Abstract
Enteropathogenic Escherichia coli (EPEC) expresses a type III secretion system (T3SS) required for pathogenesis. Regulation of the genes encoding the T3SS is complex; two major regulators control transcription, the silencer H-NS, and the related H-NS-like protein Ler. Our laboratory is interested in understanding the molecular differences that distinguish the anti-silencer Ler from H-NS, and how Ler differentially regulates EPEC virulence genes. Here, we demonstrate that mutated Ler proteins either containing H-NS α-helices 1 and 2, missing from Ler, or truncated for the 11 aa C-terminal extension compared with the related H-NS protein, did not appreciably alter Ler function. In contrast, mutating the proline at position 92 of Ler, in the conserved C-terminal DNA binding motif, eliminated Ler activity. Inserting 11 H-NS-specific amino acids, 11 alanines or 6 alanines into the Ler linker severely impaired the ability of Ler to increase LEE5 transcription. To extend our analysis, we constructed six chimeric proteins containing the N terminus, linker region or C terminus of Ler in different combinations with the complementary domains of H-NS, and monitored their in vivo activities. Replacing the Ler linker domain with that of H-NS, or replacing the Ler C-terminal, DNA binding domain with that of H-NS eliminated the ability of Ler to increase transcription at the LEE5 promoter. Thus, the linker and C-terminal domains of Ler and H-NS are not functionally equivalent. Conversely, replacing the H-NS linker region with that of Ler caused increased transcription at LEE5 in a strain deleted for hns. In summary, the interdomain linker specific to Ler is necessary for anti-silencing activity in EPEC.
INTRODUCTION
In order for bacterial pathogens to cause disease, they must possess and properly express virulence factors. For the diarrhoeal pathogens enteropathogenic and enterohaemorrhagic Escherichia coli (EPEC and EHEC), the causative agents of acute diarrhoea in infants and haemorrhagic colitis, respectively, a type III secretion system (T3SS) delivers effector proteins into the host cell cytosol via an assembled molecular syringe. The alteration of host cell signalling events leads to the formation of hallmark attaching and effacing (AE) intestinal lesions. EPEC and EHEC bacteria acquired the genes encoding their respective T3SSs by horizontal gene transfer, and by multiple, independent events placing these genes at different tRNA loci, multiple lineages of both pathotypes have arisen (Rumer et al., 2003).
The extent to which EPEC and EHEC bacteria affect their host via their T3SS is profound. At least 39 proteins are injected into the host cell cytosol through this apparatus (Garmendia et al., 2005; Tobe et al., 2006). En route to causing diarrhoea, these effector molecules alter host cell signalling events and loosen tight junctions, leading to cytoskeletal rearrangment and the hallmark pedestals associated with intimate adherence to the intestinal epithelial cell membrane (for review, see Clarke et al., 2003; Kaper et al., 2004). Several of these effector proteins are found within the locus of enterocyte effacement (LEE) encoding the T3SS, but the majority are encoded outside this locus, many within cryptic prophages (Garmendia et al., 2005; Tobe et al., 2006). Specifically, Tir, EspF, EspG, EspH, EspZ and Map are encoded within the LEE, but NleA, NleD, EspG2, EspJ and Cif, for example, are encoded outside the LEE.
Although the mechanism by which the effector molecules encoded outside the LEE are regulated remains mostly unexplained, much information exists regarding control of the LEE pathogenicity islands (PAIs) of EPEC and EHEC (for review, see Mellies et al., 2007a). Among a cadre of regulatory molecules and their perceived environmental inputs, two proteins, Ler and H-NS, have emerged as central in the coordinated expression of the T3SS of these two diarrhoeal pathogens. From genetic and biochemical evidence, H-NS has been shown to silence multiple operons of the EPEC LEE, including LEE1 (encoding Ler), LEE2, LEE3 and LEE5 (Bustamante et al., 2001; Haack et al., 2003; Umanski et al., 2002). The nucleoid-associated H-NS protein is thought to bind non-specifically to AT-rich intrinsically curved DNA and in most cases compacts the DNA. However, a specific DNA binding sequence was recently proposed for H-NS (Bouffartigues et al., 2007). Ler, a member of the H-NS family of proteins, relieves transcriptional silencing of the EPEC LEE2, LEE3, LEE4 and LEE5 operons as well as espG, escD and map of the LEE (Bustamante et al., 2001; Elliott et al., 2000; Haack et al., 2003; Li et al., 2004; Mellies et al., 1999; Sánchez-SanMartín et al., 2001; Sperandio et al., 2000; Umanski et al., 2002).
The importance of Ler to EPEC and EHEC pathogenesis is illustrated by the numerous inputs that control LEE1 transcription. Quorum-sensing signals (Sperandio et al., 1999, 2001), integration host factor (IHF) (Friedberg et al., 1999), Fis (Goldberg et al., 2001), BipA (Grant et al., 2003), PerC and PerC-like molecules (Gomez-Duarte & Kaper, 1995; Iyoda & Watanabe, 2004; Porter et al., 2005), GrlRA (Barba et al., 2005; Deng et al., 2004; Sharp & Sperandio, 2007) and GadX (Shin et al., 2001), as well as a number of environmental cues, including temperature, pH, iron, ammonium and calcium, are known to control Ler expression in vivo either directly or indirectly (Beltrametti et al., 1999; Ide et al., 2003; Kenny & Finlay, 1995; Kenny et al., 1997). Thus, multiple regulatory proteins and multiple environmental signals control the AE phenotype of EPEC and EHEC bacteria through the regulation of Ler.
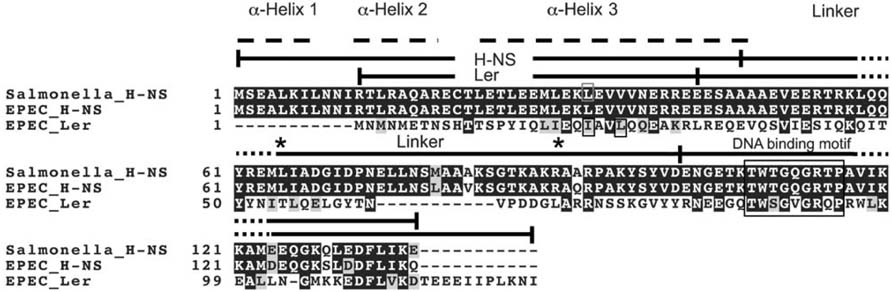
To date, Ler is the only known H-NS-like molecule that relieves transcriptional silencing and our laboratory seeks to understand the mechanism by which Ler functions. Data indicate that Ler stimulates transcriptional activity by disrupting H-NS-dependent nucleoprotein complexes (Bustamante et al., 2001; Haack et al., 2003; Sperandio et al., 2000; Umanski et al., 2002). Ler and the related H-NS molecules of E. coli and Salmonella are ~15 kDa proteins; Ler and H-NS of Salmonella share 24% amino acid identity and 44% amino acid similarity, mostly over their C-terminal DNA-binding domains (Sperandio et al., 2000). H-NS possesses an N-terminal, α-helical domain, which contains a coiled-coil motif involved in dimerization (Bloch et al., 2003). Substitution of the leucine with a proline at position 33 in α-helix 3 eliminates DNA binding and dimer formation of H-NS (Ueguchi et al., 1997). Similarly, I20R and L23R mutations within the predicted coiled-coil region of Ler eliminate the ability of this protein to bind to LEE2 regulatory DNA as well as to relieve transcriptional silencing at this locus (Sperandio et al., 2000).
Though some molecular similarities between H-NS and Ler have been experimentally established, such as the shared coiled-coil domain in their N termini (Sperandio et al., 2000), the precise molecular mechanism of Ler action remains unknown. In this study we investigated the molecular aspects of Ler that distinguish this protein from the related silencer H-NS and enable Ler to increase, as opposed to silence, transcription. We demonstrate that the interdomain linker region of Ler, connecting the N-terminal, α-helical to the C-terminal, DNA binding domain, is necessary for proper function.
METHODS
Bacterial strains, plasmids and phages
The plasmids, strains and phages used in this study are listed in Table 1. Strains were grown at 37 °C with aeration in Luria–Bertani (LB) medium supplemented with the appropriate antibiotic at the following concentrations: chloramphenicol (30 µg ml−1), kanamycin (50 µg ml−1), tetracycline (15 µg ml−1) or ampicillin (100 µg ml−1). Molecular manipulations of plasmid DNA were performed using standard methods.
Table 1.
Bacterial strains, plasmids and phages used in this study
| Strain, plasmid or phage | Genotype or description | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | supE44 Δ(argF-lac)U169 (Φ80dlacΔ(Z)M15) deoR hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Laboratory stock |
| MC4100 | araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | Casadaban (1976) |
| HN4104 | MC4100 Δ(hns tdk adhE oppABCD)118 zch-506 : : Tn10 | Colonna et al. (1995) |
| KH4100 | MC4100 ara+ | This study |
| KH4105 | KH4100 Φ LEE5–lacZ (−303 to +172) | This study |
| KH4106 | KH4100 Φ proU–lacZ (−398 to +404) | This study |
| KH4115 | KH4105 Δ(hns tdk adhE oppABCD)118 zch-506 : : Tn10 | This study |
| KH4116 | KH4106 Δ(hns tdk adhE oppABCD)118 zch-506 : : Tn10 | This study |
| KH4102 | KH4100 Ф LEE2–lacZ (−218 to +670) | This study |
| KH4112 | KH4102 Δ(hns tdk adhE oppABCD)118 zch-506 : : Tn10 | This study |
| KH4103 | KH4100 Ф LEE3-lacZ (−434 to +262) | This study |
| KH4113 | KH4103 Δ(hns tdk adhE oppABCD)118 zch-506 : : Tn10 | This study |
| SE796 | E2348/69 Δler : : aphA3 Kmr | Elliott et al. (2000) |
| Plasmids | ||
| pRS551 | Promoterless lacZ reporter fusion vector | Simons et al. (1987) |
| pKMTIR3 | LEE5–lacZ (−303 to +172) in pRS551 | Haack et al. (2003) |
| pKHpro1 | proU–lacZ (−398 to +404) in pRS551 | This study |
| pJLM166 | LEE2–lacZ (−218 to +670) in pRS551 | Mellies et al. (1999) |
| pJLM172 | LEE3–lacZ (−434 to +262) in pRS551 | Mellies et al. (1999) |
| pCR2.1 TOPO | Cloning vector | Invitrogen |
| pBAD33 | Arabinose-inducible expression vector Cmr | Guzman et al. (1995) |
| pKLHB01 | wt ler in pBAD33 | This study |
| pKHB1123 | wt hns in pBAD33 | This study |
| pMHB1 | Mutated Ler protein with α-helices 1 and 2 (MSEALKILNNIRTLRAQAREC) of H-NS fused to the N terminus in pBAD33 | This study |
| pKLHB13 | Mutated Ler protein with 11 aa, C-terminal extension TEEEIIPLKNI deleted in pBAD33 | This study |
| pLERP92A | Mutated Ler protein with alanine substitution for proline at position 92 (position 115 of H-NS) in pBAD33 | This study |
| pFL08 | pKLHB01 with residues ELLNSLAAVKS inserted at Ler position 67 in pBAD33 | This study |
| pFL09 | pKLHB01 with six alanines inserted at Ler position 67 in pBAD33 | |
| pFL10 | pKLHB01 with 11 alanines inserted at Ler position 67 in pBAD33 | This study |
| pKHB1527 | Chimera C1 (Ler 7–31 : : H-NS 48–101 : : Ler 79–123) in pBAD33 | This study |
| pKHB1163 | Chimera C2 (H-NS 1–47 : : Ler 32–78 : : H-NS 102–137) in pBAD33 | This study |
| pKHB1083 | Chimera C3 (Ler 7–78 : : H-NS 102–137) in pBAD33 | This study |
| pKHB1047 | Chimera C4 (H-NS 1–101 : : Ler 79–123) in pBAD33 | This study |
| pKHB1167 | Chimera C5 (H-NS 1–47 : : Ler 32–123) in pBAD33 | This study |
| pKHB1523 | Chimera C6 (Ler 7–78 : : H-NS 48–137) in pBAD33 | This study |
| Phages | ||
| P1vir | Generalized transducing phage | Laboratory stock |
| λRS45 | Specialized transducing phage | Simons et al. (1987) |
Generation of plasmids containing lacZ fusions
The construction of plasmid pKMTIR3 containing EPEC strain E2348/69 LEE5 regulatory DNA spanning positions −303 to +172 in relation to the transcriptional start site (+1) was described previously (Haack et al., 2003). Plasmids pJLM166 and pJLM172 for constructing the LEE2–lacZ (−218 to +670) and LEE3–lacZ (−398 to +404) fusions, respectively, were also previously described (Mellies et al., 1999). For plasmid pKHpro1, the regulatory region of proU (−398 to +404) was amplified with primers proUregF and proUregR, digested with EcoRI and BamHI, and cloned into the reporter gene vector pRS551, containing the promoterless lacZYA operon (Simons et al., 1987). All oligonucleotide PCR primers for molecular manipulations are listed in Table 2.
Table 2.
Oligonucleotides used in this study
| Primer | Sequence (5′-3′)* |
|---|---|
| proUregF | CCGGAATTCCTTTCGCCGAATGCCGAT |
| proUregR | CGCGGATCCAGGACTGGAAGACCATCGCA |
| LerForward | GCTCTAGACGGAGATTATTTATTATGAATATGGAAA CT |
| KLHlerwtR | CCCAAGCTTTTAAATATTTTTCAGCGGTATTATTTCTTCTT |
| NTERMMUT | GCTCTAGACGGAGATTATTTATTATGAGCGAAGCACTTAAAATTCTGAACAACATCCGTACTCTTCGTGCGCAGGCAAGAGAATGTAATATGGAAACTAATTCACATACAACAAGT |
| KLHlermutR | CCCAAGCTTTTAGTCCTTCACAAGAAAATCTTCTTTCT |
| ELLF | GAACTGCTGAATAGCCTTGCTGCCGTTAAATCTGTGCCTGATGATGGACTCGCTCGCCGGAAC |
| ELLR | AGATTTAACGGCAGCAAGGCTATTCAGCAGTTCATTAGTATATCCCAGCTCTTGTAAGGTTAT |
| 6AF | GCTGCAGCAGCGGCTGCAGTGCCTGATGATGGACTCGCTCGCCGGAAC |
| 6AR | TGCAGCCGCTGCTGCAGCATTAGTATATCCCAGCTCTTGTAAGGTTAT |
| 11AF | GCTGCAGCGGCAGCTGCTGCAGCGGCTGCAGCTGTGCCTGATGATGGACTCGCTCGCCGGAAC |
| 11AR | AGCTGCAGCCGCTGCAGCAGCTGCCGCTGCAGCATTAGTATATCCCAGCTCTTGTAAGGTTAT |
| 3′lerP115A | CCCAAGCTTTTAAATATTTTTCAGCGGTATTATTTCGTCTTCGGTGTCCTTCACAAGAAAATCTTCTTTCTTCATTCCATTCAACAGTGCTTCTTTAAGCCAGCGTGCCTGTCGGCCAA |
| lerRnar | GGGGCGCCCAGTCGCTTTGCTTCCTGCT |
| lerFnar | GGGGCGCCCAGGAAGTTCAAAGTGTAAT |
| lerRsal | GGGTCGACGTAGTAAACACCTTTCGATG |
| lerFsal | GGGTCGACGAAGAAGGGCAGACCTGGTC |
| 5hnsswap | GCTCTAGACGGAGATTATTTATTATGAGCGAAGCACTTAAAATTCT |
| hnsRNar | GGGGCGCCCGCGCTTTCTTCTTCGCGAC |
| hnsFNar | GGGGCGCCGCTGAAGTTGAAGAGCG |
| hnsRSal | GGGTCGACGTAGCTATATTTTGCCGGAC |
| hnsFSal | GGGTCGACGAAGAAGGGCAGACCTGGTC |
| 3hnsswap | CCCAAGCTTTTATTGCTTGATCAGGAAATC |
Restriction sites used in cloning are underlined
Construction of chromosomal single-copy LEE–lacZ and proU–lacZ transcriptional fusions
Single-copy chromosomal lacZ fusions were constructed by homologous recombination between λlRS45 and plasmids containing LEE5, proU, LEE2 and LEE3 regulatory fragments as described by Mellies et al. (1999). For strains KH4105, KH4106, KH4102 and KH4103, phage lysate was used to transduce KH4100, a spontaneous ara+ MC4100 mutant, selecting for kanamycin resistance. To generate isogenic Δhns mutants, KH4105, KH4106, KH4102 and KH4103 were transduced with P1vir lysate from strain HN4104, selecting for tetracycline resistance. These mutants were screened for a red appearance on MacConkey agar containing 1% salicin and possessing a mucoid phenotype, confirming deletion of hns (Mukerji & Mahadevan, 1997).
Generation of Ler proteins with altered N and C termini
Plasmid pKLHB01, containing wild-type (wt) ler, was generated by PCR amplification of template plasmid pSE1100 (Mellies et al., 1999) with primer pair LerForward and KLHlerwtR containing XbaI and HindIII sites, respectively. This fragment was directionally ligated into pBAD33 downstream of an arabinose-inducible promoter. Plasmid pKHB1123, containing wt hns, was similarly constructed with primers 5hnsswap and 3hnsswap, and directionally ligated into pBAD33.
Plasmid pMHB1, encoding the Ler protein with the α-helices 1 and 2 of H-NS (MSEALKILNNIRTLRAQAREC) fused to its N terminus, was created by PCR mutagenesis using primers NTERMMUT and KLHwtlerR containing XbaI and HindIII sites, respectively, and the Ler-encoding plasmid pSE1100 as template (Mellies et al., 1999). The amplicon was directionally cloned into the pBAD33 vector (Guzman et al., 1995), containing an arabinose-inducible promoter digested with the same enzymes. Plasmid pKLHB13, in which the 33 base pairs encoding the 11 aa C-terminal extension TEEEIIPLKNI are deleted, when comparing Ler to the H-NS protein (see Fig. 1), was created by PCR mutagenesis using primers LerForward and KLHlermutR. As above, the restriction sites XbaI and HindIII were used to directionally clone the DNA fragment into pBAD33 under the control of the PBAD promoter.
Fig. 1.
Sequence alignment of Ler and H-NS. The H-NS amino acid sequences of EPEC strain E2348/69 and Salmonella typhimurium are compared to the H-NS-like molecule Ler. Of these proteins, Ler shares greatest amino acid similarity with H-NS of S. typhimurium (24% identity, 44% similarity). NMR studies indicate that H-NS is made up of an N-terminal domain containing three α-helices (indicated by dashed lines; residues 1–64), a structurally undefined linker region required for higher-order oligomerization (residues 65–90), and a C-terminal DNA binding domain (residues 90–137) (Dorman, 2004). Though not experimentally defined, Ler is predicted to share similar structural modularity; Ler is predicted to possess a single α-helix, instead of three, in the N-terminal region of the protein (see Results). Identity, indicated by black shading, between Ler and H-NS is greater in the C-terminal domain, containing the core DNA binding motif TWTGXGRXP (Bertin et al., 1999; Dorman et al., 1999). Amino acids important for dimerization, found in the N termini of Ler and H-NS, are boxed (Sperandio et al., 2000; Ueguchi et al., 1997). The conserved C-terminal DNA binding motif (Bertin et al., 1999; Dorman et al., 1999) is also boxed. The regions used for the construction of Ler–H-NS chimeras (boundaries indicated by vertical lines) differ from the experimentally determined domains of H-NS (indicated by asterisks) because of technical constraints in molecular cloning protocols. Alignments were created using clustal_X (1.83) multiple sequence alignment software, and shading using Boxshade (3.21).
Plasmid pLER92A containing a Ler protein with an alanine substituted for the conserved proline at position 92 (position 115 in H-NS) was created by PCR mutagenesis using primers LerForward and 3′lerP115A, with plasmid pSE1100 as template, and directional cloning into the pBAD33 vector using the XbaI and HindIII restriction sites, as described above.
Generation of Ler linker insertion mutations
Plasmid pFL08, encoding Ler with an insertion of amino acids ELLNSLAAVKS at position 67, was generated by overlap extension mutagenesis using primer pairs LerForward and ELLF, and KLHlerwtR and ELLR, and plasmid pSE1100 as template (Mellies et al., 1999). The two partially complementary fragments were subjected to a second amplification using LerForward and KLHlerwtR as primers. Plasmids pFL09 and pFL10, encoding Ler with six and eleven alanines, respectively, inserted at position 67, were constructed in an analogous manner, using primers 6AF and 6AR and 11AF and 11AR as the mutagenic primers, respectively. Amplification products obtained in this way were inserted into pBAD33 as XbaI/HindIII fragments.
Generation of Ler–H-NS chimeras
For Ler–H-NS chimeras, individual PCR-amplified fragments corresponding to each domain of Ler were generated with the following primer pairs. The LerForward and lerRnar primers were used for the N-terminal fragment, amino acids 7–31, containing the coiled-coil domain defined by mutational analysis (Fig. 1; Sperandio et al., 2000). (For the chimera analyses, the N-terminal methionine at position 7 (accession number AF022236) corresponds to a predicted ribosome-binding site.) Primers lerFnar and lerRsal were used to construct the linker fragment, amino acids 32–78, and lerFsal and KLHlerwtR primers were used for the Ler C-terminal fragment, amino acids 79– 123. The LerForward and lerRsal primers were used for the N-terminal and linker fragments, amino acids 7–78, and primers lerFnar and KLHlerwtR for the linker and C-terminal fragments, amino acids 32–123.
For H-NS, analogous fragments were generated with the following primer pairs: 5hnsswap/hnsRnar for the N-terminal fragment, amino acids 1–47; hnsFnar and hnsRsal for the linker fragment, amino acids 48–101; hnsFsal and 3hnsswap for the C-terminal fragment, amino acids 102–138; 5′hnsswap and hnsRsal for the N-terminal and linker fragments, amino acids 1–101; hnsFnar and 3hnsswap for linker and C-terminal fragments, amino acids 49–138. Fragments were cloned into pCR2.1TOPO (Invitrogen) for DNA sequencing.
For chimera C1 (pKHB1527), the NarI- and SalI-digested linker fragment of H-NS was ligated to the NarI-digested N-terminal and SalI-digested C-terminal fragments of Ler. For the analogous C2 (pKHB1163) chimera, the NarI- and SalI-digested linker fragment of Ler was sequentially ligated to the NarI-digested N-terminal and SalI-digested C-terminal fragments of H-NS. For chimeras C4 (pKHB1047) and C3 (pKHB1083), the NarI-digested N-terminal and linker fragment of H-NS and Ler, respectively, were ligated to the NarI-digested C-terminal fragment from Ler and H-NS, respectively. Chimeras C5 (pKHB1167) and C6 (pKHB1523) combined the SalI-digested linker and N-terminal fragment from Ler and H-NS with the SalI-digested N-terminal fragment from H-NS and Ler, respectively. Following ligation of individual domain fragments, the appropriate external primer pairs complementary to either Ler or H-NS were used to amplify the entire chimeric coding region, which was inserted into pCR2.1 TOPO for verification by restriction digestion and DNA sequencing. Chimeric coding regions were then digested with XbaI/ HindIII and inserted into the XbaI/HindIII site of pBAD33.
DNA sequencing analysis
All plasmid constructs were confirmed to be correct by DNA sequencing analysis performed at the Vollum Institute at the Oregon Health Sciences University.
Enzymic assays
Strains KH4105 and KH4115 were transformed with plasmids pKLHB13, pMHB1 and pLER92A, as well as the control plasmids pKLHB01 (containing wt ler), pKH1123 (wt hns) and pBAD33. Similarly, strains KH4105, KH4115, KH4106 and KH4116 were transformed with plasmids pKHB1527, pKHB1163, pKHB1083, pKHB1047, pKHB1167 and pKHB1523 (chimeras C1– C6). Control plasmids pKLHB01, pKH1123 and pBAD33 were transformed into strains KH4106 and KH4116 as well. Cultures were grown at 37 °C in LB supplemented with chloramphenicol and either 0.1% arabinose (induced expression of Ler molecules) or 0.2% glucose (repressed expression of Ler molecules) to OD600 ~0.3–0.5 and then subjected to β-galactosidase assays, as described by Miller (1972).
To measure β-galactosidase activity derived from an E2348/69 ler deletion strain, the pKMTIR3 LEE5–lacZ fusion plasmid was transformed into SE796 and SE796 containing the following plasmids: pBAD33, pKLHB01, pKH1123, pFL08, pFL09, pKHB1527, pKHB1163, pKHB1047 and pKHB1167. Cultures were grown at 37 °C in LB supplemented with chloramphenicol and ampicillin to maintain the pBAD33 and pRS551-derived vectors, respectively. Assays were performed as described above. Statistical analysis (t test) for β-galactosidase assay values in Miller units was performed using JMP software (SAS Institute).
RESULTS
The Ler N terminus and C terminus
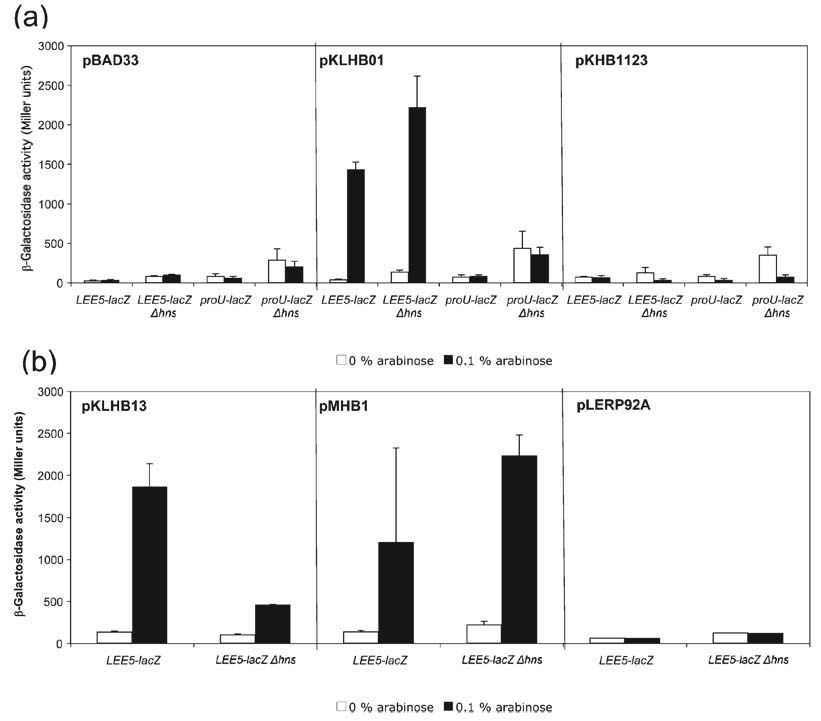
Ler is predicted to have a single α-helix in the N-terminal domain [by the nnpredict Protein Secondary Structure Prediction software (http://www.cmpharm.ucsf.edu/~nomi/nnpredict.html)], whereas the H-NS molecules of E. coli and Salmonella are predicted to possess three α-helices in this region. In H-NS, α-helix 1 spans positions 1– 8, a-helix 2 spans positions 12–19, and α-helix 3 spans positions 23–47 (Fig. 1; Dorman, 2004; Fang & Rimsky, 2008). N-terminal fragments of H-NS have been shown to exhibit dominant negative effects on transcriptional activity (Ueguchi et al., 1996; Williams et al., 1996), and thus we predicted that placing α-helices 1 and 2 of H-NS on to the N terminus of Ler would alter its function. To test this prediction, we cloned a DNA fragment encoding a mutated Ler protein containing α-helices 1 and 2 of H-NS (including the amino acids MSEALKILNNIRTLRAQAREC to position 21) under the control of PBAD, the arabinose-inducible promoter in pMHB1. The plasmid was transformed into the K-12-derived strain KH4105 containing a single copy of LEE5 regulatory DNA, from EPEC strain E2348/69, spanning positions −303 to +172 in relation to the transcriptional start site (Sánchez-SanMartín et al., 2001), fused to the lacZ reporter gene (Simons et al., 1987). As expected, control experiments demonstrated that Ler expressed from plasmid pKLHB01 increased transcription at the LEE5 promoter but not at proU, in strains wt and deleted for hns, whereas H-NS expressed from plasmid pKHB1123 silenced transcription at both LEE5 and proU (Fig. 2a).
Fig. 2.
Analysis of the Ler N terminus and C terminus. (a) β-Galactosidase activities were derived from K-12 strains containing LEE5–lacZ and proU–lacZ fusions in the presence of the pBAD33 vector, wt Ler or wt H-NS proteins, expressed from plasmids pKLHB01 and pKHB1123, respectively, under the control of the arabinose-inducible PBAD promoter. Plasmids were transformed into LEE5–lacZ fusion strains wt for hns (KH4105) or deleted for hns (KH4115); similarly, the above plasmids were transformed into proU–lacZ fusion strains wt for hns (KH4106) and deleted for hns (KH4116). The values are presented in Miller units and represent the mean of at least two independent assays performed in triplicate. Error bars, sd. (b) Plasmid pKLHB13 encodes a recombinant Ler protein containing a C-terminal truncation, missing the last 11 aa compared with H-NS. Plasmid pMHB1 encodes a protein containing the first 21 aa of H-NS, containing α-helices 1 and 2, fused to the N terminus of Ler (see Fig. 1). Plasmid pLERP92A contains a mutated Ler protein with the point mutation P92A within the core DNA binding motif.
The mutated Ler protein containing the α-helices 1 and 2 of H-NS expressed from plasmid pMHB1 increased transcriptional activity of the LEE5–lacZ fusion in a manner similar to that observed for the wt Ler protein (Fig. 2b), and did not alter expression of the proU–lacZ fusion under any of the conditions tested (data not shown). Therefore, we concluded that even gross alteration of the N terminus of Ler did not affect its ability to differentially stimulate transcription of LEE virulence genes.
The C termini of Ler and H-NS contain the core DNA binding motif TWTGXGRXP, where X is any amino acid (Bertin et al., 1999; Dorman et al., 1999). The Ler core DNA binding motif, indicated by the boxed region in Fig. 1, is TWSGVGRQP, a perfect match except for the serine residue in the third position. The proline at position 116 of H-NS, at the furthest position in the binding motif is highly conserved amongst all H-NS molecules (Dorman, 2004, 2007; Fang & Rimsky, 2008). A P116A mutation abolishes the ability of H-NS to silence transcription in vivo, disrupts oligomerization activity (Spurio et al., 1997) and eliminates the ability of the protein to distinguish curved from non-curved DNA (Spurio et al., 1997). We therefore determined whether this conserved proline was necessary for Ler anti-silencing activity, and constructed an analogous mutation in Ler, P92A (Fig. 1). As predicted, this point mutation at the C terminus of the conserved DNA binding motif eliminated the ability of Ler to increase LEE5–lacZ transcriptional activity, in strains containing the chromosomal hns or deleted for this gene (Fig. 2b). We concluded that the proline in the predicted core DNA binding motif of Ler was necessary for increasing LEE transcriptional activity, and presumably plays an analogous role in both Ler and H-NS.
Ler contains an 11 aa extension on its C terminus that is not present in H-NS (Fig. 1). Based on the prediction that H-NS forms head-to-tail oligomers (Esposito et al., 2002) and the knowledge that Ler binds over an extended region to regulate LEE virulence genes (Haack et al., 2003), we predicted that this 11 aa, C-terminal extension was necessary for Ler to increase LEE transcriptional activity. To test this prediction, we cloned a DNA fragment encoding a mutated Ler protein missing the C-terminal amino acids TEEEIIPLKNI under the control of PBAD, the arabinose-inducible promoter in pKLHB13, transformed into the K-12-derived strain KH4105 and KH4106, as described above.
In the presence of wt hns the truncated Ler protein encoded in the pKLHB13 plasmid increased β-galactosidase activity from the LEE5–lacZ fusion to a level similar to that observed for the wt Ler protein, ~1500 Miller units (compare Fig. 2a, b). In the KH4115 strain deleted for hns, the truncated Ler protein still increased β-galactosidase activity, from ~100 to 500 Miller units (P<0.0001), but not to ~2000 Miller units as observed for wt Ler under the same conditions (Fig. 2a). The truncated Ler protein in plasmid pKLHB13 did not alter expression of the proU–lacZ control fusion under any of the conditions tested (data not shown). We concluded that the 11 aa C-terminal extension found in Ler, but absent from the related H-NS protein, did not contribute to the ability of Ler to increase transcription from the LEE5–lacZ fusion, though the level of expression was reduced compared with wt Ler when assayed in a strain deleted for hns, ~500 Miller units. As a control, transcriptional activity derived from the proU–lacZ fusion increased in the strain deleted for hns independently of the presence of arabinose or Ler, and this activity was abolished when H-NS was supplied in trans (Fig. 2a). Thus, the 11 aa C-terminal extension might play a role, but is not essential for relieving repression by a non-H-NS factor acting at LEE5.
Non-H-NS factor acting at LEE5
Our data indicate that, in addition to H-NS, a second negative-acting factor controls LEE5 expression at host body temperature, 37 °C (Haack et al., 2003; Fig. 2a). In Fig. 2(a), transcriptional activity derived from the LEE5–lacZ fusion strain was modestly de-repressed in the absence of the inducer arabinose, from ~50 to 100 Miller units (P=0.002), when comparing β-galactosidase activity in the presence of wt hns with that derived from the hns deletion strain. However, the LEE5–lacZ reporter gene fusion was still inducible by Ler, to ~2000 Miller units (P<0.0001), in the presence of arabinose in a strain containing an hns deletion. Previously published reports investigating Ler and H-NS control of the LEE5 operon are consistent with these data, demonstrating evidence for a non-H-NS factor acting at LEE5 in E. coli K-12 and EPEC strains (Haack et al., 2003; Umanski et al., 2002). Intriguingly, the mutated Ler protein lacking the 11 aa C-terminal extension in plasmid pKLHB13 exhibited differential ability to increase LEE5 transcriptional activity depending on whether it was assayed in the presence or absence of the chromosomal hns gene (Fig. 2b). Because the mutated Ler protein in pKLHB13 only modestly increased activity in the strain deleted for hns in comparison with wt Ler expressed from the pKLHB01 plasmid, this result suggested that the truncated protein was deficient in counteracting the repressing activity of the second, unidentified factor. As a control, it was determined that the recombinant Ler protein expressed from the pKLHB13 plasmid did not affect transcriptional activity from the proU–lacZ control fusion in strains either wt for or deleted for hns (data not shown). These data indicated that at least two negative-acting factors silence LEE5, and that Ler is able to counteract the activities of both H-NS and the unidentified factor to increase transcriptional activity. Again, these data indicated that the unidentified factor, whether it is a protein or a specific nucleoid structure, is not unique to EPEC, but is also found in K-12-derived strains.
The Ler linker
Stella et al. (2005) demonstrated that the H-NS linker is essential for the N-terminal and C-terminal domains to mediate tetramerization, and thus silencing, a first report for a function of the linker besides simply connecting the N-terminal and C-terminal domains. The central linker regions of H-NS and Ler are the least similar regions of the molecules and, coincidentally, 11 aa found in H-NS are missing from the Ler linker region (Fig. 1). We therefore hypothesized that inserting the 11 aa sequence ELLNSLAAVKS from H-NS into position 67 (between amino acids N and V) of the Ler linker would alter Ler function, and more specifically, change Ler into a transcriptional silencer.
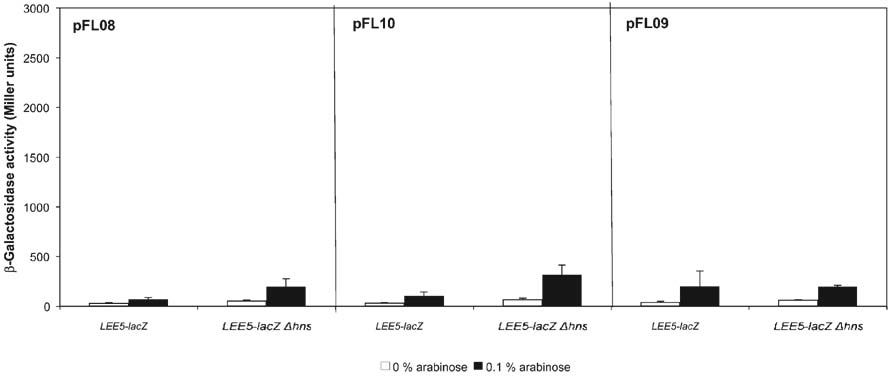
We found that the Ler molecule containing the 11 aa H-NS linker sequence (pFL08) was impaired in its ability to increase transcription from the LEE5–lacZ fusion in strains wt or deleted for hns, but the mutated Ler molecule did not silence LEE5 transcription (Fig. 3). In the strain deleted for hns, the Ler molecule containing the ELLNSLAAVKS insertion modestly increased LEE5 transcriptional activity, from ~50 to 200 Miller units (P<0.0001). To investigate further we constructed mutated Ler proteins containing either 11 or six alanines inserted into position 67 of Ler. These recombinant proteins were constructed to test whether the spacing between the N terminus and C terminus, irrespective of the specific amino acid sequence of the interdomain linker, was essential for Ler function. With results nearly identical to those for the pFL08 plasmid, inserting either 11 or six alanines (pFL10 and pFL09, respectively) impaired the ability of Ler to increase LEE5 transcription, with only modest increases in transcription occurring in the strain deleted for hns (compare Fig 3 and Fig 2a). These mutated Ler proteins did not decrease transcriptional activity, as observed for the wt H-NS protein acting on the LEE5 and proU fusions in the strain with a deleted chromosomal hns gene (Fig. 2a). We concluded that altering the spacing between the N-terminal and C-terminal domains severely affected the ability of Ler to increase LEE gene expression.
Fig. 3.
Inserting the H-NS linker sequence into Ler reduced, but did not eliminate, the ability of Ler to increase LEE5 activity. β-Galactosidase activities were derived from LEE5–lacZ fusions in the presence of Ler proteins containing either the H-NS linker sequence ELLNSLAAVKS (pFL08), 11 alanines (pFL10) or six alanines (pFL09) inserted at amino acid position 67. β- Galactosidase assays were performed as described for Fig. 2.
Replacing the Ler linker and/or C-terminal DNA-binding domain with the corresponding domains of H-NS eliminated Ler anti-silencing activity
Though Ler possesses the coiled-coil domain (Sperandio et al., 2000) and the conserved H-NS DNA binding motif (Fig 1 and Fig 2b) in its N terminus and C terminus, respectively, the modular nature of Ler is not well established experimentally. Therefore, we investigated the contribution of the N-terminal α-helical, linker region, and C-terminal DNA binding domains of Ler and H-NS in the control of LEE5 transcriptional activity. Chimeric Ler–H-NS molecules, containing all combinations of the two functional domains and linker regions were constructed. The α-helical, N-terminal fragments contained the coiled coils and the C-terminal fragments contained the conserved DNA binding motifs of Ler and H-NS, determined by mutational analyses (Sperandio et al., 2000; Ueguchi et al., 1997; Fig. 1). DNA fragments encoding these chimeric proteins were placed under the control of the arabinose-inducible promoter in derivatives of the plasmid pBAD33. The plasmids encoding chimeras, and the wt Ler and H-NS molecules were transformed into the single-copy LEE5–lacZ and proU–lacZ fusion strains as described above, and β-galactosidase activities were monitored in strains wt and deleted for hns. In Fig 4 and Fig 5, domains of the Ler protein are depicted as white rectangles, whereas those of H-NS are depicted as black rectangles.
Fig. 4.
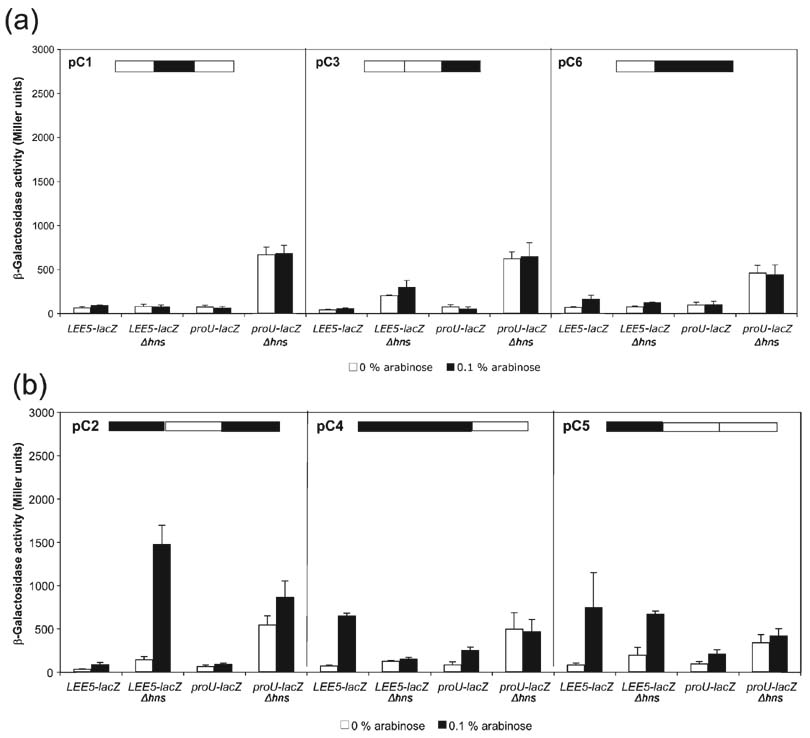
Analysis of Ler–H-NS chimeric proteins. (a) Replacing the Ler linker and/or C terminus with those of H-NS eliminated Ler and H-NS activity. Chimeric proteins C1, C3 and C6 encoded on plasmids pKHB1527 (pC1), pKHB1083 (pC3) and pKHB1523 (pC6) were transformed into the LEE5–lacZ strains wt and deleted for hns, KH4105 and KH4115, respectively. Similarly, the three plasmids were transformed into the proU–lacZ fusion strains. (b) When paired with the N terminus of H-NS, the linker region of Ler and C-terminal domains conferred distinct effects on LEE5 transcription; the chimeric proteins C2, C4 and C5 in plasmids pKHB1163 (pC2), pKBH1047 (pC4) and pKHB1167 (pC5), respectively, retained the differential activity of Ler at LEE5. β-Galactosidase assays were performed as described for Fig. 2.
Fig. 5.
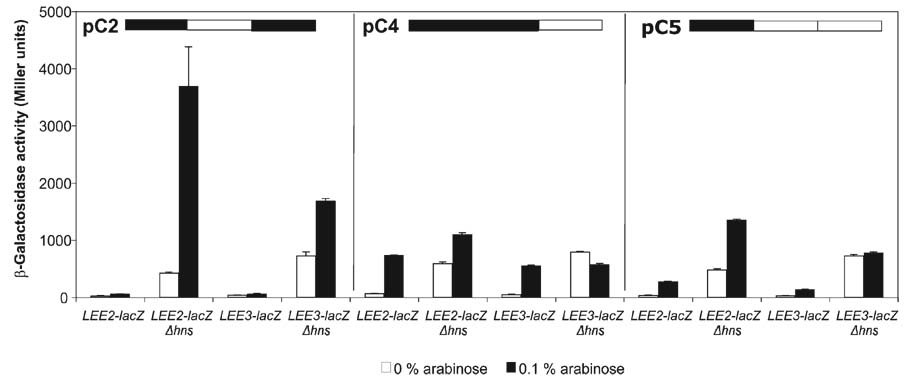
The chimeric proteins C2, C4 and C5 showed similar effects on transcription at LEE2 and LEE3 to those for LEE5. Chimeric proteins C2, C4 and C5 expressed from plasmids pKHB1163 (pC2), pKBH1047 (pC4) and pKHB1167 (pC5), respectively, were transformed into LEE2–lacZ and LEE3–lacZ fusion strains, and β-galactosidase activities were determined as described for Fig. 2. The LEE2–lacZ fusion strains KH4102 and KH4112 are wt and deleted for hns, respectively, while the LEE3–lacZ fusion strains KH4103 and KH4113 are wt and deleted for hns, respectively.
Expression of chimeras C1, C3 and C6, containing the H-NS linker, H-NS C terminus, and H-NS linker and C-terminal domain, respectively, did not alter transcription at LEE5 or proU promoters under any of the conditions tested (Fig. 4a). These data indicated that the Ler linker and C-terminal domain are critical for anti-silencing activity. Thus, the Ler linker and C terminus are not functionally equivalent to those of H-NS.
The C2 chimeric protein containing the Ler linker increased LEE gene expression in a strain deleted for hns
The opposite of the C1 protein, the C2 protein, exchanged the linker region of H-NS with that of Ler. This chimeric protein did not silence expression from either the LEE5–lacZ or the proU–lacZ fusions (Fig. 4b). However, the C2 chimera increased transcription 10-fold at LEE5 in the absence of hns, from ~150 to 1500 Miller units (P<0.0001), suggesting that it was still able to disrupt a non-H-NS regulator of LEE5. The C2 protein increased proU–lacZ expression slightly, less than twofold, in a strain containing an hns deletion. These data were consistent with those observed for the C1 protein and our directed mutagenesis of the Ler linker (Fig 3 and Fig 4a), indicating that the Ler and H-NS linker regions are not functionally equivalent.
The C4 protein containing the N terminus and linker of H-NS and the C terminus of Ler increased the expression of the LEE5–lacZ fusion sixfold, from ~100 to 600 Miller units (P<0.0001) (Fig. 4b). However, in contrast to C2, the C4 chimeric protein did not affect expression of the LEE5–lacZ fusion in a strain containing an hns deletion.
Compared with the C4 protein, the C5 chimeric protein contained the N terminus of H-NS and the linker and C terminus of Ler. This protein increased LEE5–lacZ expression eightfold in the presence of hns, from ~100 to 800 Miller units (P<0.0001) (Fig. 4b). Unlike the C4 protein, however, the C5 chimera also increased LEE5–lacZ transcription in a strain deleted for the hns gene. Overall, the C5 chimeric protein affected the LEE5–lacZ fusion similarly to the wt Ler protein (see Fig. 2a), except that the induced levels of expression were reduced. These data were also consistent with the effect of the pMHB1 construct (α-helices 1 and 2 of H-NS placed on the N terminus of Ler) on LEE5 activity (see Fig. 2b), in that both mutated proteins still relieved silencing of LEE transcription in strains wt and deleted for hns. Since the only difference between the C4 and C5 chimeras was that C5 contained the linker of Ler, whereas C4 had the linker of H-NS, these data further suggested that the Ler linker is important for interaction with or disruption of the non-H-NS factor acting at LEE5.
Interestingly, none of the chimeric proteins appreciably affected expression of the H-NS-regulated proU operon. The C4 and C5 proteins increased proU–lacZ expression approximately twofold in a strain wt for hns (Fig. 4b), whereas wt Ler had no effect on this fusion in either the presence or the absence of hns (Fig. 2a). However, the approximately twofold increase in proU–lacZ expression in the presence of the C4 and C5 chimeras was far less than the ~15-fold increase in LEE5–lacZ expression in the presence of wt Ler (Fig. 2a). Thus, we concluded that though the C4 and C5 chimeras modestly affected proU–lacZ expression, most likely due to dominant negative effects on the wt H-NS protein, our analysis did not elucidate a molecular explanation for differential regulation of LEE virulence genes by Ler.
As an important control, we expressed the wt Ler, wt H-NS and chimeric proteins from the arabinose-inducible pBADMycHISA expression vector in our fusion strains, monitoring LEE5–lacZ and proU–lacZ activities and protein expression by the anti-Myc epitope antibody (Invitrogen). We observed similar β-galactosidase activities to those of the pBAD33-based constructs presented in Fig 2 and Fig 4, and found that all of the wt and chimeric proteins were visible by immunoblot analysis under growth conditions identical to those used for monitoring activities derived from the lacZ fusions (data not shown).
C2, C4 and C5 chimeric proteins altered expression of the LEE2 and LEE3 operons
To ensure that the observed activities associated with the mutated Ler proteins were generalized regulatory phenomena, and not simply specific to the LEE5 operon of EPEC, we monitored transcriptional activity derived from the EPEC LEE2 and LEE3 operons in the presence of the C2, C4 and C5 chimeric proteins. For the C2 chimera containing the linker region of Ler and the N terminus and C terminus of H-NS, expression derived from the LEE2–lacZ and LEE3–lacZ fusions increased approximately sevenfold (P<0.0001) and twofold (P<0.0001), respectively, in the strain deleted for the chromosomal hns gene (Fig. 5), a similar phenotype to that observed at LEE5 (Fig. 4b). The C4 protein containing the N terminus and linker of H-NS and the C terminus of Ler increased LEE2 and LEE3 expression in the strain wt for hns ~10-fold (P<0.0001; P<0.0001), whereas expression from both of these fusions in the absence of chromosomal hns was only modestly affected (Fig. 5). The presence of the C5 chimeric protein increased transcriptional activity derived from the LEE2–lacZ fusion in strains with and without hns approximately fivefold (P=0.0002) and threefold (P<0.0001), respectively, a result similar to that observed for the LEE5–lacZ fusion (Fig 4b and Fig 5). The presence of the C5 chimera did not alter expression derived from the LEE3–lacZ fusion under any of the conditions tested, but differences in LEE2 and LEE3 regulation have been observed elsewhere (Russell et al., 2007). Thus, we concluded that those chimeric proteins with demonstrated effect at the LEE5 operon functioned similarly at the LEE2 and LEE3 operons, with the exception of the C5 chimeric protein not affecting LEE3 expression (Fig. 5).
Activity of mutated and chimeric Ler proteins in EPEC
In order to demonstrate regulatory observations in the specific pathotype EPEC, we assayed the effects of the mutated Ler proteins on LEE5 transcription using the multi-copy LEE5–lacZ fusion plasmid pKMTIR3 (Haack et al., 2003). For this analysis, the ler deletion strain SE796 with pBAD33-derived vectors expressing Ler, H-NS, the mutated Ler proteins and the empty vector control were transformed with plasmid pKMTIR3. Strains were grown and harvested in conditions identical to those for β-galactosidase assays in K-12-derived strains, except that 0.1% glucose was added to cultures to repress expression of wt lac in strain SE796.
As expected, in the presence of the wt Ler protein, LEE5 transcription increased 20-fold (P<0.0001), whereas the H-NS protein caused a decrease, 0.5-fold (P=0.03), in strain SE796, which is wt for hns (Table 3). As presented in Fig. 3, mutated Ler proteins containing either the H-NS sequence ELLNSLAAVKS (pFL08) or six alanines (pFL09) inserted at position 67 of the linker were deficient compared with wt Ler, but were still able to relieve silencing of LEE5 transcription by 2.1-fold (P=0.002) and 2.6-fold (P=0.001), respectively (Table 3). Consistent with the observed effects of the C1 and C2 chimeras in the K-12-derived strain in the presence of wt hns (see Fig. 4a, b), these mutated proteins had negligible effect on LEE5 transcription, 1.1-fold and 1.0-fold, respectively (Table 3). Surprisingly, the C4 chimeric protein did not show anti-silencing activity towards LEE5 in strain SE796 (Table 3, Fig. 4b), perhaps due to altered stability in the EPEC strain or interaction with EPEC-specific regulators. The C5 protein caused a 4.6-fold increase in LEE5 transcription in strain SE796 (P=0.0001), consistent with data presented in Fig. 4(b). We therefore concluded that, as previously reported (Bustamante et al., 2001; Mellies et al., 2007b; Umanski et al., 2002), with the exception of the C4 chimera, Ler-associated regulatory phenomena were similar in EPEC- and K-12-derived strains.
Table 3.
Effect of mutated Ler proteins on LEE5 expression in EPEC strain SE796
The EPEC E2348/69-derived strain SE796 contains an in-frame deletion of ler.
| Fusion plasmid | Protein plasmid | Glucose* | Arabinose* | Fold-induction† |
|---|---|---|---|---|
| – | – | 19 (6) | 8 (3) | – |
| pRS551 | – | 31 (1) | 43 (2) | – |
| pKMTIR3 | pBAD33 | 692 (19) | 731 (26) | 1.1 |
| pKMTIR3 | pKHLB01(Ler) | 526 (13) | 10 218 (185) | 20 |
| pKMTIR3 | pKHB1123 (H-NS) | 439 (23) | 216 (12) | 0.5 |
| pKMTIR3 | pFL08 | 752 (19) | 1588 (196) | 2.1 |
| pKMTIR3 | pFL09 | 717 (74) | 1887 (334) | 2.6 |
| pKMTIR3 | pKHB1527(C1) | 996 (68) | 1067 (41) | 1.1 |
| pKMTIR3 | pKHB1163(C2) | 1139 (63) | 1128 (38) | 1.0 |
| pKMTIR3 | pKHB1047(C4) | 1015 (20) | 891 (13) | 0.9 |
| pKMTIR3 | pKHB1167(C5) | 822 (24) | 3767 (113) | 4.6 |
β-Galactosidase activity derived from the pKMTIR3 5–lacZ reporter gene vector in the strain SE796 was monitored in the presence of the listed protein expression and control plasmids under repressing (0.1% glucose) and inducing (0.1% arabinose) conditions. The values are presented in Miller units and represent the mean of representative assays performed in triplicate with SDs in parentheses.
Represents the ratio of transcriptional activity in the presence of the inducer (arabinose) to that in the presence of the repressor (glucose) for the indicated strains.
DISCUSSION
The Ler linker
In this report we investigated the roles of the Ler N-terminal α-helical, coiled-coil-containing domain, linker region and C-terminal DNA-binding domain in their ability to increase transcription at the LEE PAI of EPEC. Our investigation focused on the molecular differences between the silencing protein H-NS and the related anti-silencer Ler that allow for opposite functions. The data clearly indicated that the linker region unique to Ler is necessary for function: inserting 11 amino acids specific to the H-NS linker, 11 alanines or six alanines at position 67 severely limited Ler anti-silencing activity (Fig. 3). Additionally, replacing the H-NS linker with the linker of Ler created a chimeric protein (C2) that increased LEE transcription in a strain deleted for the chromosomal hns gene (Fig 4b and Fig 5). Combined with evidence of a second, non-H-NS factor presented here and in a previous report (Haack et al., 2003), we propose that the C2 chimera retains the ability to counteract repression by the unknown factor, but has lost the ability to disrupt H-NS-mediated silencing at LEE operons. The C5 chimera, containing the linker and C terminus of Ler, relieved silencing of LEE genes similarly to the wt Ler protein (Fig 4b and Fig 5), albeit to a lesser extent (Fig. 2a), and the activities of the C2 and C5 chimeric proteins were confirmed in the EPEC strain SE796 (Table 3).
Previous reports have indicated that LEE regulatory observations in K-12-derived strains are faithfully reproduced in the EPEC pathotype (Bustamante et al., 2001; Mellies et al., 2007b; Umanski et al., 2002). Here, we made similar observations measuring LEE5 transcriptional activity on multi-copy plasmids in the E2348/69-derived ler deletion strain SE796 as a function of expressed Ler and H-NS molecules (Table 3). Our attempts to construct an EPEC ler hns double mutant were unsuccessful, most likely due to disruption of complex regulatory networks within this pathogenic bacterium in the absence of H-NS. Consistently, we observed that hns deletion derivatives of the EDL933 E. coli serotype O157 : H7 possess growth defects, having a 40% increase in doubling time (g), compared with their isogenic parent strains (Torres et al., 2007; our unpublished results). We were able to measure the effects of the mutated Ler proteins in the absence of wt Ler in the EPEC E2348/69 derivative SE796, but were unable to measure effects in the absence of the wt H-NS protein expressed from the chromosomal hns locus.
Interdomain linkers in other transcriptional regulators
Reports from other laboratories have indicated an important role for interdomain linkers in transcriptional regulators. For example, the response regulator OmpR of E. coli that controls expression of the outer membrane porins OmpF and OmpC in response to changes in osmolarity contains a linker important for inter-domain communication (Mattison et al., 2002). In OmpR, the C-terminal DNA binding domain is affected by signals mediated by phosphorylation of the N-terminal domain, which are dependent on the interdomain linker (Delgado et al., 1993; Kanamaru et al., 1990). Both OmpR and the related response regulator PhoB contain N-terminal phosphorylation domains and C-terminal DNA binding domains connected by a flexible linker. Even though OmpR and PhoB are closely related members of the OmpR family of response regulators, their interdomain linkers are not interchangeable (Walthers et al., 2003). The OmpR protein requires its linker for activation of the C-terminal winged helix–turn–helix DNA binding domain because the OmpR protein containing the linker of the related PhoB protein is defective in DNA binding and signalling. Thus, the interdomain linkers of related proteins can contribute to differential function of closely related proteins, as OmpR and its cognate sensor kinase EnvZ respond to medium osmolarity, while PhoB and its cognate sensor kinase PhoR respond to extracellular levels of inorganic phosphate. Though Ler and H-NS share N-terminal coiled-coil motifs (Sperandio et al., 2000) and a conserved C-terminal DNA binding motif (Fig 1 and Fig 2b; Bertin et al., 1999; Dorman et al., 1999), The interdomain linker of Ler varies greatly from that of H-NS and partially explains why the activity of Ler is opposite to that of H-NS.
Ler–H-NS chimeric protein activity
One observation that might explain how the C2, C4 and C5 chimeric proteins altered LEE transcription is that all contained H-NS N-terminal domains, while those without any demonstrable activity possessed the N terminus of Ler (Fig 4 and Fig 5). This observation suggested dominant negative effects with H-NS, or perhaps interactions with other DNA binding/regulatory proteins, since N-terminal fragments of H-NS have been observed to affect transcription in a dominant negative manner (Ueguchi et al., 1996; Williams et al., 1996). The C2 chimera increased LEE5 transcriptional activity in a strain containing an hns deletion, while the C4 chimera increased LEE5 trancriptional activity in a strain with wild-type hns (Fig 4 and Fig 5). Thus, the C4 chimera might interact with H-NS in a dominant negative manner, while the Ler linker region of the C2 chimera functions in the absence of H-NS. We purified the C2, C4 and C5 chimeras and found that their ability to bind LEE5 regulatory DNA was severely impaired (data not shown). This observation is consistent with the C2, C4 and C5 chimeras altering transcriptional activity via interaction with other proteins as opposed to minor alterations in DNA binding affinities. Though we observed expression of all six chimeric proteins by immunoblot analysis, we were not able to purify the C1, C3 and C6 chimeras (data not shown). Thus, their inactivity might have been due to instability or because they were sequestered within inclusion bodies inside the bacterium, unable to gain access to DNA. Clearly other, yet to be identified regulatory factors also play an important role in regulation of LEE virulence genes.
Silencing of LEE5
Though evidence suggests that Ler does not function as a classical activator proteins at other LEE operons (Bustamante et al., 2001; Umanski et al., 2002), we cannot rule out the possibility that Ler functions as such at LEE5, either by recruiting RNA polymerase directly or by altering nucleoid structure. Precedent exists for this type of dual regulatory function, as SlyA of Salmonella and a homologue, RovA of Yersinia, and ToxT of Vibrio cholerae can act as H-NS antagonists as well as classical transcriptional activator proteins (Ellison & Miller, 2006; Heroven et al., 2004; Yu & DiRita, 2002). It is also possible that Ler increases the expression of a yet to be described activator to stimulate LEE5 transcription. If a non-H-NS, negative-acting protein silencing LEE5 does exist, a likely candidate is StpA, because its expression increases in an hns mutant and there is evidence that it can compensate for the loss of H-NS (Müller et al., 2006).
Conclusions
Recent work has demonstrated that a major function of H-NS is to silence horizontally acquired segments of DNA, particularly those genes involved in bacterial virulence (Navarre et al., 2006). Anti-silencing, or countersilencing of H-NS leading to gene expression occurs by a variety of mechanisms. Examples of countersilencing include changes in the concentration of H-NS itself (Hansen et al., 2005), direction of transcription by the alternate sigma factors σS (Typas et al., 2007), antagonizing H-NS binding or remodelling nucleoprotein structure by specific regulatory proteins such as SlyA (Corbett et al., 2007; Lithgow et al., 2007), changes in local DNA topology (Falconi et al., 1998), and disruption of silencing by H-NS homologues, which include Ler and the H-NST molecules of EPEC (Haack et al., 2003; Mellies et al., 1999; Williamson & Free, 2005).
Homologues of H-NS, such as Ler, countersilence by either competing for binding sites or interfering with multi-merization (reviewed by Fang & Rimsky, 2008). At the LEE5 operon of EPEC, Ler and H-NS bind in the same location upstream of the promoter, whereas H-NS also acts downstream at LEE5 (Haack et al., 2003; our unpublished results). Because Ler possesses a lower KD of binding to LEE regulatory DNA than H-NS (Umanski et al., 2002), Ler clearly can compete for binding sites to increase LEE gene expression. However, we do not know whether Ler also affects H-NS multimerization. Additional questions remain. Might the distinct linker region of Ler play a role in disruption of H-NS multimerization? What is the molecular explanation for the greater affinity of Ler for DNA than the related H-NS molecule? Finally, why does Ler regulate LEE virulence genes but not other H-NS-silenced operons such as proU, and what other factors contribute to the differential regulation of Ler?
ACKNOWLEDGEMENTS
We thank Dr Bianca Colonna for providing strains, and Dr Ken Haack for construction of chimeric Ler–H-NS proteins. This work was supported by NIH AREA grant R15 AI047802-02 awarded to J. L. M., and a Miller Foundation grant awarded to the Biology Department of Reed College.
Abbreviations
- AE
attaching and effacing
- EHEC
enterohaemorrhagic E. coli
- EPEC
enteropathogenic E. coli
- LEE
locus of enterocyte effacement
- T3SS
type III secretion system
- wt
wild-type
REFERENCES
- Barba J, Bustamante VH, Flores-Valdez MA, Deng W, Finlay BB, Puente JL. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J Bacteriol. 2005;187:7918–7930. doi: 10.1128/JB.187.23.7918-7930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrametti F, Kresse AU, Guzmán CA. Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli. J Bacteriol. 1999;181:3409–3418. doi: 10.1128/jb.181.11.3409-3418.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin P, Benhabiles N, Krin E, Laurent-Winter C, Tendeng C, Turlin E, Thomas A, Danchin A, Brasseur R. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in Gram-negative bacteria. Mol Microbiol. 1999;31:319–329. doi: 10.1046/j.1365-2958.1999.01176.x. [DOI] [PubMed] [Google Scholar]
- Bloch V, Yang Y, Margeat E, Chavanieu A, Auge MT, Robert B, Arold S, Rimsky S, Kochoyan M. The H-NS dimerization domain defines a new fold contributing to DNA recognition. Nat Struct Biol. 2003;10:212–218. doi: 10.1038/nsb904. [DOI] [PubMed] [Google Scholar]
- Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat Struct Mol Biol. 2007;14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- Bustamante VH, Santana FJ, Calva E, Puente JL. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol Microbiol. 2001;39:664–678. doi: 10.1046/j.1365-2958.2001.02209.x. [DOI] [PubMed] [Google Scholar]
- Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Clarke SC, Haigh RD, Freestone PP, Williams PH. Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin Microbiol Rev. 2003;16:365–378. doi: 10.1128/CMR.16.3.365-378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna B, Casalino M, Fradiani PA, Zagaglia C, Naitza S, Leoni L, Prosseda G, Coppo A, Ghelardini P, Nicoletti M. H-NS regulation of virulence gene expression in enteroinvasive Escherichia coli harboring the virulence plasmid integrated into the host chromosome. J Bacteriol. 1995;177:4703–4712. doi: 10.1128/jb.177.16.4703-4712.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett D, Bennett HJ, Askar H, Green J, Roberts IS. SlyA and H-NS regulate transcription of the Escherichia coli K5 capsule gene cluster, and expression of slyA in Escherichia coli is temperature-dependent, positively autoregulated, and independent of H-NS. J Biol Chem. 2007;282:33326–33335. doi: 10.1074/jbc.M703465200. [DOI] [PubMed] [Google Scholar]
- Delgado J, Forst S, Harlocker S, Inouye M. Identification of a phosphorylation site and functional analysis of conserved aspartic acid residues of OmpR, a transcriptional activator for ompF and ompC in Escherichia coli. Mol Microbiol. 1993;10:1037–1047. doi: 10.1111/j.1365-2958.1993.tb00974.x. [DOI] [PubMed] [Google Scholar]
- Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vázquez A, Barba J, Ibarra JA, O’Donnell P, et al. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman CJ. H-NS: a universal regulator for a dynamic genome. Nat Rev Microbiol. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- Dorman CJ. H-NS, the genome sentinel. Nat Rev Microbiol. 2007;5:157–161. doi: 10.1038/nrmicro1598. [DOI] [PubMed] [Google Scholar]
- Dorman CJ, Hinton JC, Free A. Domain organization and oligomerization among H-NS-like nucleoid-associated proteins in bacteria. Trends Microbiol. 1999;7:124–128. doi: 10.1016/s0966-842x(99)01455-9. [DOI] [PubMed] [Google Scholar]
- Elliott SJ, Sperandio V, Girón JA, Shin S, Mellies JL, Wainwright L, Hutcheson SW, McDaniel TK, Kaper JB. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 2000;68:6115–6126. doi: 10.1128/iai.68.11.6115-6126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison DW, Miller VL. Regulation of virulence by members of the MarR/SlyA family. Curr Opin Microbiol. 2006;9:153–159. doi: 10.1016/j.mib.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Esposito D, Petrovic A, Harris R, Ono S, Eccleston JF, Mbabaali A, Haq I, Higgins CF, Hinton JC, et al. H-NS oligomerization domain structure reveals the mechanism for high order self-association of the intact protein. J Mol Biol. 2002;324:841–850. doi: 10.1016/s0022-2836(02)01141-5. [DOI] [PubMed] [Google Scholar]
- Falconi M, Colonna B, Prosseda G, Micheli G, Gualerzi CO. Thermoregulation of Shigella and Escherichia coli EIEC pathogenicity. A temperature-dependent structural transition of DNA modulates accessibility of virF promoter to transcriptional repressor H-NS. EMBO J. 1998;17:7033–7043. doi: 10.1093/emboj/17.23.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang FC, Rimsky S. New insights into transcriptional regulation by H-NS. Curr Opin Microbiol. 2008;11:113–120. doi: 10.1016/j.mib.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg D, Umanski T, Fang Y, Rosenshine I. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol Microbiol. 1999;34:941–952. doi: 10.1046/j.1365-2958.1999.01655.x. [DOI] [PubMed] [Google Scholar]
- Garmendia J, Frankel G, Crepin VF. Enteropathogenic and enterohemorrhagic Escherichia coli infections: translocation, translocation, translocation. Infect Immun. 2005;73:2573–2585. doi: 10.1128/IAI.73.5.2573-2585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MD, Johnson M, Hinton JC, Williams PH. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol Microbiol. 2001;41:549–559. doi: 10.1046/j.1365-2958.2001.02526.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Duarte OG, Kaper JB. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant AJ, Farris M, Alefounder P, Williams PH, Woodward MJ, O’Connor CD. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC) Mol Microbiol. 2003;48:507–521. doi: 10.1046/j.1365-2958.2003.t01-1-03447.x. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack KR, Robinson CL, Miller KJ, Fowlkes JW, Mellies JL. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect Immun. 2003;71:384–392. doi: 10.1128/IAI.71.1.384-392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AM, Qiu Y, Yeh N, Blattner FR, Durfee T, Jin DJ. SspA is required for acid resistance in stationary phase by downregulation of H-NS in Escherichia coli. Mol Microbiol. 2005;56:719–734. doi: 10.1111/j.1365-2958.2005.04567.x. [DOI] [PubMed] [Google Scholar]
- Heroven AK, Nagel G, Tran HJ, Parr S, Dersch P. RovA is autoregulated and antagonizes H-NS-mediated silencing of invasin and rovA expression in Yersinia pseudotuberculosis. Mol Microbiol. 2004;53:871–888. doi: 10.1111/j.1365-2958.2004.04162.x. [DOI] [PubMed] [Google Scholar]
- Ide T, Michgehl S, Knappstein S, Heusipp G, Schmidt MA. Differential modulation by Ca2+ of type III secretion of diffusely adhering enteropathogenic Escherichia coli. Infect Immun. 2003;71:1725–1732. doi: 10.1128/IAI.71.4.1725-1732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyoda S, Watanabe H. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157 : H7 to HEp-2 cells. Microbiology. 2004;150:2357–2571. doi: 10.1099/mic.0.27100-0. [DOI] [PubMed] [Google Scholar]
- Kanamaru K, Aiba H, Mizuno T. Transmembrane signal transduction and osmoregulation in Escherichia coli: I. Analysis by site-directed mutagenesis of the amino acid residues involved in phosphotransfer between the two regulatory components, EnvZ and OmpR. J Biochem. 1990;108:483–487. doi: 10.1093/oxfordjournals.jbchem.a123225. [DOI] [PubMed] [Google Scholar]
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- Kenny B, Finlay BB. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc Natl Acad Sci U S A. 1995;92:7991–7995. doi: 10.1073/pnas.92.17.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny B, Abe A, Stein M, Finlay BB. Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect Immun. 1997;65:2606–2612. doi: 10.1128/iai.65.7.2606-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Rosenshine I, Tung SL, Wang XH, Friedberg D, Hew CL, Leung KY. Comparative proteomic analysis of extracellular proteins of enterohemorrhagic and enteropathogenic Escherichia coli strains and their ihf and ler mutants. Appl Environ Microbiol. 2004;70:5274–5282. doi: 10.1128/AEM.70.9.5274-5282.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lithgow JK, Haider F, Roberts IS, Green J. Alternate SlyA and H-NS nucleoprotein complexes control hlyE expression in Escherichia coli K-12. Mol Microbiol. 2007;66:685–698. doi: 10.1111/j.1365-2958.2007.05950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison K, Oropeza R, Kenney LJ. The linker region plays an important role in the interdomain communication of the response regulator OmpR. J Biol Chem. 2002;277:32714–32721. doi: 10.1074/jbc.M204122200. [DOI] [PubMed] [Google Scholar]
- Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per regulon of enteropathogenic Escherichia coli:identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- Mellies JL, Barron AM, Carmona AM. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infect Immun. 2007a;75:4199–4210. doi: 10.1128/IAI.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellies JL, Haack KR, Galligan DC. SOS regulation of the type III secretion system of enteropathogenic Escherichia coli. J Bacteriol. 2007b;189:2863–2872. doi: 10.1128/JB.01859-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Mukerji M, Mahadevan S. Characterization of the negative elements involved in silencing the bgl operon of Escherichia coli:possible roles for DNA gyrase, H-NS, and CRP-cAMP in regulation. Mol Microbiol. 1997;24:617–627. doi: 10.1046/j.1365-2958.1997.3621725.x. [DOI] [PubMed] [Google Scholar]
- Müller CM, Dobrindt U, Nagy G, Emèody L, Uhlin BE, Hacker J. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J Bacteriol. 2006;188:5428–5438. doi: 10.1128/JB.01956-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- Porter ME, Mitchell P, Free A, Smith DG, Gally DL. The LEE1 promoters from both enteropathogenic and enterohemorrhagic Escherichia coli can be activated by PerC-like proteins from either organism. J Bacteriol. 2005;187:458–472. doi: 10.1128/JB.187.2.458-472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumer L, Jores J, Kirsch P, Cavignac Y, Zehmke K, Wieler LH. Dissemination of pheU- and pheV-located genomic islands among enteropathogenic (EPEC) and enterohemorrhagic (EHEC) E. coli and their possible role in the horizontal transfer of the locus of enterocyte effacement (LEE) Int J Med Microbiol. 2003;292:463–475. doi: 10.1078/1438-4221-00229. [DOI] [PubMed] [Google Scholar]
- Russell RM, Sharp FC, Rasko DA, Sperandio V. QseA and GrlR/GrlA regulation of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli. J Bacteriol. 2007;189:5387–5392. doi: 10.1128/JB.00553-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-SanMartín C, Bustamante VH, Calva E, Puente JL. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J Bacteriol. 2001;183:2823–2833. doi: 10.1128/JB.183.9.2823-2833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp FC, Sperandio V. QseA directly activates transcription of LEE1 in enterohemorrhagic Escherichia coli. Infect Immun. 2007;75:2432–2440. doi: 10.1128/IAI.02003-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, Castanie-Cornet MP, Foster JW, Crawford JA, Brinkley C, Kaper JB. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol Microbiol. 2001;41:1133–1150. doi: 10.1046/j.1365-2958.2001.02570.x. [DOI] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Mellies JL, Nguyen W, Shin S, Kaper JB. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperandio V, Mellies JL, Delahay RM, Frankel G, Crawford JA, Nguyen W, Kaper JB. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol Microbiol. 2000;38:781–793. doi: 10.1046/j.1365-2958.2000.02168.x. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Giron JA, Kaper JB. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157 : H7. J Bacteriol. 2001;183:5187–5197. doi: 10.1128/JB.183.17.5187-5197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurio R, Falconi M, Brandi A, Pon CL, Gualerzi CO. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stella S, Spurio R, Falconi M, Pon CL, Gualerzi CO. Nature and mechanism of the in vivo oligomerization of nucleoid protein H-NS. EMBO J. 2005;24:2896–2905. doi: 10.1038/sj.emboj.7600754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobe T, Beatson SA, Taniguchi H, Abe H, Bailey CM, Fivian A, Younis R, Matthews S, Marches O, et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci U S A. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Lopez-Sanchez GN, Milflores-Flores L, Patel SD, Rojas-Lopez M, Martinez de la Pena CF, Arenas-Hernandez MM, Martinez-Laguna Y. Ler and H-NS, regulators controlling expression of the long polar fimbriae of Escherichia coli O157 : H7. J Bacteriol. 2007;189:5916–5928. doi: 10.1128/JB.00245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Becker G, Hengge R. The molecular basis of selective promoter activation by the σS subunit of RNA polymerase. Mol Microbiol. 2007;63:1296–1306. doi: 10.1111/j.1365-2958.2007.05601.x. [DOI] [PubMed] [Google Scholar]
- Ueguchi C, Suzuki T, Yoshida T, Tanaka K, Mizuno T. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J Mol Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- Ueguchi C, Seto C, Suzuki T, Mizuno T. Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J Mol Biol. 1997;274:145–151. doi: 10.1006/jmbi.1997.1381. [DOI] [PubMed] [Google Scholar]
- Umanski T, Rosenshine I, Friedberg D. Thermoregulated expression of virulence genes in enteropathogenic Escherichia coli. Microbiology. 2002;148:2735–2744. doi: 10.1099/00221287-148-9-2735. [DOI] [PubMed] [Google Scholar]
- Walthers D, Tran VK, Kenney LJ. Interdomain linkers of homologous response regulators determine their mechanism of action. J Bacteriol. 2003;185:317–324. doi: 10.1128/JB.185.1.317-324.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RM, Rimsky S, Buc H. Probing the structure,function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J Bacteriol. 1996;178:4335–4343. doi: 10.1128/jb.178.15.4335-4343.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson HS, Free A. A truncated H-NS-like protein from enteropathogenic Escherichia coli acts as an H-NS antagonist. Mol Microbiol. 2005;55:808–827. doi: 10.1111/j.1365-2958.2004.04421.x. [DOI] [PubMed] [Google Scholar]
- Yu RR, DiRita VJ. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol Microbiol. 2002;43:119–134. doi: 10.1046/j.1365-2958.2002.02721.x. [DOI] [PubMed] [Google Scholar]