Abstract
Networks of no-take marine protected areas (MPAs) have been widely advocated for the conservation of marine biodiversity. But for MPA networks to be successful in protecting marine populations, individual MPAs must be self-sustaining or adequately connected to other MPAs via dispersal. For marine species with a dispersive larval stage, populations within MPAs require either the return of settlement-stage larvae to their natal reserve or connectivity among reserves at the spatial scales at which MPA networks are implemented. To date, larvae have not been tracked when dispersing from one MPA to another, and the relative magnitude of local retention and connectivity among MPAs remains unknown. Here we use DNA parentage analysis to provide the first direct estimates of connectivity of a marine fish, the orange clownfish (Amphiprion percula), in a proposed network of marine reserves in Kimbe Bay, Papua New Guinea. Approximately 40% of A. percula larvae settling into anemones in an island MPA at 2 different times were derived from parents resident in the reserve. We also located juveniles spawned by Kimbe Island residents that had dispersed as far as 35 km to other proposed MPAs, the longest distance that marine larvae have been directly tracked. These dispersers accounted for up to 10% of the recruitment in the adjacent MPAs. Our findings suggest that MPA networks can function to sustain resident populations both by local replenishment and through larval dispersal from other reserves. More generally, DNA parentage analysis provides a direct method for measuring larval dispersal for other marine organisms.
Keywords: Amphiprion percula, connectivity, parentage analyses, self-recruitment
Networks of no-take marine protected areas (MPAs) have been widely recommended for both biodiversity protection and fishery management (1–3), and an increasing number of networks are being planned and implemented (4–6). To promote population persistence, MPAs must be simultaneously self-sustaining (7, 8) and linked to other protected areas to promote recovery from local extinctions (9–12). Most marine animals produce tiny larvae with pelagic durations ranging from days to months. Prevailing oceanographic currents may transport these propagules over large distances to form demographically “open” populations that are linked by larval dispersal (2, 13). Recent evidence from such diverse fields as physical oceanography (14, 15), molecular genetics (16, 17), and otolith chemistry (18, 19) suggest that at least some larvae return to the same subpopulation as their parents. The spatial scale over which marine populations are connected by larval dispersal continues to generate controversy, however, due to a lack of empirical data on how far larvae travel (20, 21). This information is critical in practical terms, because the degree of connectivity among geographic areas sets the scale at which management strategies for exploited marine species need to be applied.
One direct method of measuring larval dispersal distances relies on identifying the location of an individual's parents using highly polymorphic genetic markers and probability-based assignment techniques. Several recent studies using microsatellite DNA markers for paternity analysis have focused on reproductive behavior and variability in male reproductive success (22, 23). The parents of an individual also can be identified based on likelihood statistics using hypervariable microsatellite markers. DNA parentage analysis also allows the determination of an individual's natal origin if the parents' location at the time of conception is known. But although parentage analysis has been used to determine levels of local replenishment, this approach has not yet been used to estimate connectivity among subpopulations of marine fishes.
We have conducted the first large-scale application of DNA parentage analysis to examine larval connectivity among marine reserves in Kimbe Bay, New Britain, Papua New Guinea (Fig. 1 A and B). Our field study focused on orange clownfish (Amphiprion percula) at Kimbe Island, an isolated oceanic island lying 30 km from the nearest point of land in the Bismark Sea (Fig. 1 C and D). At Kimbe Island, a node in a proposed network of MPAs that extends 180 km across Kimbe Bay (6), we previously found evidence of high local replenishment to a population of ≈200 adults living in anemones on shallow reefs near the island (24). Here we generate the first direct estimates of larval connectivity in a marine fish species by identifying the parents of A. percula larvae that returned to Kimbe Island and those that dispersed to adjacent subpopulations up to 35 km away. These data provide evidence that larval subsidies from a single reserve may contribute to the resilience of subpopulations at other reserves within a network of MPAs.
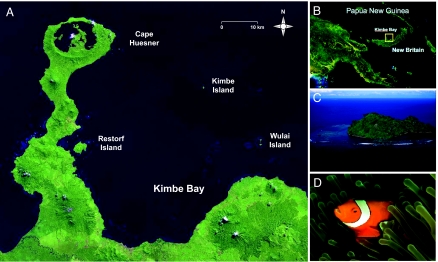
Fig. 1.
Location maps and focal species. (A) LANDSAT satellite image of western Kimbe Bay showing the study sites. (B) Location of Kimbe Bay on the north side of New Britain, Papua New Guinea. (C) Aerial photograph of Kimbe Island showing lagoonal habitats in which A. percula are concentrated in the study area. (Photo courtesy of Tami Pelusi.) (D) A. percula sheltering in an anemone, Kimbe Bay. (Photo courtesy of Simon Thorrold.)
Results
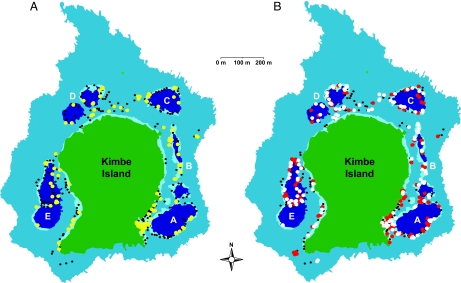
We screened 16 polymorphic microsatellite DNA markers from a total of 506 potential A. percula parents at Kimbe Island sampled in December 2004, which were assumed to represent the entire population. We then collected 400 newly settled juvenile A. percula from Kimbe Island in December 2004 and April 2005 and also from anemones at Wulai Island, Cape Huessner, and Restorf Island in April 2005 (Fig. 1A). Of these 400 juveniles, 122 were identified as being progeny of adults at Kimbe Island, based on a parentage assignment by a maximum likelihood procedure (see Materials and Methods). We tested the accuracy of the DNA parentage method by comparing results with those from transgenerational isotope labeling of the same adults, and found 100% agreement between the 2 techniques (see Materials and Methods). Our estimates of local replenishment to adult A. percula colonies around Kimbe Island in December 2004 and April 2005 were remarkably similar. Of the 133 juveniles collected at Kimbe Island in December 2004, 56 (42%) were identified as being progeny of Kimbe Island adults, compared with 51 of 121 juveniles (42%) collected in April 2005 (Fig. 2). The species of host anemone that the parents inhabited had little observable effect on the propensity for larvae to return to Kimbe Island. The percentage of larvae originating from adults living in Heteractus magnifica and Stichodactyla haddoni (57% and 43%, respectively) was similar to the relative abundance of adult occupancy in each anemone (62% and 38%, respectively). Finally, unlike in our earlier study (24), here DNA parentage analysis allowed us to document levels of local replenishment at the individual lagoon scale (Table 1). In 2004, 23 of the 56 recruits (41%) that were spawned by adults at Kimbe Island settled back into the lagoon from which they originated, compared with 18 of 51 (33%) in 2005. But the Kimbe Island connectivity matrix revealed no relationship between the magnitude of connectivity and the distance between the lagoons (Table 1); that is, an individual lagoon was not necessarily more closely connected to lagoons that were close by compared with those that were farther away.
Fig. 2.
Map of locations of all anemones in each of 5 lagoons (A– E) that harbored adult or juvenile A. percula around Kimbe Island. (A) Location of anemones with adult A. percula that either produced larvae that subsequently settled into anemones around Kimbe Island (yellow symbols) or did not produce larvae that returned to Kimbe Island (black symbols). (B) Location of anemones with recently settled juvenile A. percula that either were progeny of Kimbe Island adults (red symbols) or had dispersed from reefs at least 6 km away from Kimbe Island (white circles).
Table 1.
A. percula connectivity matrix among 5 lagoons surrounding Kimbe Island, calculated by identifying the natal origins of juveniles collected during 2 sampling trips in December 2004 and April 2005
| Settlement lagoon, December 2004 collection |
|||||
|---|---|---|---|---|---|
| A (n = 32) | B (n = 20) | C (n = 11) | D (n = 32) | E (n = 38) | |
| Natal lagoon | |||||
| A | 12 | 3 | 1 | 1 | 1 |
| B | 3 | 1 | 0 | 3 | 3 |
| C | 0 | 1 | 0 | 0 | 2 |
| D | 2 | 2 | 3 | 4 | 5 |
| E | 0 | 1 | 1 | 0 | 6 |
| Settlement lagoon, April 2005 collection | |||||
| A (n = 29) | B (n = 23) | C (n = 18) | D (n = 18) | E (n = 37) | |
| Natal lagoon | |||||
| A | 5 | 1 | 1 | 1 | 5 |
| B | 4 | 2 | 2 | 0 | 2 |
| C | 2 | 2 | 2 | 1 | 0 |
| D | 3 | 1 | 0 | 1 | 1 |
| E | 5 | 1 | 1 | 1 | 7 |
Lagoon A, 69 anemones (nane), 125 adults screened (nscr); lagoon B, nane = 38, nscr = 73; lagoon C, nane = 29, nscr = 58; lagoon D, nane = 59, nscr = 105; lagoon E, nane = 75, nscr = 145.
All juveniles in the analysis were identified as being the progeny of Kimbe Island adults using DNA parentage analysis. Bold figures on the matrix diagonal indicate the number of juveniles that returned to their natal lagoon.
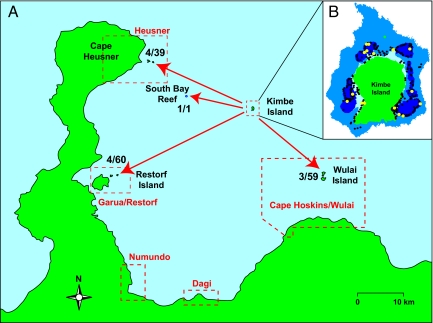
We also identified 15 juveniles from Kimbe Island parents at surrounding reefs in Kimbe Bay (Fig. 3). The parents of the only juvenile collected on South Bay Reef were located on Kimbe Island, confirming that larvae are capable of dispersing at least 6 km and successfully settling into adult habitat. Perhaps more surprisingly, we also found juveniles that had been spawned on Kimbe Island reefs at the other 3 Kimbe Bay locations, indicating a larval dispersal of 15–35 km. These individuals represented a significant proportion of the total number of juveniles collected at each location. Kimbe Island contributed 10% (5 of 50) of the juveniles collected at Restorf Island, 6% (6 of 105) of those collected at Cape Huessner, and 5% (3 of 56) of those collected at Wulai Island.
Fig. 3.
Larval dispersal of A. percula from Kimbe Island to other designated marine reserves in western Kimbe Bay. (A) Proportion of recently settled juvenile A. percula collected at each of 4 locations that were progeny of Kimbe Island A. percula. The red boxes outline proposed reserve boundaries (6). (B) Location of adult A. percula that produced larvae that successfully dispersed and settled on anemones away from Kimbe Island (yellow symbols).
Discussion
Our approach, based on quantifying the spatial relationships between parents and their progeny, provides a powerful tool for assessing local replenishment and connectivity in marine populations that may be applied to other species as well. Although other mass-marking techniques have demonstrated that a significant proportion of juveniles return to their natal population (8, 24, 25), DNA parentage analysis provides information on the individual parents responsible for successful recruitment and the actual larval dispersal distances of individuals. Levels of local replenishment to Kimbe Island (≈40%) were remarkably stable over time and within the ranges reported for Pomacentrus amboinensis (25) and Amphiprion polymnus (8). We also found the longest direct measure of larval dispersal distance for any marine fish species reported to date. We located juveniles known to have traveled up to 35 km from their natal population. These long-distance dispersers contributed up to 10% of the individuals that had recently settled into the subpopulations that they joined.
The full geographic extent of this metapopulation remains to be determined, but given that A. percula has a relatively short pelagic larval duration for a reef fish (≈11 days) (24), significant demographic connectivity between the nodes of the Kimbe Bay MPA network appears to be likely for most other reef fishes as well. Lying ≈30 km offshore of the north coast of New Britain in the Bismarck Sea, Kimbe Island is located within a dynamic hydrodynamic regime subject to eddies originating from instabilities in the South Equatorial Current and New Guinea Coastal Current (26). Biophysical modeling for the tropical western Pacific region also suggests high levels of connectivity in regions where reefs are only 20–30 km apart, including species with a wide range of pelagic larval durations (27). Levels of both local replenishment and connectivity for A. percula appear to be demographically significant and likely contribute to the persistence of discrete populations within the larger metapopulation. Furthermore, the finding of a similar level of local replenishment at Kimbe Island in butterflyfish, with a pelagic larval duration of ≈35 days (24), suggests that our approach also applies to larger reef fish species with longer pelagic larval durations.
DNA parentage analysis offers new and largely unexplored opportunities to directly measure larval dispersal in marine ecosystems. This information is vital to the spatial management of marine species, particularly for evaluating the optimal size and spacing of reserves in MPA networks (21, 28). The dispersal pattern that we found supports the contention that individual marine reserves can be of a size that offers protection of resident populations and spaced within a network to allow for significant exchange of dispersers among reserves (24, 29). Although the number of dispersing larvae detected in our study was small, the focal reserve accounted for a significant proportion of the recruitment in the adjacent reserves in Kimbe Bay. Theory suggests that low rates of migration often are sufficient to rescue individual populations from local extinction (10, 11); thus, it is encouraging to find that for an MPA network designed with limited information on larval dispersal (6), one iconic reef fish species appears to experience the conservation benefits of both local replenishment and larval connectivity among reserves. Given the increasing evidence that populations of many small reef fish are in decline (30–33), parentage analysis has the potential to play a key role in developing conservation strategies for other species and in different regional settlings.
Materials and Methods
The orange clownfish (A. percula) lives in a mutualistic association with 2 anemone species, H. magnifica and S. haddoni (34). Female A. percula spawn benthic eggs that hatch after ≈7 days of parental care. Larvae then spend ≈11 days in the pelagic environment before settling into an anemone, where they will remain for the rest of their lives. We located a total of 270 anemones occupied by colonies of A. percula around Kimbe Island in December 2004. The 2 largest fish in each colony were captured, fin-clipped, and injected with an enriched Ba isotope solution underwater (24). Another collection, in April 2005, was targeted at juvenile A. percula that were recruited between December 2004 and April 2005. The finclips were preserved in 90% ethanol, and the adult fish were immediately returned to their anemones. We screened a total of 506 adults and 469 juveniles for 16 microsatellite DNA loci that satisfied Hardy-Weinberg equilibrium assumptions. Missing values accounted for 7.3% of the data distributed over all loci but were concentrated in only a few individuals, likely due to the original quality of DNA. We used the FAMOZ platform (35) to assign juveniles back to Kimbe Island adults. Of the 122 new recruits assigned to parents in the Kimbe Island population, 68 (56%) were independently assigned to a female and a male within a single anemone, and 54 (44%) were assigned to a single parent (male or female), likely due to blanks during the scoring of microsatellite loci or to missed adults during the collection process. Nonetheless, we never assigned a juvenile to parents of 2 different anemones or 2 males or 2 females. We evaluated the DNA parentage analysis by comparing the results with those from a previous study based on transgenerational isotope labeling (TRAIL) using an enriched Ba isotope (24, 36). The TRAIL technique identified 9 of the 15 juveniles collected in February 2005 as being spawned by Kimbe Island adults. Our DNA parentage analysis confirmed that only these same 9 juveniles identified by the TRAIL technique originated from Kimbe Island.
Acknowledgments.
This research was supported by the Australian Research Council, the Coral Reef Initiatives for the Pacific (CRISP), the Global Environmental Facility CRTR Connectivity Working Group, the National Science Foundation, the ARC Centre of Excellence for Coral Reef Studies, the Nature Conservancy, Total Foundation, James Cook University, and the Woods Hole Oceanographic Institution. We thank Glenn Almany, Michael Berumen, Caroline Hervet, Vanessa Messmer, Craig Syms, and Maya Srinivasan for providing assistance in the field; the Mahonia Na Dari Research and Conservation Centre, Walindi Plantation Resort, the Nature Conservancy, and the crew of M.V. FeBrina for providing essential logistic support; and the traditional owners for allowing us access to their reefs.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Agardy MT. Advances in marine conservation: The role of marine protected areas. Trends Ecol Evol. 1994;9:267–270. doi: 10.1016/0169-5347(94)90297-6. [DOI] [PubMed] [Google Scholar]
- 2.Roberts CM. Connectivity and management of Caribbean coral reefs. Science. 1997;278:1454–1457. doi: 10.1126/science.278.5342.1454. [DOI] [PubMed] [Google Scholar]
- 3.Lubchenco J, Palumbi SR, Gaines SD, Andelman S. Plugging a hole in the ocean: The emerging science of marine reserves. Ecol Appl. 2003;13:S3–S7. [Google Scholar]
- 4.Airame S, et al. Applying ecological criteria to marine reserve design: A case study from the California Channel Islands. Ecol Appl. 2003;13:S170–S184. [Google Scholar]
- 5.Fernandes L, et al. Establishing representative no-take areas in the Great Barrier Reef: Large-scale implementation of theory on marine protected areas. Conserv Biol. 2005;19:1733–1744. [Google Scholar]
- 6.Green A, et al. Designing a resilient network of marine protected areas for Kimbe Bay, Papua New Guinea. Oryx. in press. [Google Scholar]
- 7.Hastings A, Botsford LW. Persistence of spatial populations depends on returning home. Proc Natl Acad Sci U S A. 2006;103:6067–6072. doi: 10.1073/pnas.0506651103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones GP, Planes S, Thorrold SR. Coral reef fish larvae settle close to home. Curr Biol. 2005;15:1314–1318. doi: 10.1016/j.cub.2005.06.061. [DOI] [PubMed] [Google Scholar]
- 9.Clinchy M. Does immigration “rescue” populations from extinction? Implications regarding movement corridors and the conservation of mammals. Oikos. 1997;80:618–622. [Google Scholar]
- 10.Gonzales A, Lawton JH, Gilbert FS, Blackburn TM, Evans-Freke I. Metapopulation dynamics, abundance, and distribution in a microecosystem. Science. 1998;281:2045–2047. doi: 10.1126/science.281.5385.2045. [DOI] [PubMed] [Google Scholar]
- 11.Hill MF, Hastings A, Botsford LW. The effects of small dispersal rates in extinction times in structured metapopulations models. Am Nat. 2002;160:389–402. doi: 10.1086/341526. [DOI] [PubMed] [Google Scholar]
- 12.Allison GW, Gaines SD, Lubchenco J, Possingham HP. Ensuring persistence of marine reserves: Catastrophes require adopting an insurance factor. Ecol Appl. 2003;13:S8–S24. [Google Scholar]
- 13.Roughgarden J, Gaine S, Possingham H. Recruitment dynamics in complex life cycles. Science. 1988;241:1460–1466. doi: 10.1126/science.11538249. [DOI] [PubMed] [Google Scholar]
- 14.Cowen RK, Lwiza KM, Sponaugle S, Paris CB, Olson DB. Connectivity of marine populations: Open or closed? Science. 2000;287:857–859. doi: 10.1126/science.287.5454.857. [DOI] [PubMed] [Google Scholar]
- 15.Cowen RK, Paris CB, Srinivasan A. Scaling of connectivity in marine populations. Science. 2006;311:522–527. doi: 10.1126/science.1122039. [DOI] [PubMed] [Google Scholar]
- 16.Barber PH, Moosa MK, Palumbi SR. Rapid recovery of genetic diversity of stomatopod population on Krakatau: Temporal and spatial scales of marine larval dispersal. Proc R Soc London Ser B. 2002;269:1591–1597. doi: 10.1098/rspb.2002.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilg MR, Hilbish TJ. The geography of marine larval dispersal: Coupling genetics with fine-scale physical oceanography. Ecology. 2003;84:2989–2998. [Google Scholar]
- 18.Swearer SE, Caselle JE, Lea DW, Warner RR. Larval retention and recruitment in an island population of a coral reef fish. Nature. 1999;402:799–802. [Google Scholar]
- 19.Thorrold SR, Latkoczy C, Swart PK, Jones CM. Natal homing in a marine fish metapopulation. Science. 2001;291:297–299. doi: 10.1126/science.291.5502.297. [DOI] [PubMed] [Google Scholar]
- 20.Mora C, Sale PF. Are populations of coral reef fish open or closed ? Trends Ecol Evol. 2002;9:422–428. [Google Scholar]
- 21.Sale PF, et al. Critical science gaps impede use of no-take fishery reserves. Trends Ecol Evol. 2005;20:74–80. doi: 10.1016/j.tree.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Krützen M, Barre LM, Connor RC, Mann J, Sherwin B. “O father, where art thou?”: Paternity assessment in an open fission–fusion society of wild bottlenose dolphins (Tursiops sp.) in Shark Bay, Western Australia. Mol Ecol. 2004;13:1975–1990. doi: 10.1111/j.1365-294X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- 23.MacKiewicz M. A genetic assessment of parentage in a natural population of dollar sunfish (Lepomis marginatus) based on microsatellite markers. Mol Ecol. 2002;11:1877–1833. doi: 10.1046/j.1365-294x.2002.01577.x. [DOI] [PubMed] [Google Scholar]
- 24.Almany GR, Berumen ML, Thorrold SR, Planes S, Jones GP. Local replenishment of coral reef fish populations in a marine reserve. Science. 2007;316:742–744. doi: 10.1126/science.1140597. [DOI] [PubMed] [Google Scholar]
- 25.Jones GP, Milicich MJ, Emslie MJ, Lunow C. Self-recruitment in a coral reef fish population. Nature. 1999;402:802–804. [Google Scholar]
- 26.Steinberg CR, Choukroun SM, Slivkoff MM, Mahoney MV, Brinkman RM. Brisbane: The Nature Conservancy; 2006. Currents in the Bismarck Sea and Kimbe Bay, Papua New Guinea. Australian Institute of Marine Science and The Nature Conservancy. TNC Pacific Island Countries Report 6/06. [Google Scholar]
- 27.Treml EA, Halpin PN, Urban DL, Pratson LF. Modeling population connectivity by ocean currents: A graph-theoretic approach for marine conservation. Land Ecol. 2008;23:19–36. [Google Scholar]
- 28.Hastings A, Botsford LW. Comparing designs of marine reserves for fisheries and for biodiversity. Ecol Appl. 2003;13:S65–S70. [Google Scholar]
- 29.Halpern BS, Warner RR. Matching marine reserve function to stakeholder needs. Proc R Soc London Ser B. 2003;270:1871–1878. doi: 10.1098/rspb.2003.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones AM, Gardner S, Sinclair W. Losing “Nemo”: Bleaching and collection appear to reduce inshore populations of anemonefishes. J Fish Biol. 2008;73:753–761. [Google Scholar]
- 31.Jones GP, McCormick MI, Srinivasan M, Eagle JV. Coral decline threatens fish biodiversity in marine reserves. Proc Natl Acad Sci U S A. 2004;101:8251–8253. doi: 10.1073/pnas.0401277101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munday PL. Habitat loss, resource specialization, and extinction on coral reefs. Global Change Biol. 2004;10:1642–1647. [Google Scholar]
- 33.Graham NAJ, et al. Dynamic fragility of oceanic coral reef ecosystems. Proc Natl Acad Sci U S A. 2006;103:8425–8429. doi: 10.1073/pnas.0600693103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fautin DG, Allen GR. Anemonefishes and Their Host Anemones. Perth, Australia: Western Australian Museum; 1997. [Google Scholar]
- 35.Gerber S, Chabrier P, Kremer A. FAMOZ: A software analysis using dominant, codominant and uniparentally inherited markers. Mol Ecol Notes. 2003;3:479–481. [Google Scholar]
- 36.Thorrold SR, Jones GP, Planes S, Hare JA. Transgenerational marking of embryonic otoliths in marine fishes using barium stable isotopes. Can J Fish Aquat Sci. 2006;63:1193–1197. [Google Scholar]