—acidic aspartic acid and glutamic acid and basic lysine and arginine. Although this domain is located at the surface, dynamic studies have shown that parts of the C-terminus have a high degree of mobility. This suggests that other parts of the protein may also be involved in the adsorption interaction. Further studies are being done to study the interactions of the N-terminus and other parts of the protein with hydroxyapatite surfaces.
We found that different structures adsorbed onto the CH3 surface compared to the charged COOH and FAP surfaces. This was shown most strikingly in the thickness data in Figure 2 and Table 1. We propose that the CH3 surface is promoting the adsorption of monomers and the FAP and COOH surfaces are promoting the adsorption of dimer to hexamer-sized oligomers because the sizes of these structures estimated from space filling models10 are close to the ellipsometric and XPS-determined thicknesses observed, as summarized in Table 1. Another interpretation of the adsorbates on the CH3 surfaces is that they are small oligomers that are present at lower coverage, resulting in a lower ellipsometric thickness. The adsorbate coverage on the CH3 surfaces appeared similar to the COOH and FAP surfaces, however, and the average AFM sizes of the structures were smaller. There was a distribution of AFM diameters and heights for the structures on all of the surfaces, suggesting that there may be some oligomers mixed in with the monomers on the CH3 surfaces and some monomers mixed in with the oligomers on the COOH and FAP surfaces.
The various structures observed (such as monomers on the CH3 surface and dimers or trimers on the COOH and FAP surfaces) may break away from the nanospheres because these sizes promote attractive binding interactions between the protein cluster and the surface. TEM studies have shown that the nanospheres consist of spherical substructures that are 4−8 nm in diameter.10 This suggests that amelogenin monomers initially assemble into dimer, trimer, and/or hexamer subunits which in turn assemble into the larger nanosphere structure. It has been proposed that the monomers may assemble with the hydrophobic core oriented toward the center and hydrophilic C-terminal domain exposed at the surface of the oligomer,10 as shown schematically in Figure 8. The charged C-terminal domains of the oligomers may interact with the hydrophilic COOH and FAP surfaces, resulting in breaking away of the entire 8 nm oligomer, overcoming the interactions holding the oligomers together within the nanosphere. This mechanism is shown in the schematic in Figure 8. For the CH3 surface, interactions of the hydrophobic region of the protein with the hydrophobic surfaces may actually break up the oligomers, resulting in the adsorption of the ∼4 nm monomer components of the oligomer. Interactions with the CH3 surface, therefore, may overcome the hydrophobic interactions holding the monomers together within the larger oligomer. We suggest these disassembly mechanisms based on our current understanding of our research and the literature; however, further studies of the internal hierarchical structure of the nanosphere as well as the structure of the adsorbed monomers and oligomers will be necessary to better understand the disassembly process.
Several previous studies have used AFM to show nanosphere-like structures adsorbed onto enamel crystals,19 HAP nanoparticles,20 and FAP surfaces.21 Our studies suggest that these structures may be remnant nanospheres and may have been overlying smaller disassembled adsorbates. In some cases, the nanospheres were present at high coverage and would mask any underlying adsorbates.19,21 In addition, roughness of the surfaces (e.g., enamel19 and HAP nanoparticles20) may have made it difficult to resolve the smaller structures. We found that it was challenging to resolve the structures on rough surfaces like SAMs on polycrystalline gold. The ability to observe these structures was greatly enhanced by using SAMs on atomically smooth gold on mica and polished FAP crystals. The work by Habelitz et al.21 showed small adsorbate structures from 1.6 mg/mL amelogenin solutions on polished glass and FAP surfaces (Figure 1f) which may have been disassembled adsorbates. The underestimation of the protein heights by AFM also contributed to the difficulty in detecting these structures. It was necessary to be able to detect height relief in the range 0.5−2 nm in order to be able to see these structures by AFM. We also found it important to use additional techniques such as ellipsometry and XPS in order to obtain further evidence for the disassembled structures and more accurate size estimates.
Previous studies used optical waveguide light-mode spectroscopy (OWLS) to obtain thicknesses of amelogenin adsorbed to positively charged polyelectrolyte multilayers.49 They found that the adsorbed protein thicknesses ranged from 11 to 60 nm depending on the surface used and whether the samples were rinsed or not. These thickness values were interpreted to represent adsorbed nanospheres. Our results, however, suggest that the thicknesses may also be interpreted as the adsorption of disassembled adsorbates with overlayers of remnant nanospheres. Imaging studies would be necessary to give a definitive interpretation of the adsorbed protein structures in that study. It was found that absorbates were removed by rinsing the surfaces with buffer, suggesting that the nanosphere multilayers are weakly adsorbed and can be removed to a certain degree by rinsing. Amelogenin adsorbate structures, therefore, may be influenced by the amount of rinsing used. Without rinsing, a high coverage of nanosphere adsorbates may mask the underlying disassembled structures.
AFM Imaging
We found that the AFM-determined heights underestimated the actual heights of the monomeric and dimer/trimer structures in comparison to the ellipsometric and XPS-determined thicknesses and the known amelogenin sizes. This underestimation is well-known in the AFM measurement of soft structures such as oligonucleotides and proteins and has only recently been explained as a consequence of the nonlinear dynamic response of the oscillating cantilever.31,34,35 Under typical tapping mode conditions, biomolecule heights have been underestimated by 4−6 times.34,50 Even using optimized measurement conditions at low attractive interaction between tip and substrates, sizes of structures such as DNA and IgA were underestimated by at least 2 times.34,35 The underestimation makes it difficult to detect the subnanosphere adsorbate structures of amelogenin experimentally. The ability to detect these structures in this work was greatly aided by using substrates with atomic smoothness.
In some cases, the monomer to trimer adsorbates appeared indistinct and not clearly resolved. This type of appearance is very common for AFM images of physisorbed proteins because of the deformations induced by tip—protein forces during imaging.31,36 The protein moves while being imaged, resulting in blurriness. In addition, protein is picked up onto the tip during imaging which can affect the resolution. In several cases, the feature resolution appeared higher than others which we attribute to reductions in the degree to which the tip interacts with the surface.
In Vivo Structures
This work shows that amelogenin can exist at interfaces in in vitro studies as relatively small structures such as monomers and small oligomers. The protein has a range of quaternary structures, from nanospheres to monomers, depending on whether it is in solution or interacting with surfaces. Our work suggests that amelogenin may also have a range of quaternary structures in vivo. A number of studies have examined the structure of the organic matrix within developing enamel using TEM.13,51-53 Protein regions appeared as electron deficient negative images, while the surrounding regions appeared electron dense. Parts of the electron deficient regions appeared to be organized as 20 nm spherical structures which were suggested to be nanospheres. The nanospheres were organized in rows in between and parallel to the highly elongated developing enamel crystallites. In addition to spherical nanosphere-like structures, the TEM images in these studies also showed smaller electron deficient structures within the intercrystalline spaces in developing enamel. Many of these small structures were located at the surface of the rod-shaped enamel crystallites. Although it is not known if these structures are amelogenin or other hydrophobic protein, some of the structures had sizes consistent with the amelogenin monomer and oligomer. We suggest, therefore, that these structures may be amelogenin monomers and small oligomers. Although it is very challenging to conclusively determine the identify of a single protein within a tissue, it would be very interesting to take a second look at the small subnanosphere-sized electron deficient structures found in developing enamel and develop techniques to determine the identity of these proteins.
The nanosphere quaternary structure is thought to be very important in the development of highly organized enamel, as suggested by numerous in vivo studies. Transgenic mice containing amelogenin transgenes with the N-terminal or A-domain deleted showed alterations in the degree of organization and ordering of the enamal rods.51,52 The wild type animals had well preserved boundaries between the rod and interrod components of enamel, resulting in a woven morphology. The A-domain deficient mice, on the other hand, had an increase in interrod enamel and some degree of fusion between rod and interrod components. In vitro studies showed that removal of the A-domain inhibited the assembly of amelogenin into nanospheres, resulting in solutions composed of monomers.28 It was suggested that the removal of A domains of amelogenin disrupted the formation of uniform nanospheres which in turn affected the organization and structure of the resulting enamel rod and interrod crystals. Nanospheres, therefore, were proposed to function by promoting the organization of enamel crystals—controlling the spacing of the crystallites, preventing early crystal–crystal fusion, and preserving rod and interrod boundaries.13,51
Even though the A-domain deleted amelogenin did not self-assemble to form nanospheres in vitro,28 the A-domain deleted transgenic mice had enamel tissue containing long, rod-shaped enamel crystallites with the same high aspect ratio habit as crystallites in wild type mice.51 This suggests that although the nanosphere quaternary structure may be important for the organization and assembly of enamel crystallites, it may not be necessary for the growth of the elongated calcium phosphate crystals. Our work showed that amelogenin can adsorb onto calcium phosphate surfaces as small oligomers, suggesting that the protein may function in the monomer or small oligomer state in vivo by adsorbing onto enamel crystals, controlling crystal growth. It has been proposed that amelogenin preferentially adsorbs onto the b face of octacalcium phosphate, inhibiting growth of that face but favoring growth along the c-axis, leading to the unique high aspect ratio crystallites of enamel.54
We speculate, therefore, that amelogenin may have several different quaternary structures in vivo and these structures may have different biological functions. Amelogenin may function in the monomer and small oligomer state to control the growth and aspect ratio of the crystallites, while amelogenin in the nanosphere state may function by controlling the spacing and organization of the crystallites. Although these potential roles for different quaternary structures of amelogenin are largely speculative, we believe it is important to consider the possibility of a number of quaternary structures in vivo. It would be very interesting if amelogenin existed in a range of structures from monomer to nanosphere and if each quaternary structure had its own biological role. This would certainly make amelogenin a very unique biomineralization protein, one that can be a model for understanding relationships between biological structure and function.
Conclusions
We studied the adsorption behavior of amelogenin onto self-assembled surfaces with COOH and CH3 functionality and fluoroapatite surfaces. Solution quaternary structures were examined using DLS and AFM of fixed solutions, and the surface structures were studied using AFM imaging, XPS, ellipsometry, and ERFTIR. Although the solutions contained nanospheres and clusters of nanospheres, the surfaces had adsorbates that were much smaller structures than the original nanospheres, from monomers to small oligomers in size. In some cases, remnants of the original nanospheres adsorbed as multilayers on top of the underlying subnanosphere layers. We suggest that the small protein structures disassembled from the nanospheres in solution onto the surfaces, reversing the process of nanosphere assembly. Future work will be necessary to confirm this mechanism, understand the structure of the small adsorbates, and develop a more detailed understanding of the adsorption behavior onto HAP and FAP surfaces.
Acknowledgment
This work was supported by NIH-NIDCR Grant DE-015347. This research was performed at Pacific Northwest National Laboratory, operated by Battelle for the US-DOE. A portion of the research was performed in the EMSL, a national scientific user facility sponsored by the DOE-OBER at PNNL.
Footnotes
Supporting Information Available: Further experimental details, ellipsometry, AFM, and XPS data. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Norde W. Adv. Colloid Interface Sci. 1986;25:267. doi: 10.1016/0001-8686(86)80012-4. [DOI] [PubMed] [Google Scholar]
- 2.Brash JL, Horbett TA. Proteins at Interfaces: Physicochemical and Biochemical Studies. Anaheim, CA: 1987. Proteins at Interfaces: Current Issues and Future Prospects. [Google Scholar]
- 3.Norde W, Haynes CA. Proteins at Interfaces II: Fundamentals and Applications. San Diego, CA: 1995. Reversibility and the mechanism of protein adsorption. [Google Scholar]
- 4.Lowenstam HA, Weiner S. On Biomineralization. Oxford University Press; New York: 1989. [Google Scholar]
- 5.Horbett TA. Colloids Surf., B. 1994;2:225. [Google Scholar]
- 6.Kleijn M, Norde W. Heterogeneous Chem. Rev. 1995;2:157. [Google Scholar]
- 7.Brash JL, Horbett TA. Proteins at Interfaces II: Fundamentals and Applications. San Diego, CA: 1995. Proteins at Interfaces: An Overview. [Google Scholar]
- 8.Fincham AG, Moradian-Oldak J, Simmer JP. J. Struct. Biol. 1999;136:270. doi: 10.1006/jsbi.1999.4130. [DOI] [PubMed] [Google Scholar]
- 9.Moradian-Oldak J, Simmer JP, Lau EC, Sarte PE, Slavkin HC, Fincham AG. Biopolymers. 1994;34:1339. doi: 10.1002/bip.360341006. [DOI] [PubMed] [Google Scholar]
- 10.Du C, Falini G, Fermani S, Abbott C, Moradian-Oldak J. Science. 2005;307:1450. doi: 10.1126/science.1105675. [DOI] [PubMed] [Google Scholar]
- 11.Moradian-Oldak J, Tan J, Fincham AG. Biopolymers. 1998;46:225. doi: 10.1002/(SICI)1097-0282(19981005)46:4<225::AID-BIP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 12.Moradian-Oldak J, Goldberg M. Cells Tissues Organs. 2005;181:202. doi: 10.1159/000091382. [DOI] [PubMed] [Google Scholar]
- 13.Fincham AG, Moradian-Oldak J, Diekwisch T, Lyaruu DM, Wright JT, Jr., Slavkin HC. J. Struct. Biol. 1995;115:50. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- 14.Tarasevich BJ, Howard CJ, Larson JL, Snead ML, Simmer JP, Paine M, Shaw WJ. J. Cryst. Growth. 2007;304:407. doi: 10.1016/j.jcrysgro.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beniash E, Simmer JP, Margolis HC. J. Struct. Biol. 2005;149:182. doi: 10.1016/j.jsb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Iijima M, Moriwaki Y, Takagi T, Moradian-Oldak JJ. Cryst. Growth. 2001;222:615. [Google Scholar]
- 17.Wen HB, Moradian-Oldak J, Fincha AG. J. Dent. Res. 2000;79:1902. doi: 10.1177/00220345000790111501. [DOI] [PubMed] [Google Scholar]
- 18.Moradian-Oldak J, Tan J, Fincham AG. Biopolymers. 1998;46:225. doi: 10.1002/(SICI)1097-0282(19981005)46:4<225::AID-BIP4>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 19.Wallwork ML, Kirkham J, Zhang J, Smith DA, Brookes SJ, Shore RC, Wood SR, Ryu O, Robinson C. Langmuir. 2001;17:2508. [Google Scholar]
- 20.Bouropoulos N, Moradian-Oldak J. Calcif. Tissue Int. 2003;72:599. doi: 10.1007/s00223-002-1099-1. [DOI] [PubMed] [Google Scholar]
- 21.Habelitz S, Kullar A, Marshall SJ, DenBesten PK, Balooch M, Marshall GW, Li W. J. Dent. Res. 2004;83:698. doi: 10.1177/154405910408300908. [DOI] [PubMed] [Google Scholar]
- 22.Prime KL, G. MW. J. Am. Chem. Soc. 1993;115:10714. [Google Scholar]
- 23.Simmer JP, Lau EC, Hu CC, Aoba M, Lacey D, Nelson M, Zeichner-David ML, Snead ML, Slavkin HC, Fincham AG. Calcif. Tissue Int. 1994;54:312. doi: 10.1007/BF00295956. [DOI] [PubMed] [Google Scholar]
- 24.Lestelius M, Liedberg B, Tengvall P. Langmuir. 1997;13:5900. [Google Scholar]
- 25.Ortega-Vinuesa JL, Tengvall P, Lundstrom I. J. Colloid Interface Sci. 1998;207:228. doi: 10.1006/jcis.1998.5624. [DOI] [PubMed] [Google Scholar]
- 26.Wen HB, Fincham AG, Moradian-Oldak J. Matrix Biol. 2001;20:387. doi: 10.1016/s0945-053x(01)00144-5. [DOI] [PubMed] [Google Scholar]
- 27.Cumpson PJ, Seah MP. Surf. Interface Anal. 1997;25:430. [Google Scholar]
- 28.Moradian-Oldak J, Paine ML, Lei YP, Fincham AG, Snead ML. J. Struct. Biol. 2000;131:27. doi: 10.1006/jsbi.2000.4237. [DOI] [PubMed] [Google Scholar]
- 29.Aichmayer B, Margolis HC, Sigel R, Yamakoshi Y, Simmer JP, Fratzl P. J. Struct. Biol. 2005;151:239. doi: 10.1016/j.jsb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Petta V, Moradian-Oldak J, Yannopoulos SN, Bouropoulos N. Eur. J. Oral Sci. 2006;114:308. doi: 10.1111/j.1600-0722.2006.00325.x. [DOI] [PubMed] [Google Scholar]
- 31.Garcia R, Perez R. Surf. Sci. Rep. 2002;47:197. [Google Scholar]
- 32.Grabar KC, Brown KR, Keating CD, Stranick SJ, Tang SL, Natan MJ. Anal. Chem. 1997;69:471. doi: 10.1021/ac9605962. [DOI] [PubMed] [Google Scholar]
- 33.Yang D, Xiong Y, Guo Y, Da D, Lu W. J. Mater. Sci. 2001;36:263. [Google Scholar]
- 34.Round AN, Miles MJ. Nanotechnology. 2004;15:S176. [Google Scholar]
- 35.San Paulo A, Garcia R. Biophys. J. 2000;78(3):1599. doi: 10.1016/S0006-3495(00)76712-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.San Paulo A, Garcia R. Phys. ReV.B. 2002;66:041406(R). [Google Scholar]
- 37.Moradian-Oldak J, Du C, Falini G. Eur. J. Oral Sci. 2006;114:289. doi: 10.1111/j.1600-0722.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- 38.Renugopalakrishnan V, Strawich ES, Horowitz PM, Glimcher MJ. Biochemistry. 1986;25:4879. doi: 10.1021/bi00365a023. [DOI] [PubMed] [Google Scholar]
- 39.Plant AL. Langmuir. 1993;9:2764. [Google Scholar]
- 40.Ohki S, Doyle D, Flanagan TD, Hui SW, Mayhew E. Molecular mechanisms of membrane fusion. Plenum Press; New York: 1988. [Google Scholar]
- 41.Richter R, Mukhopadhyay A, Brisson A. Biophys. J. 2003;85:3035. doi: 10.1016/S0006-3495(03)74722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stelzle M, Weissmuller G, Sackmann E. J. Phys. Chem. 1993;97:2974. [Google Scholar]
- 43.Meuse CW, Niaura G, Lewis ML, Plant A. Langmuir. 1998;14:1504. [Google Scholar]
- 44.Moradian-Oldak J, Leung W, Fincham AG. J. Struct. Biol. 1998;122:320. doi: 10.1006/jsbi.1998.4008. [DOI] [PubMed] [Google Scholar]
- 45.Matsushima N, Yoshinobu I, Aoba T. J. Biochem. 1998;123:150. doi: 10.1093/oxfordjournals.jbchem.a021902. [DOI] [PubMed] [Google Scholar]
- 46.Bell LC, Posner AM, Quirk JP. J. Colloid Interface Sci. 1973;42:250. [Google Scholar]
- 47.Shaw WJ, Campbell AA, Paine ML, Snead ML. J. Biol. Chem. 2004;279:40263. doi: 10.1074/jbc.C400322200. [DOI] [PubMed] [Google Scholar]
- 48.Shaw WJ, Ferris K, Tarasevich B, Larson JL. Biophys. J. 2008;94:3247. doi: 10.1529/biophysj.107.119636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gergely C, Szalontai B, Moradian-Oldak J, Cuisinier FJG. Biomacromolecules. 2007;8:2228. doi: 10.1021/bm070088+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marchant RE, Barb MD, Shainoff JR, Eppell SJ, Wilson DL, Siedleck CA. Thromb. Haemostasis. 1997;77:1048. [PubMed] [Google Scholar]
- 51.Paine ML, White SN, Luo W, Fong HK, Sarikaya M, Snead ML. Matrix Biol. 2001;20:273. doi: 10.1016/s0945-053x(01)00153-6. [DOI] [PubMed] [Google Scholar]
- 52.Paine ML, Zhu D-H, Luo W, Bringas J,P, Goldberg M, White SN, Lei Y-P, Sarikaya M, Fong HK, Snead ML. J. Struct. Biol. 2000;132:191. doi: 10.1006/jsbi.2000.4324. [DOI] [PubMed] [Google Scholar]
- 53.Robinson C, Fuchs P, Weatherell JA. J. Cryst. Growth. 1981;53:160. [Google Scholar]
- 54.Iijima M, Moriwaki Y, Takagi T, Moradian-Oldak J. J. Cryst. Growth. 2001;222:615. [Google Scholar]