Abstract
IL-10 is a key regulator of the immune system that critically determines health and disease. Its expression is finely tuned both at the transcriptional and posttranscriptional levels. Although the importance of posttranscriptional regulation of IL-10 has been previously shown, understanding the underlying mechanisms is still in its infancy. In this study, using a combination of bioinformatics and molecular approaches, we report that microRNA (hsa-miR-106a) regulates IL-10 expression. The hsa-miR-106a binding site in the 3′ UTR of IL10 has been identified by site-directed mutagenesis studies. Also, the involvement of transcription factors, Sp1 and Egr1, in the regulation of hsa-miR-106a expression and concomitant decrease in the IL-10 expression, has also been demonstrated. In summary, our results showed that IL-10 expression may be regulated by miR-106a, which is in turn transcriptionally regulated by Egr1 and Sp1.
Keywords: Egr1, IL-10, microRNA, SP1, miR-106a promoter
IL-10 is a key orchestrator of the immune system that has been shown to be antiinflammatory in many model systems (1). It is secreted by various cell types like different T cell subsets, macrophages, dendritic cells (DC), B-cells, mast cells, and eosinophils. It mediates a plethora of immunoregulatory events such as maturation and activation of macrophages and DC, expression of MHC-II and B-7, activation of T cells, synthesis of cytokines, and antibody production (2, 3). Also, dysregulation of IL-10 leads to various immunological diseases, such as cancer, rheumatoid arthritis, asthma, infectious disorders, etc. (2). Therefore, it is likely that IL-10 expression is tightly regulated.
Regulation of IL-10 expression has been studied at the transcriptional and posttranscriptional levels. Although Sp1 and Sp3 have been found to regulate transcription, it has been shown that IL10 mRNA is constitutively transcribed in many cells; however, the availability of its protein level is significantly determined by posttranscriptional mechanisms (4, 5). AU-rich elements (ARE) in the 3′ UTR of mouse IL10 that lead to the degradation of its mRNA have been shown (6). Half life of IL10 mRNA in normal melanocytes is much shorter than in melanoma cell lines (7). Also, genetic variations in the 3′ UTR of IL10 have been shown to be associated with IL-10 levels that could lead to disease pathogenesis (8, 9). Therefore, a pellucid understanding of the posttranscriptional regulation of IL10 will be of scientific and clinical significance.
A growing class of noncoding RNAs called microRNAs (miRNAs) is involved in posttranscriptional regulation of genes (10). MicroRNAs have been reported to modulate hematopoietic lineage differentiation (11), angiogenesis (12), cell adhesion (13), etc., indicating that they could have important roles in numerous biological processes (14). There is also a growing body of literature supporting the potential role of miRNAs in regulation of inflammatory processes (15). The importance of IL-10 in the orchestration of the immune response, and the strong evidence for its posttranscriptional regulation, makes it an attractive candidate for miRNA mediated regulation of inflammation.
Here, we report the identification of a miRNA, hsa-miR-106a, that regulates IL-10 expression. We also find that Sp1 and Egr1 have an important role in hsa-miR-106a transcription and, thus, indirectly regulate the expression of IL-10 posttranscriptionally.
Results
Prediction of miRNA Regulators of IL-10.
To predict the miRNA that may regulate IL-10, we used a consensus approach by employing 3 different miRNA target prediction software, as described in Materials and Methods. We found that 8 miRNAs have potential binding site in the 3′ UTR of IL10 (Table 1). Also, using miRex, we found that 5 miRNAs (namely hsa-miR-106a, miR-106b, miR-20a, miR-20b, and miR-93) were expressed in cells of lymphoid and myeloid origin, which are known to express IL-10. Because information regarding the expression of the remaining 3 miRNAs was not available, we focused our study on hsa-miR-106a, miR-106b, miR-20a, miR-20b, and miR-93.
Table 1.
A list of predicted miRNAs (in the decreasing order of binding free energy) having potential binding sites in the 3′ UTR of IL10
| MicroRNA | Start site | End site | Binding free energy, Kcal/mol | Expression status in lymphoid and myeloid origin cells |
|---|---|---|---|---|
| hsa-miR-20b_MIMAT0001413 | 625 | 647 | −24.32 | Yes |
| hsa-miR-640_ MIMAT0003310 | 654 | 670 | −24.16 | No data |
| hsa-miR-93_ MIMAT0000093 | 625 | 646 | −23.88 | Yes |
| hsa-miR-20a_ MIMAT0000075 | 625 | 647 | −22.21 | Yes |
| hsa-miR-106b_ MIMAT0003310 | 627 | 647 | −20.77 | Yes |
| hsa-miR-597_ MIMAT0003265 | 249 | 270 | −20.52 | No data |
| hsa-miR-372_ MIMAT0000724 | 619 | 646 | −20.46 | No data |
| hsa-miR-106a_ MIMAT0003310 | 621 | 647 | −20.35 | Yes |
Identification of hsa-miR-106a As a Regulator of IL-10 Expression.
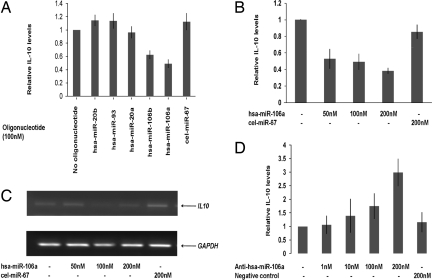
To identify the miRNA(s) that may be involved in the regulation of IL-10, initially Raji cell line was chosen, because it constitutively expresses high level of IL-10 and, thus, presumably may have lower level of regulatory miRNA(s). When these cells were transfected with the 5 predicted miRNA oligonucleotides, hsa-miR-106a oligonucleotide significantly down-regulated the expression of IL-10 present in the culture supernatant (Fig. 1A). Interestingly, hsa-miR-106b, which differs from hsa-miR-106a in only 2 nucleotide positions, also affected IL-10 expression, albeit at a lower level. In contrast, transfection with other 3 miRNA or cel-miR-67, a miRNA from Caenorhabditis elegans having no sequence identity with the predicted human miRNAs (used as negative control), had no effect on the IL-10 expression (Fig. 1A). Also, when increasing concentration of hsa-miR-106a was used, IL-10 expression was found to be reduced in a dose-dependent manner (Fig. 1B).
Fig. 1.
Identification of hsa-miR-106a as a regulator of IL-10 expression. (A) Raji cells were transfected with or without the 5 candidate miRNA oligonucleotides individually, or with a negative control oligonucleotide, cel-miR-67 (having no sequence identity with any known human miRNAs); 24 h after transfection, IL-10 was measured from the culture supernatants by ELISA, and plotted as fold change in IL-10 with respect to untransfected control. (B) Raji cells were transfected without or with increasing concentrations of hsa-miR-106a or cel-miR-67, and IL-10 level was measured, as described in Materials and Methods. (C) Raji cells were transfected with or without increasing concentrations of hsa-miR-106a or cel-miR-67 as described in B, and RNA was prepared followed by RT-PCR by using IL10 specific primers. GAPDH RT-PCR was performed as loading control. Representative of 3 independent experiments. (D) Jurkat cells were transfected with or without increasing concentration of anti-hsa-miR-106a oligonucleotide or a negative control oligonucleotide followed by IL-10 measurement in the culture supernatant by ELISA, 24 h after transfection. Results in A, B, and D are presented as mean ± SE.
To check whether hsa-miR-106a affects the stability of IL10 mRNA, RT-PCR was performed by using RNA prepared from Raji cells transfected with increasing concentration of hsa-miR-106a oligonucleotide. It was observed that it down-regulated IL10 mRNA in a concentration-dependent manner (Fig. 1C), whereas cel-miR-67 had no effect. Thus, it is likely that hsa-miR-106a decreases IL-10 expression by degrading its mRNA.
To confirm the involvement of miR-106a further, we chose Jurkat cells that produce low basal level of IL-10, and, thus, may produce higher level of hsa-miR-106a. When Jurkat cells were transfected with increasing concentration of anti-hsa-miR-106a oligonucleotide and IL-10 levels were measured in the culture supernatant, a dose-dependent increase in the levels of IL-10 was observed (Fig. 1D). In contrast, a random oligonucleotide had no effect on IL-10 expression. Thus, these experiments demonstrate that hsa-miR-106a regulates IL-10 expression.
Expression of hsa-miR-106a and Its Involvement in IL-10 Regulation in Different Cells.
To specifically detect the expression of hsa-miR-106a, we standardized Northern blotting conditions so that we specifically detect hsa-miR-106a, not hsa-miR-106b (Fig. 2A). Next, when blot was performed using RNA prepared from A549, Jurkat, Raji, HeLa, Hep-G2, and THP-1 cells, hsa-miR-106a was found to be expressed significantly in Jurkat, Raji, and THP-1 cells (Fig. 2B). However, Hep-G2, HeLa and A549 cells expressed lower amounts of hsa-miR-106a. To correlate the expression of hsa-miR-106a with 3′ UTR regulatory activity, Jurkat, Raji, THP-1, and A549 cells were transfected with pMIR-REPORT-IL10 3′ UTR (intact). A substantial fall in luciferase activity was observed in Jurkat, Raji, and THP-1 cells. In comparison, the decrease in A549 cells was not profound. Thus, the down-regulatory effect of IL10 3′ UTR was found to be correlated with the levels of mature hsa-miR-106a present in these cells (Fig. 2C).
Fig. 2.
Expression of hsa-miR-106a and its involvement in IL-10 regulation in different cells. (A) DNA oligonucleotides of hsa-miR-106a, hsa-miR-106b, and a random sequence of equal length were run on an acrylamide gel and probed with hsa-miR-106a radiolabeled probe. The probe was able to specifically discriminate between 2 forms of the miRNA. (B) Northern blotting was performed with total RNA from A549, Jurkat, Raji, HeLa, Hep-G2, and THP-1 cells by using radiolabeled probe specific for hsa-miR-106a. The membrane was stripped, and Northern blotting was performed for U6, which was used for normalization. Lower panel indicates U6 band. Representative of 3 independent experiments. (C) The pMIR-REPORT-IL10 3′ UTR (intact) was transfected in A549, Jurkat, Raji, and THP-1 cells, and luciferase expression was measured. Relative fold change in luciferase activity for each cell line was plotted with respect to pMIR-REPORT. A dark bar indicates cells transfected with pMIR-REPORT vector, whereas a light bar indicates cells transfected with pMIR-REPORT-IL10 3′ UTR (intact). Results are presented as mean ± SE.
Confirmation of Target Site for hsa-miR-106a in IL10 3′ UTR.
Our bioinformatics analysis predicted the binding of hsa-miR-106a to the 3′ UTR of IL10 in a region encompassing +4451 to +4478 bases (Fig. 3A). When luciferase reporter vector containing the intact 3′ UTR was transfected in Jurkat or Raji cells, a significant down-regulation in the luciferase expression in both cells with respect to control was observed (Fig. 3B). This down-regulation was not seen in mutant 3′ UTR construct lacking the predicted 27 base hsa-miR-106a binding site. Also, when A549 cells, which expresses minimal level of hsa-miR-106a, were cotransfected with hsa-miR-106a oligonucleotide and pMIR-REPORT-IL10 3′ UTR (intact), we observed >2-fold reduction in luciferase expression (Fig. 3C). However, cotransfection with cel-miR-67 did not have any appreciable effect on luciferase expression. Most importantly, when hsa-miR-106a was cotransfected along with pMIR-REPORT-IL10 3′ UTR (mutant), there was minimal reduction in the luciferase expression. Also, cotransfection of pMIR-REPORT-IL10 3′ UTR (mutant) with cel-miR-67 had no effect on luciferase expression (Fig. 3C). These experiments indicate that mature hsa-miR-106a regulates IL-10 expression by interacting with its 3′ UTR.
Fig. 3.
Confirmation of target site for hsa-miR-106a in IL10 3′ UTR. (A) Schematic representation of IL10 3′ UTR indicating the binding site of hsa-miR-106a as predicted. This region encompassing +4451 to +4478 bases was deleted by PCR based site-directed mutagenesis. (B) Luciferase reporter vectors pMIR-REPORT -IL10 3′ UTR (intact) or pMIR-REPORT-IL10 3′ UTR (mutant) were transfected in Jurkat or in Raji cells. The fold change in relative luciferase activity was plotted. Dark and light bars indicate luciferase activity in Jurkat cells and Raji cells, respectively. (C) A549 cell line was cotransfected with hsa-miR-106a oligonucleotide or cel-miR-67 along with either pMIR-REPORT-IL10 3′ UTR (intact) or pMIR-REPORT-IL10 3′ UTR (mutant). The luciferase activity relative to pMIR-REPORT-IL10 3′ UTR (intact) was plotted. Results in B and C are presented as mean ± SE for 3 independent experiments.
Transcriptional Regulation of hsa-miR-106a.
Because the level of hsa-miR-106a was found to vary in different cell lines (Fig. 2B), and this variation could be due to differential regulation of its expression, we sought to investigate the transcriptional regulation of hsa-miR-106a locus present on the X chromosome. Thus, a region encompassing 1 kilobase upstream of the putative transcription start site (TSS) of hsa-miR-106a was analyzed by using Transfac Alibaba prediction software (16). Two transcription factors, Egr1 and Sp1, were predicted to bind to this region (Fig. 4A). When this 1 kilobase putative promoter region was cloned upstream of the luciferase gene (pGL3 basic, a promoter less vector) and transfected in Jurkat cells, we observed a 4-fold induction in the luciferase expression (Fig. 4B); thus, indicating that the putative promoter sequence possessed regulatory activity.
Fig. 4.
Transcriptional regulation of hsa-miR-106a. (A) Schematic representation of the region encompassing the cluster encoding hsa-miR-106a that codes for 6 different miRNAs. TSS is indicated as a dark box is ≈2 kb upstream. Transcription factors, Egr1 (dark circle) and Sp1 (dark oval), were predicted to bind at a region between −305 to −317 base pair. (B) The hsa-miR-106a promoter construct (in pGL3 basic) or pGL3 basic vector was transfected in Jurkat cells, and relative luciferase activity was plotted. Results are presented as mean ± SE of 3 independent experiments. (C) Western blotting was performed by using unstimulated or PHA/PMA (combination) stimulated nuclear extracts from Jurkat cells to determine the expression of Egr1 (panel 1) and Sp1 (panel 2), and equal loading was determined by Lamin (panel 3). (D) EMSA were performed by using end-labeled hsa-miR-106a promoter oligonucleotide. Sp1 binds to the hsa-miR-106a promoter in unstimulated conditions. (E) Egr1 binds to the promoter region on stimulation with PHA/PMA combination at an overlapping binding site. NE, nuclear extract; un, unstimulated; st, stimulated; ODN, oligonucleotide; 106a, hsa-miR-106a promoter oligonucleotide; Egr (M), Egr mutant oligonucleotide; Egr-1 SS, Egr1 super shifted band. (F) Northern blotting (panel 1) was performed by using the RNAs from Jurkat cells stimulated with PMA/ PHA combination for various time points (in hours as H), as indicated. The membrane was stripped and Northern blotting was performed for U6 for normalization; (panel 2) the extreme left lane represents radiolabeled Decade Marker (Ambion). C–F are representative of 3 independent experiments.
To establish the binding of putative transcription factors Sp1 and Egr1, we determined the levels of these proteins using Western blotting of nuclear extracts prepared from unstimulated or stimulated Jurkat cells (PHA/PMA combination for 2 h). Although the basal level of Egr1 in Jurkat cells was undetectable, there was a significant increase after stimulation with PHA/PMA combination (panel 1 in Fig. 4C). However, the level of Sp1 was unaffected by stimulation (panel 2). To demonstrate the binding of Sp1 and Egr1, EMSAs were performed by using end-labeled oligonucleotide probe from hsa-miR-106a potential promoter region (−285 to −317 bases) and nuclear extract from unstimulated (for Sp1) Jurkat cells. Out of the shifted bands (Fig. 4D, lane 2), our cold chase experiments with miR-106a promoter oligonucleotide (lane 3), irrelevant oligonucleotides NF-κB (lane 4), and Sp1 oligonucleotide (lane 5) indicated the uppermost band (with an arrow) could be Sp1. The specificity of Sp1 band was further confirmed by antibody experiments, where anti-Sp1 antibody abolished this uppermost band (lane 6), whereas anti-Sp3 (lane 7) or normal goat IgG (lane 8) could not.
For Egr1, when nuclear extract from stimulated Jurkat cells was incubated with labeled hsa-miR-106a promoter oligonucleotide, only 1 shifted band was observed (Fig. 4E, lane 3). However, the band was absent when unstimulated nuclear extract was used (Fig. 4E, lane 2). Specificity of the shifted band was confirmed by cold chase with excess of Egr consensus oligonucleotide (lane 4), which abolished the specific band. However, cold chase with Egr mutant oligonucleotide was not able to chase the specific band (lane 5). This observation was further confirmed by antibody super shift experiments, where antibody against Egr1 was able to shift the band (lane 6), whereas non specific IgG did not (lane 7). These experiments indicate that both Sp1 and Egr1 bind at an overlapping sequence in the promoter region, and could regulate the transcription of hsa-miR-106a.
To determine the effect of induced Egr1 expression on hsa-miR-106a level, Jurkat cells were stimulated with PHA/PMA combination for different time intervals, and Northern blotting was performed. It showed that there was an increase in the levels of hsa-miR-106a up to 8 h (Fig. 4F), and its levels declined gradually with time (16 and 24 h). It was also observed that the levels of pre-hsa-miR-106a increased with time up to 4 h, and then slowly declines. Interestingly, the decline of pre-hsa-miR-106a preceded the decline of mature hsa-miR-106a (compare 4 versus 8 h). These results indicate that induced Egr1 increased the expression of hsa-miR-106a.
Activation of hsa-miR-106a Promoter Regulates IL-10 Expression.
To correlate Egr1 mediated activation of hsa-miR-106a promoter with IL-10 expression, Jurkat cells were stimulated with PMA, PHA, or PMA/PHA combination for 2 h, and expression of Egr1 was detected by Western blotting (Fig. 5A). It was observed that although PMA (50 ng/mL) alone did not affect Egr1 levels, PHA (5 μg/mL) was able to induce it. However, it was the combination of both PMA/PHA that was able to maximally induce Egr1 levels (Fig. 5A). Next, to determine the level of induced hsa-miR-106a, Jurkat cells were treated with PMA, PHA, or PMA/PHA combination for 8 h. Stimulation with PMA/PHA combination but not PMA alone induced hsa-miR-106a expression (Fig. 5B). This upregulation could be inhibited by specifically knocking down Egr1 expression by using Egr1 siRNA before stimulation (Fig. 5 C and D).
Fig. 5.
Activation of hsa-miR-106a promoter regulates IL-10 expression. (A) Western blotting was performed for Egr1 expression by using Jurkat cells nuclear extracts prepared after stimulation with or without PHA, PMA, or PMA/PHA combination for 2 h, as indicated. (B) Northern blotting was performed for hsa-miR-106a by using RNA from Jurkat cells treated with or without PHA, PMA, or PMA/PHA combination for 8 h. (C) Western blotting was performed for Egr1 expression by using Jurkat cells transfected with Egr1 siRNA or a random control oligonucleotide followed by stimulation without or with PMA/PHA combination for 2 h, as indicated. (D) Northern blotting was performed for hsa-miR-106a by using RNA from Jurkat cells treated as in C, followed by treatment with PMA/PHA combination for 8 h. For loading control, the membranes from B and D were stripped, and Northern blotting was performed for U6. (E) The Jurkat cells were treated with or without PHA, PMA, or PHA/PMA combination for 24 h, and IL-10 was measured in the culture supernatant by ELISA. The figure represents fold change in IL-10 levels with respect to the unstimulated control, and is presented as mean ± SE of 3 independent experiments. A–D are representative of 3 independent experiments.
To correlate the level of Egr1 induction with the levels of IL-10, Jurkat cells were treated with PHA, PMA, or PHA/PMA together for 24 h, and IL-10 was measured in the culture supernatant by ELISA (Fig. 5E). It was observed that Jurkat cells induced with PMA alone resulted in 3.5-fold increase in IL-10 expression (Fig. 5E, lane 3), whereas PHA alone or PMA/PHA together significantly reduced IL-10 levels (Fig. 5E, lanes 2 and 4). Thus, the level of IL-10 was inversely correlated with the level of Egr1-induced hsa-miR-106a expression.
Discussion
The posttranscriptional regulation of IL-10 is a subject of paramount clinical interest because of the crucial role of IL-10 as a modulator of inflammation. In this study, we found that hsa-miR-106a has a key role in regulating IL-10 expression. We provide a combination of in silico and in vitro evidence including a series of transfection experiments with various potential miRNAs in support of this conclusion.
Our finding of differential level of hsa-miR-106a (Fig. 2B) in various cells supports miRNA mediated control of IL-10 is operational in cells that actively produce it. Recently, it has been demonstrated that regulatory T (Treg) cells have a miRNA profile distinct from conventional CD4 T cells, and some miRNAs, including hsa-miR-106a, were found to be down-regulated (17). In our study, the identification of hsa-miR-106a that regulates IL-10 expression further supports this observation. It can also be seen that there is a thin line between regulation and dysregulation of IL-10 expression. It is observed that in 46% of human T cell leukemias, pri-miR-106–363 cluster is overexpressed (18), presumably modulating IL-10 expression that could promote clonal expansion and leukemic cell survival. Our current results with the promoter revealed the presence of overlapping binding sites for Sp1 and Egr1 that may have potential regulatory function for hsa-miR-106a, a member of this cluster.
Sp1 is a well characterized ubiquitous transcription factor that regulates a vast array of genes, including IL-10 (4, 5). The early growth response gene product (Egr1) is a zinc finger transcription factor with a characteristic brisk kinetics of induction. It can either positively or negatively regulate gene transcription in response to stimulation, because Egr1 contains both transactivation and repression domains (19). It has been proposed that Egr1 might be up-regulated in response to environmental challenge. Overlapping binding sites for Egr1 and Sp1 have been previously seen in the promoter of several genes. For example, it has been previously demonstrated that under quiescent conditions the promoters of PDGF-A chain (A) and β-(1)-adrenergic receptor are occupied by Sp1, which regulates basal expression of these genes (20, 21). However, on stimulation, levels of Egr1 rise, allowing Egr1 to displace Sp1 from this region; thus, inducing the expression of PDGF-A as a response to the induction (21). Physical and functional cooperation between Sp1 and Egr1 has also been seen at IL-2Rβ promoter in T cells (22). Here, constitutive level of IL-2Rβ is maintained by Sp1, whereas on induction, Egr1 physically interacts with Sp1 to maximize transcription. Similarly, in our study, Egr1 expression in Jurkat cells resulted in the induction of pre-hsa-miR-106a followed by mature hsa-miR-106a. This finding seems to be in consonance with the observation that Sp1 is present in Jurkat cells at basal levels and may be responsible for basal transcription of this miRNA. Perturbation of this equilibrium by Egr1 would then up-regulate hsa-miR-106a; thus, negatively regulating IL-10. This notion was further supported by our observation that Egr1 stimulated hsa-miR-106a expression was negatively correlated with IL-10 levels. However, the mechanism by which Sp1 and Egr1 cooperate for regulation of hsa-miR-106a expression remains to be elucidated.
PHA and PMA have been known to induce vast array of genes by activating p38 (23), p42/44 ERK (24), and JNK MAP kinases (25). These 3 types of MAP kinases can be activated individually or simultaneously; thus, suggesting their independent signaling roles. Although PMA alone is known to induce the activation of Sp1 in T cells (4), both PMA and PHA have been shown to activate Egr1 in CEM cells (T cell line) (26). IL-10 has been previously shown to negatively regulate LPS-stimulated Egr1 expression in mouse macrophages (27). Thus, it would be interesting to study the effect IL-10 on Egr1 induced expression of hsa-miR-106a.
The regulatory loops and feedback mechanism for miRNA mediated posttranscriptional regulation of IL10 may be critical in maintaining immune homeostasis, and its dysregulation may lead to disease. It seems plausible that 3 regulatory mechanisms like transcriptional regulation, ARE mediated 3′ UTR regulation, and hsa-miR-106a mediated 3′ UTR regulation may critically regulate IL-10 expression. In fact, recently, it has been shown that inhibitory function of NKp46+CD49b+CD3− NK cells recruited in the hepatic granulomas in experimental model of visceral leishmaniasis was correlated with high IL-10 production as a result of increased stability of IL10 mRNA (28). It is very likely that hsa-miR-106a mediated posttranscriptional control could potentially be involved in fine tuning the critical level of IL-10 expression in a context-dependent manner, and hence dictates the outcome of immune response to specific external stimuli.
In summary, our results suggest that IL-10 expression is regulated by hsa-miR-106a, which is in turn transcriptionally regulated by Egr1 and Sp1. Further studies examining the relevance of this regulatory mechanism to human disease are warranted. It would also be interesting to investigate the effect of pathological conditions (like rheumatoid arthritis, cancer, and viral infections, where an altered IL-10 expression is an important aspect of disease pathogenesis) on this regulatory pathway involving IL-10, hsa-miR-106a, and Egr1. Also, identifying other targets of hsa-miR-106a and the effect of activation of Egr1 on other members of the miRNA cluster would be of considerable interest.
Materials and Methods
In Silico Identification of miRNA Binding Sites.
Based on experimental observations of miRNA binding to the 3′ UTR, several target prediction software have been developed. Although each of them has been used extensively for target site detection, they are associated with many false positive results due to lack of specificity of prediction. To avoid the overprediction, we used a consensus approach employing 3 widely-used software to perform the target prediction. Only those miRNA target pairs were identified that were detected by all 3 software to bind to the same location in the target 3′ UTR sequence (29). All known 470 human miRNAs (miRBase v9) were queried against human IL10 3′ UTR using miRanda (30), RNAhybrid (31), and TargetScan (32).
Analysis of miRNA Expression Profiles.
To check which among the 8 miRNAs, predicted to target IL10, were expressed in cells of lymphoid and myeloid origin, we collected data from the miRNA expression resource (miRex; http://miracle.igib.res.in/mirex). This database allows cross-comparison of miRNA expression profile data across various platforms collected from public repositories like Gene Expression Omnibus (National Center for Biotechnology Information-GEO) and ArrayExpress. The prenormalized data retrieved from these repositories are internally normalized to a standard score (z-score) by using an in-house developed platform (miRex; http://miracle.igib.res.in/mirex). Expression of each miRNA in a particular tissue is represented as a fold change. The database currently contains a total set of 1,507 experiments spanning 18 datasets.
Plasmids.
Plasmids pMIR-REPORT-IL10 3′ UTR (intact), pMIR-REPORT-IL10 3′ UTR (mutant) and hsa-miR-106a promoter in pGL3 (basic) constructs were prepared as described in detail in SI Materials and Methods.
Cell Culture and Transfection.
A549, Raji, Jurkat, and THP-1 cells were grown and transfected as described in SI Materials and Methods.
IL-10 ELISA.
Because IL-10 is a secretory cytokine, its levels were detected from culture supernatant of transfected cells by performing ELISA (R&D Systems) following the manufacturer's manual.
RNA Extraction and RT-PCR.
RNA was prepared from cells treated with or without PHA (5 μg/mL), PMA (50 ng/mL), or combination of PHA/PMA (5 μg/mL and 50 ng/mL) for different time intervals by using TRIzol reagent (Invitrogen) following the product manual. RNA was stored at −70 °C until further use. RT-PCR was performed by using specific primers for IL10 and GAPDH following the standard protocol.
The hsa-miR-106a Northern Blotting.
RNA isolated from A549, Jurkat, Raji, THP-1, HeLa, and HepG2 cells were resolved on acrylamide gel, transferred on nylon membrane (Gene screen plus; PerkinElmer), and hybridized with Gamma 32P-labeled hsa-miR-106a oligonucleotide probe, as described in SI Materials and Methods.
EMSA.
Nuclear extracts were prepared from Jurkat cells as previously described (33). These extracts were incubated with radio-labeled double stranded oligonucleotide probes for hsa-miR-106a potential promoter region. Protein-DNA complexes were separated on acrylamide gels. Super shift experiments were performed as described in SI Materials and Methods.
Western Blotting.
Nuclear protein was prepared as described previously (33), and Western blotting was performed for Egr1, Sp1, and Lamin, as described in SI Materials and Methods.
Statistical Analysis.
Results are given as mean of 3 independent experiments ± SEM. An independent 2-tailed Student's t test was performed. Differences were considered statistically significant for P ≤ 0.05. For normalization of transfection efficiency, cells were cotransfected with vector pMIR-REPORT beta-Gal as indicated.
Supplementary Material
Acknowledgments.
We thank Mr. Nomesh Goplani for technical help. A.S., M.K., and M.H. are supported by fellowships from the Council of Scientific and Industrial Research (CSIR). This work is supported by a grant from CSIR (Task Force Project Grant NWP0033) India.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808743106/DCSupplemental.
References
- 1.Asadullah K, Sterry W, Volk HD. Interleukin-10 therapy-review of a new approach. Pharmacol Rev. 2003;55:241–269. doi: 10.1124/pr.55.2.4. [DOI] [PubMed] [Google Scholar]
- 2.O'Garra A, Barrat FJ, Castro AG, Vicari A, Hawrylowicz C. Strategies for use of IL-10 or its antagonists in human disease. Immunol Rev. 2008;223:114–131. doi: 10.1111/j.1600-065X.2008.00635.x. [DOI] [PubMed] [Google Scholar]
- 3.O'Garra A, Vieira P. T(H)1 cells control themselves by producing interleukin-10. Nat Rev Immunol. 2007;7:425–428. doi: 10.1038/nri2097. [DOI] [PubMed] [Google Scholar]
- 4.Tone M, Powell MJ, Tone Y, Thompson SA, Waldmann H. IL-10 gene expression is controlled by the transcription factors Sp1 and Sp3. J Immunol. 2000;165:286–291. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- 5.Ma W, et al. The p38 mitogen-activated kinase pathway regulates the human interleukin-10 promoter via the activation of Sp1 transcription factor in lipopolysaccharide-stimulated human macrophages. J Biol Chem. 2001;276:13664–13674. doi: 10.1074/jbc.M011157200. [DOI] [PubMed] [Google Scholar]
- 6.Powell MJ, Thompson SA, Tone Y, Waldmann H, Tone M. Posttranscriptional regulation of IL-10 gene expression through sequences in the 3′-untranslated region. J Immunol. 2000;165:292–296. doi: 10.4049/jimmunol.165.1.292. [DOI] [PubMed] [Google Scholar]
- 7.Brewer G, Saccani S, Sarkar S, Lewis A, Pestka S. Increased interleukin-10 mRNA stability in melanoma cells is associated with decreased levels of A + U-rich element binding factor AUF1. J Interferon Cytokine Res. 2003;23:553–564. doi: 10.1089/107999003322485053. [DOI] [PubMed] [Google Scholar]
- 8.Lyon H, et al. IL10 gene polymorphisms are associated with asthma phenotypes in children. Genet Epidemiol. 2004;26:155–165. doi: 10.1002/gepi.10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurreeman FA, Schonkeren JJ, Heijmans BT, Toes RE, Huizinga TW. Transcription of the IL10 gene reveals allele-specific regulation at the mRNA level. Hum Mol Genet. 2004;13:1755–1762. doi: 10.1093/hmg/ddh187. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP, et al. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.varez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 15.Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: Tiny players in a big field. Immunity. 2007;26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Grabe N. AliBaba2: Context specific identification of transcription factor binding sites. In Silico Biol. 2002;2:S1–S15. [PubMed] [Google Scholar]
- 17.Cobb BS, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106–363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 19.Thiel G, Cibelli G. Regulation of life and death by the zinc finger transcription factor Egr-1. J Cell Physiol. 2002;193:287–292. doi: 10.1002/jcp.10178. [DOI] [PubMed] [Google Scholar]
- 20.Bahouth SW, Beauchamp MJ, Vu KN. Reciprocal regulation of beta(1)-adrenergic receptor gene transcription by Sp1 and early growth response gene 1: Induction of EGR-1 inhibits the expression of the beta(1)-adrenergic receptor gene. Mol Pharmacol. 2002;61:379–390. doi: 10.1124/mol.61.2.379. [DOI] [PubMed] [Google Scholar]
- 21.Khachigian LM, Williams AJ, Collins T. Interplay of Sp1 and Egr-1 in the proximal platelet-derived growth factor A-chain promoter in cultured vascular endothelial cells. J Biol Chem. 1995;270:27679–27686. doi: 10.1074/jbc.270.46.27679. [DOI] [PubMed] [Google Scholar]
- 22.Lin JX, Leonard WJ. The immediate-early gene product Egr-1 regulates the human interleukin-2 receptor beta-chain promoter through non canonical Egr and Sp1 binding sites. Mol Cell Biol. 1997;17:3714–3722. doi: 10.1128/mcb.17.7.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loza MJ, Foster S, Peters SP, Penn RB. Beta-agonists modulate T-cell functions via direct actions on type 1 and type 2 cells. Blood. 2006;107:2052–2060. doi: 10.1182/blood-2005-08-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li YQ, Hii CS, Der CJ, Ferrante A. Direct evidence that ERK regulates the production/secretion of interleukin-2 in PHA/PMA-stimulated T lymphocytes. Immunology. 1999;96:524–528. doi: 10.1046/j.1365-2567.1999.00724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kesherwani V, Sodhi A. Involvement of tyrosine kinases and MAP kinases in the production of TNF-alpha and IL-1beta by macrophages in vitro on treatment with phytohemagglutinin. J Interferon Cytokine Res. 2007;27:497–505. doi: 10.1089/jir.2007.0166. [DOI] [PubMed] [Google Scholar]
- 26.Cogswell PC, Mayo MW, Baldwin AS., Jr Involvement of Egr-1/RelA synergy in distinguishing T cell activation from tumor necrosis factor-alpha-induced NF-kappa B1 transcription. J Exp Med. 1997;185:491–497. doi: 10.1084/jem.185.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamimura M, et al. Interleukin-10 suppresses tissue factor expression in lipopolysaccharide-stimulated macrophages via inhibition of Egr-1 and a serum response element/MEK-ERK1/2 pathway. Circ Res. 2005;97:305–313. doi: 10.1161/01.RES.0000177893.24574.13. [DOI] [PubMed] [Google Scholar]
- 28.Maroof A, et al. Posttranscriptional regulation of IL10 gene expression allows natural killer cells to express immunoregulatory function. Immunity. 2008;29:295–305. doi: 10.1016/j.immuni.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hariharan M, Scaria V, Pillai B, Brahmachari SK. Targets for human encoded microRNAs in HIV genes. Biochem Biophys Res Commun. 2005;337:1214–1218. doi: 10.1016/j.bbrc.2005.09.183. [DOI] [PubMed] [Google Scholar]
- 30.John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 33.Kumar S, Sharma A, Madan B, Singhal V, Ghosh B. Isoliquiritigenin inhibits IkappaB kinase activity and ROS generation to block TNF-alpha induced expression of cell adhesion molecules on human endothelial cells. Biochem Pharmacol. 2007;73:1602–1612. doi: 10.1016/j.bcp.2007.01.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.