Abstract
The obligate intracellular bacterium Wolbachia pipientis infects around 20% of all insect species. It is maternally inherited and induces reproductive alterations of insect populations by male killing, feminization, parthenogenesis, or cytoplasmic incompatibility. Here, we present the 1,445,873-bp genome of W. pipientis strain wRi that induces very strong cytoplasmic incompatibility in its natural host Drosophila simulans. A comparison with the previously sequenced genome of W. pipientis strain wMel from Drosophila melanogaster identified 35 breakpoints associated with mobile elements and repeated sequences that are stable in Drosophila lines transinfected with wRi. Additionally, 450 genes with orthologs in wRi and wMel were sequenced from the W. pipientis strain wUni, responsible for the induction of parthenogenesis in the parasitoid wasp Muscidifurax uniraptor. The comparison of these A-group Wolbachia strains uncovered the most highly recombining intracellular bacterial genomes known to date. This was manifested in a 500-fold variation in sequence divergences at synonymous sites, with different genes and gene segments supporting different strain relationships. The substitution-frequency profile resembled that of Neisseria meningitidis, which is characterized by rampant intraspecies recombination, rather than that of Rickettsia, where genes mostly diverge by nucleotide substitutions. The data further revealed diversification of ankyrin repeat genes by short tandem duplications and provided examples of horizontal gene transfer across A- and B-group strains that infect D. simulans. These results suggest that the transmission dynamics of Wolbachia and the opportunity for coinfections have created a freely recombining intracellular bacterial community with mosaic genomes.
Keywords: horizontal transfer, recombination, ankyrin repeat gene, genome evolution, insect symbiosis
Wolbachia pipientis are intracellular α-proteobacteria of the order Rickettsiales that infect insects as well as isopods, spiders, scorpions, mites, and filarial nematodes (1, 2). These bacteria represent a single species, with strains classified into supergroups, of which the most abundant are supergroups A and B. A lack of concordance between host and bacterial strain phylogenies indicate frequent host shifts in addition to maternal inheritance within the individual host (1–4). In insect populations, Wolbachia induce reproductive manipulations to enhance their own spreading. The most frequently observed reproductive abnormality is cytoplasmic incompatibility (CI) (1, 2, 5), where uninfected females are unable to produce offspring with infected males, whereas infected females can produce offspring with both infected and uninfected males, thus creating a reproductive advantage for infected females. Other spectacular effects of Wolbachia infections are male embryo killing, feminization, and parthenogenesis induction (1, 2).
Three genomes of Wolbachia have been published to date. The A-group strain wMel from Drosophila melanogaster (6) and the B-group strain wPip from the mosquito Culex quinquefasciatus (7) are both reproductive parasites that cause CI, whereas the D-group strain wBm is an obligate mutualist in the nematode Brugia malayi (8). The 1.27-Mb genome of wMel and the 1.48-Mb genome of wPip contain several prophages and high frequencies of repeated sequences, including many IS-elements. These genomes have a large repertoire of genes with ankyrin repeat motifs, 23 in wMel and 60 in wPip, several of which are associated with mobile elements (6, 7). In contrast, the 1.08-Mb genome of strain wBm contains no prophage, only a few ankyrin repeat genes and a much lower fraction of repeated sequences, possibly reflecting its mutualistic adaptation to a single-host species.
Sequence comparisons of single genes from multiple strains have provided evidence for recombination (9–12) and horizontal transfer of prophages and insertion sequence (IS)-elements across Wolbachia strains (13–16). However, the distant relationship of the 3 sequenced Wolbachia genomes, manifested as an almost complete lack of gene-order conservation and synonymous substitution frequencies that are close to saturation, has precluded attempts to infer patterns and rates of recombination at the whole-genome level. Thus, with the exception of a few single-gene studies, the extent to which recombination distorts the evolutionary coherence of Wolbachia genomes is currently unknown.
We report here the complete genome sequence of the supergroup-A Wolbachia strain wRi that naturally infects Drosophila simulans and induces almost complete CI in its host (17, 18). Additionally, we present partial genome data of Wolbachia strain wUni from Muscidifurax uniraptor, likewise a supergroup-A strain that induces parthenogenesis in its host (19). A comparison of these 2 A-group Wolbachia strains with the previously sequenced genome of the A-group strain wMel reveals the most highly recombining obligate intracellular bacterial community examined to date.
Results and Discussion
General Features of the Wolbachia wRi Genome.
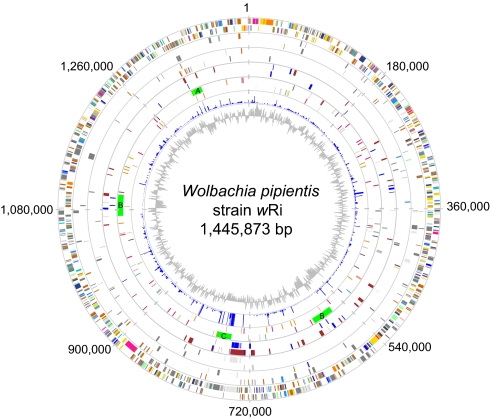
The complete genome sequence of Wolbachia pipientis wRi from Drosophila simulans is a single circular chromosome of 1,445,873 bp, with 1,150 potential protein-coding sequences and 114 pseudogenes (Fig. 1). We identified 4 prophage segments, here called wRi-WOA, wRi-WOB (present in identical duplicates), and wRi-WOC. Additionally, we found 35 genes coding for proteins containing one or more ankyrin repeat (ANK) domain [Tables S1–S3] as compared to 23 such genes annotated in the 1.27-Mb genome of wMel (6). Of the genes solely present in either the wRi or the wMel genome for which a function or domain hit can be identified, the large majority encodes phage proteins, transposases and ANK proteins (Tables S4–S6).
Fig. 1.
Circular map of the Wolbachia pipientis wRi genome. Each circle confined by the gray lines except for the 2 innermost circles illustrates different features on the plus (outer region) and minus (inner region) strands. Lines and boxes in the 3 outermost circles are colored according to the Clusters of Orthologous Groups (COG) categories. First (Outer) circle: protein-coding genes (CDSs). Second circle: pseudogenes. Third circle: unique CDSs compared to wMel. Fourth circle: ANK genes in blue and gene synteny breakpoints compared to wMel in red. Fifth circle: prophage regions in green (not affiliated to a strand) and IS elements color-coded as described in Table S5. Sixth circle: a diagram showing the synonymous substitution frequency (Ks) between wRi and wMel potential orthologs; the maximum cut-off was set to Ks = 1. Seventh circle: GC-skew of the wRi genome.
Overall, the wRi genome contains 22.1% repeated sequences, as compared to 8.9% in wMel (> 200 bp, 95% sequence identity). About 10% of the wRi genome is covered by IS-elements; these represent 11 different types, including 67 complete copies, 48 copies with frameshifts or internal stop codons, and 11 truncated copies. A total of 46 genes have been disrupted by IS-element insertions, of which 19 are hypothetical proteins, 10 are transposases, and 5 are ANK genes. Furthermore, more than 200 insertions of the Wolbachia palindromic element (20) or remnants thereof were found in the genome of wRi and of these, 43 were inserted into genic sequences. The Wolbachia palindromic element is also present in wMel and wUni, but not always in the same genes or at the same locations. The most abundant 23-bp hairpin-loop structure is present in 79 copies in the wRi genome, in 91 copies in wMel, but only once in wBm.
Genome Integrity Following Host Shifts.
A comparison of the structures of the wRi and wMel genomes revealed 35 gene-order breakpoints (Fig. S1), 17 of which are flanked by IS-elements, 11 are located within or flanked by prophage sequences, and 6 are flanked by long repeats. The final breakpoint contains a 22-bp hairpin structure with a 4-bp loop. Among these breakpoints, 6 are close to genes encoding reverse trancriptase and 3 to genes encoding DNA recombinase. This shows that mobile genetic elements and repeated sequences are hot spots for rearrangements in Wolbachia. To examine the stability of these recombination hot spots, we used PCR to analyze all breakpoints from genomic DNA of wRi isolated from naturally infected and transinfected symbiotic associations of D. simulans, Drosophila yakuba, Drosophila teisseiri, and Drosophila santomea, which have been kept in laboratory conditions from 8 to 15 years (Table S7). The results of this survey showed that the wRi genome remains stable in structure over these sites and suggests that the wRi genome does not oscillate between different genomic structures, nor do host switches trigger rearrangements at these sites. The observed short-term stability indicates that the recombination frequencies at these repeated sequences are lower than could be detected over this time period.
The A-Group Wolbachia Strains are Evolutionary Genome Mosaics.
We estimated the nonsynonymous (Ka) and synonymous (Ks) substitution frequency per site to quantify sequence divergences across strains, although these values may not correspond to actual substitutions if genes evolve mainly by recombination. For 851 positional homologs in the wRi and wMel genomes identified by reciprocal BLAST searches (excluding phage genes), the median Ka and Ks values were estimated to be 6.2 × 10−3 and 3.2 × 10−2 substitutions per site, respectively, with a more than 500-fold variation in Ks-values across genes (Fig. S2a). A similar spectrum of divergences between wRi and wMel was observed for 343 core genes that are conserved across 3 Wolbachia strains (wRi, wMel, and wBm), Orientia tsutsugamushi, and 8 Rickettsia species (as defined in ref. 21) (Fig. S2b), suggesting that these estimates are not inflated by the inadvertent inclusion of inactivated gene fragments.
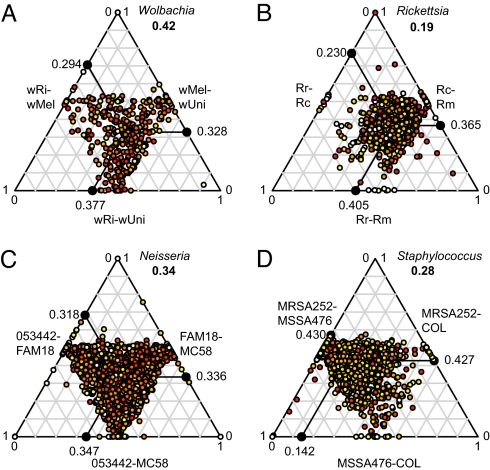
To investigate the mechanisms underlying the remarkable variation in Ks-values across genes, we sequenced orthologous genes in wUni, another A-group Wolbachia strain, using wMel as the reference genome. A 3-strain comparison of 450 sequenced orthologs showed a broad variability of Ks-values (Fig. 2A) that did not correlate with functional categories (Mann-Whitney and Kolmogorov-Smirnov tests using Bonferroni-Holm correction, P > 0.05 for all COG categories) (Fig. S2c). One-third of the genes were most similar in wMel and wRi, including 52 orthologs with no synonymous sustitutions. Another one-third indicated the highest sequence similarity for wMel and wUni, of which 26 have no synonymous substitutions. Finally, one-fifth of the genes were most similar in wRi and wUni, with 22 lacking synonymous substitutions. Only 40 genes showed no substitutions at synonymous sites in any of the 3 pairs. Such a complex pattern of sequence divergences indicates extensive recombination.
Fig. 2.
Ternary plot showing substitution frequency variation across (A) 410 genes in A-group Wolbachia (40 genes with no synonymous substitutions in all 3 strains were excluded); (B) 818 genes in Spotted-Fever Group Rickettsia; (C) 1,529 genes in Neisseria meningitidis; and (D) 2,207 genes in Staphylococcus aureus. Each dot in the diagram represents 1 gene. Absolute Ks-values have been transformed to relative values between 0 and 1. The mean relative Ks-value for each pair is shown on each axis. Numbers in bold represent the median distance to the average point. The color of each dot represents the maximum absolute Ks-value among the 3 pairs, ranging from light yellow (low values) to red (high values). Median Ks-values for all pairs are reported in Table S8. Rr, Rickettsia rickettsii; Rc, Rickettsia conorii; Rm, Rickettsia massiliae.
To obtain a simple measure of the relative levels of recombination for comparisons across species, we calculated the spread of the relative Ks-values as the median distance to the average relative Ks-values in the ternary plots (see Fig. 2). Notably, the level of recombination thus inferred was higher in Wolbachia (spread = 0.42) (see Fig. 2A) than in Neisseria meningitidis (spread = 0.34) (see Fig. 2C), which is naturally competent for transformation and highly recombining (22). The level was twice as high as in its close relative Rickettsia (spread = 0.19) (see Fig. 2B), where the lowest Ks-values were typically associated with 1 pair of strains, as expected in nonrecombining bacteria.
Additionally, of the 450 orthologous genes found in wRi, wMel, and wUni, intragenic recombination was detected in 129 genes by at least 2 different methods implemented in RDP3 (23). Genes for which intragenic recombination was detected by recombination detection program (RDP) were concentrated to the middle of the inner triangle (spread = 0.27), whereas genes with no detected recombination were more spread out (spread = 0.56) in the ternary plot (Fig. S2d). Genes located near different corners of the inner triangle in the ternary plot provide the strongest evidence for inconsistency in the phylogenetic signal. As no conflict in signal was detected by RDP within these genes, it is likely that recombination has occurred over the complete sequences in many cases. Taken together, this indicates that at least three-fourths of the genes that were analyzed are affected by recombination.
The switches in sequence similarity patterns within gene alignments is here illustrated with the gene for Leucyl-tRNA synthetase (LeuRS), where 1 region was found to be identical in wMel and wUni, but contained many substitutions in wRi, followed by a divergent segment in wUni that was identical in wRi and wMel (Fig. 3A). Another example is the virB10 gene, where the 5′-part of the wRi gene clustered with the A-group strain wAtab3 from the braconid wasp Asobara tabida (Fig. 3B), whereas the 3′-part of the same gene clustered with the B-group strains wTai, from the Taiwan cricket, Teleogryllus taiwanemma, and wPip, from the mosquito Culex quinquefasciatus (Fig. 3C).
Fig. 3.
Recombination within leuRS and virB10. (A) The diagram shows the Ks-values calculated for the leuRS gene pair segments consisting of a 99-base window sliding over the gene alignment with 15-base steps. The boxes under the x-axis indicate regions where wRi (red box) and wUni (black box) are highly diverged from the other two strains. (B and C) Inference of sequence relationships based on segments in the virB10 gene between (B) positions 72 and 892 and (C) positions 967 and 1526, respectively, of the gene alignment. The gray circle shows the position of wRi in the tree. The strains wKueYo, wMel, wUni, wAtab3, and wRi belong to supergroup A (names in red), whereas strains wPip and wTai belong to supergroup B (names in blue).
Rapid Diversification by Expansion-Contraction of Tandem Repeats.
We classified the 35 ANK genes identified in the wRi genome into 3 different groups based on the extent of sequence divergence from their homologs in wMel (see Tables S1–S3). One-third, 13 genes, are highly conserved in length, domain organization, and nucleotide sequence (Ks <0.05). Another 12 genes showed variability in the number of ANK domains and gene length plus high sequence divergence (Ks 0.1 to >1.0), although they are located in segments with otherwise conserved gene-order structures. The final 10 genes lacked homologs in wMel.
To study the relationships of the ANK domains within and among genomes, we performed a Bayesian phylogenetic analysis of 319 ANK domains identified in the wRi, wMel, and wUni genomes (Fig. 4). The ANK domains found in the highly conserved genes clustered with the corresponding domains in the other strains. In contrast, several ANK domains in the variable gene set clustered with other domains in the same gene. In these cases, variability in domain numbers and organization is generated by expansion or contraction of repeated sequences, with the repeated unit often spanning across the borders of the ANK domains (Fig. S3). The ANK protein WRi_003070 is particularly interesting; the N-terminal repeats are most similar to wMel and the repeats in the middle part to wUni, with recent duplications of ANK domains in both wRi and wUni (see Fig. S3). Additionally, this is the only wRi ANK gene that is solely transcribed in female adults and ovaries, but not in male adults and testes (Tables S9 and S10).
Fig. 4.
Clustering of ankyrin repeat domains from wRi, wMel, and wUni using Bayesian phylogenetic inference. The colors on the branches indicate different intervals of posterior probability: (red) 0.95–1.0, (yellow) 0.90–0.94, and (blue) 0.89–0.80 (nonsignificant), and (black) <0.80.
Gene Transfer Across Supergroups.
Of the combined 58 ANK genes in the wRi and wMel genomes, 31 are located near to prophages and many of these are unique to the individual strain and may have been acquired recently. One such gene, WRi_006870, is located near phage wRi-WOC. Although the gene is absent from the other 2 A-group strains, homologs were identified in wNo, wMa, and wMau B-group strains that like wRi use D. simulans as their natural host. The gene is not transcribed in early or late (overnight) embryos in wRi, not in testes and early embryos in wNo and not in adult males in wMau: that is, this ANK gene exhibits stage-specific expression patterns in both A- and B-group genomic backgrounds (see Tables S9 and S10). A phylogeny based on the minor capsid protein shows that wRi-WOC clusters with one of the prophages in wNo (Fig. S4), indicating that prophage wRi-WOC, along with some of its associated ANK genes, may have been acquired from the B-group strains. Transmission of phages between Wolbachia strains of different supergroups that infect the same host has been seen in moths (14) and has also been shown to occur between wNo and wHa during coinfections of D. simulans in the laboratory (15).
In contrast, no examples of horizontal gene transfer between the A-group strains and the D-group Wolbachia strain wBm were observed, consistent with the absence of prophages and low levels of recombination in mutualistic Wolbachia strains (24). Likewise, mutualistic endosymbionts of aphids show low recombination frequencies and have the most stable bacterial genomes identified to date (25). This dramatic difference in recombination may reflect variability in host-adaptation strategies, access to mobile elements, and different sets of recombination genes (15), as well as increased possibilities for rare genome variants to reach fixation in such populations (26, 27).
Wolbachia Sequences in Drosophila Genome Assemblies.
Wolbachia sequences were previously identified in the trace archive files of both the D. simulans and the Drosophila ananassae genome projects and assumed to originate from contaminating bacterial DNA (28), a discovery that was followed by a debate about whether any corresponded to wRi (29, 30). The more recent finding that Wolbachia genes have been transferred into the nuclear genomes of D. ananassae and other hosts (31–34) further added complexity with regards to the origin of these sequences.
Using BLAST, we recovered 73 scaffolds from the FlyBase assembly of the D. ananassae genome containing 177 kb (excluding gaps) that partially or fully matched wRi sequences, some of which are currently annotated as D. ananassae genes in both GenBank and FlyBase. We did not identify any wRi sequences that are shared with wMel, presumably because the wMel genome was used to filter out Wolbachia reads. A comparison to the wRi genome revealed the absence of 2 IS-elements within scaffolds of otherwise conserved gene-order structures, suggesting that the Wolbachia sequences in the D. ananassae genome are similar but not identical to the wRi genome. In contrast, no wRi sequences were identified in the FlyBase assembly of the D. simulans genome. Furthermore, amplification of wRi ANK and VIR sequences by PCR from D. simulans treated with tetracycline failed. We conclude that a corresponding transfer of Wolbachia genes into the nuclear genome of D. simulans is not likely.
Patchy Wolbachia Populations.
Pervasive recombination in parasitic Wolbachia destroys the anticipated correlation between gene history, genome history, and strain phenotype. The wsp surface protein has been extensively used for genotyping but was found to be especially prone to recombination (9, 35) and 2 different sets of housekeeping genes, gatB, coxA, hcpA, fbpA, ftsZ (11) and aspC, atpD, sucB, and pdhB (12) were proposed as an alternative. However, not even these genes are protected from recombination events (11) and our comparisons of wUni, wMel, and wRi show divergences at synonymous sites ranging from Ks = 0 in aspC to Ks = 0.1–0.2 for gatB, with different genes and segments of genes supporting different strain relationships. Hence, no single gene sequence will accurately describe the relationships of these A-group Wolbachia strains.
The global Wolbachia population is likely to consist of many subpopulations, or patches (36), where the boundaries are defined by, for example, geography or host specificity. Recombination among strains is expected to be frequent within patches, but less so between patches. For example, mutualistic adaptations to a single host may lead to the isolation and evolution of a subpopulation with limited recombination, as is possibly the case in nematode Wolbachia. While multilocus sequence typing may be useful to characterize supergroups, the intense recombination seen between A-group strains indicates that characterization of genotypes might require analysis at the whole genome level. However, as selection is expected to act on traits involved in host-adaptation processes within a patch, such genes may be useful to identify “fitness types,” although not conferring any information that is meaningful in a phylogenetic sense.
Future Perspectives.
The availability of complete genome sequence data for the model organisms D. melanogaster and D. simulans, as well as for their respective Wolbachia endosymbionts, offers an excellent opportunity to study host-adaptation processes by monitoring the coevolution of host and endosymbiont gene interactions in natural and transinfected hosts. Rapid diversification of the ANK genes by segmental gene duplication may reflect diversifying selection to match a divergent set of target molecules in different cells, tissues, and hosts. To test this hypothesis, target proteins should be searched for among host genes that are also rapidly evolving, with prime candidates in gametogenesis, meiosis, reproduction (37), and innate immunity responses (38). An exciting avenue for future research is to identify the interacting endosymbiont-host proteins and determine whether these evolve by purifying, positive, or diversifying selection within Wolbachia subpopulations.
The association between wRi and D. simulans is one of a few Wolbachia infections that have been studied in natural populations. Using wRi as the reference genome, it is now possible to initiate comparative studies of wRi genomes extracted from natural D. simulans populations with different phenotypes. Although we have demonstrated stability over the breakpoints during a period of 15 years in wRi strains kept in the laboratory, other genetic changes, such as transposition of IS-elements, gene inactivation by IS-element insertions, and novel gene acquisition could occur rapidly in natural populations. For example, changes in fecundity have been observed during a 20-year period in a natural D. simulans population infected with wRi in southern California (18). The genetic basis of this and other rapidly changing phenotypes can now be investigated.
Materials and Methods
Sequencing Strategies.
wRi: Drosophila simulans Riverside eggs were collected after 2 h and 1 to 2 ml of embryos were homogenized. A continuous renografin gradient (28%–45%) was used to concentrate Wolbachia cells. The 28% to 32% zone was collected and placed in agarose plugs that were treated with bacterial cell lysis and proteinase K solution. To remove contaminating host DNA, the plugs were run on a 1% Seakem Gold agarose (FMC BioProducts) gel for 24 h and the isolated DNA was subsequently used for library construction in a modified M13 vector as described previously (39). From the M13 library, 34,322 reads were sequenced, of which 19,727 were present in the final assembly, resulting in an overall 8.2-times coverage. An additional 18,031 reads were generated during gap closure and finishing.
wUni: DNA was isolated from 300 dissected ovaries from adult females Muscidifurax uniraptor using a CTAB protocol, followed by lysozyme treatment and chloroform extraction. Primers were designed based on the genome sequence of Wolbachia strain wMel, to amplify 1,100-bp products with 300-bp overlap on both ends to the adjacent product. Primers that successfully amplified short PCR products were selected and combined to generate long-range PCR products, where short products did not amplify. Next, 26,834 reads were sequenced and assembled into 287 contigs, of which 106 were longer than 2 kb. Short PCR-products were sequenced directly and long products were sheared by nebulization and cloned into the pSMART-HCKan vector before sequencing.
Verifying the wRi Genome Assembly.
The wRi assembly was confirmed over each IS-element or inferred breakpoint using genomic DNA from wRi isolated from D. simulans and other infected hosts using PCR with specific primers. The size of the assembled genome wRi is slightly lower than the 1.66 Mb previously estimated from pulse-field gel electrophoresis (40). However, the relative order of the observed restriction fragments matches those predicted from the genome sequence, except that the sizes of the individual fragments appear to have been systematically overestimated in the pulsed-field gel electrophoretic -analysis.
Informatics.
Assembly was performed with PHRED-PHRAP-CONSED (41–43). Protein-coding genes were identified with GLIMMER (44) and CRITICA (45) and tRNA genes by tRNAscan-SE (46). Putative functions were inferred using BLAST against the National Center for Biotechnology Information databases and InterProScan (47). Repeat identification was made using MUMmer (48). Codeml, PAML 3.14 (49) was used to calculate substitution rates. Orthologs used for Ks calculations were retrieved by reciprocal best blast with additional cutoffs. RDP3 (23) was used to check nucleotide alignments for intragenic recombination using 6 methods, RDP, Geneconv, Bootscan, MaxChi, Chimaera, and 3Seq, with default settings except for window and step sizes. Sequences of the minor capsid gene were aligned with CLUSTALW (50) on the protein level and back-translated to nucleotide sequences. The phylogeny was reconstructed using MrBayes 3.12 (51) with the GTR+G model and run for 10,000,000 generations. Ankyrin repeats were found with the ANK HMM from PFAM (52) running HMMER 2.0 (53). An amino acid alignment was produced with hmmalign and then back-translated to nucleotides. The phylogeny was reconstructed using MrBayes3.12 (51) under the GTR+I+G model and run for 27,000,000 generations. For both trees, sampling was made every one-hundredth generation with 2 runs of 4 chains and default priors and a consensus trees were constructed using a “burnin” of 25%.
Transcription Analyses.
For each of the tested Wolbachia-Drosophila associations, 300 testis and 150 ovaries were dissected from adults (1-day-old males and 3-day-old females). Embryos were collected every 2 h and late embryos every 16 h. Total RNA was extracted using TRIzol (Invitrogen) and treated with RNase-free DNase (Invitrogen). First-strand cDNA was synthesized from 5 μg of total RNA using reverse transcriptase (SuperScript III; Invitrogen) and random primers (Promega), and thereafter treated with RNase H. For each gene, specific primers were designed based on the corresponding wRi gene nucleotide sequence and used for PCR amplification
Supplementary Material
Acknowledgments.
We thank Richard Stouthamer and Fabrice Vavre for providing Muscidifurax uniraptor, Gabor Nyiro for technical assistance, and Lionel Guy for helpful suggestions. This work was supported by Grant QLK3-CT2000-01079, “The European Wolbachia Project: Towards novel biotechnological approaches for control of arthropod pests and modification of beneficial arthropod species by endosymbiotic bacteria” from the European Union (to K.B., H.R.B., R.G. and S.G.E.A.), from the European Community's Seventh Framework Program CSA-SA_REGPROT-2007–1 under Grant agreement 203590 and intramural funding from University of Ioannina (to K.B.), and from the Swedish Agricultural Research Council, the Swedish Research Council, the Göran Gustafsson Foundation, the Swedish Foundation for Strategic Research and the Knut and Alice Wallenberg Foundation (to S.G.E.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences described in this paper have been deposited in GenBank database [accession nos. CP001391 (W. pipientis wRi) and ACFP01000000 (Wolbachia wUni); the version described in this article is ACFP01000000].
This article contains supporting information online at www.pnas.org/cgi/content/full/0810753106/DCSupplemental.
References
- 1.Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 2.Stouthamer R, Breeuwer JA, Hurst GD. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu Rev Microbiol. 1999;53:71–102. doi: 10.1146/annurev.micro.53.1.71. [DOI] [PubMed] [Google Scholar]
- 3.Werren JH, Zhang W, Guo LR. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc Biol Sci. 1995;261:55–63. doi: 10.1098/rspb.1995.0117. [DOI] [PubMed] [Google Scholar]
- 4.McGraw EA, O'Neill SL. Evolution of Wolbachia pipientis transmission dynamics in insects. Trends Microbiol. 1999;7:297–302. doi: 10.1016/s0966-842x(99)01531-0. [DOI] [PubMed] [Google Scholar]
- 5.Bourtzis K, Braig HR, Karr TL. In: Insect Symbiosis. Bourtzis K, Miller TA, editors. Boca Raton: CRC Press; 2003. pp. 217–246. [Google Scholar]
- 6.Wu M, et al. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2004;2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klasson L, et al. Genome evolution of Wolbachia strain wPip from the Culex pipiens group. Mol Biol Evol. 2008;25:1877–1887. doi: 10.1093/molbev/msn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster J, et al. The Wolbachia genome of Brugia malayi: endosymbiont evolution within a human pathogenic nematode. PLoS Biol. 2005;3:e121. doi: 10.1371/journal.pbio.0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldo L, Lo N, Werren JH. Mosaic nature of the Wolbachia surface protein. J Bacteriol. 2005;187:5406–5418. doi: 10.1128/JB.187.15.5406-5418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldo L, Bordenstein S, Wernegreen JJ, Werren JH. Widespread recombination throughout Wolbachia genomes. Mol Biol Evol. 2006;23:437–449. doi: 10.1093/molbev/msj049. [DOI] [PubMed] [Google Scholar]
- 11.Baldo L, et al. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 2006;72:7098–7110. doi: 10.1128/AEM.00731-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paraskevopoulos C, Bordenstein SR, Wernegreen JJ, Werren JH, Bourtzis K. Toward a Wolbachia multilocus sequence typing system: discrimination of Wolbachia strains present in Drosophila species. Curr Microbiol. 2006;53:388–395. doi: 10.1007/s00284-006-0054-1. [DOI] [PubMed] [Google Scholar]
- 13.Cordaux R, et al. Intense transpositional activity of insertion sequences in an ancient obligate endosymbiont. Mol Biol Evol. 2008;25:1889–1896. doi: 10.1093/molbev/msn134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masui S, Kamoda S, Sasaki T, Ishikawa H. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J Mol Evol. 2000;51:491–497. doi: 10.1007/s002390010112. [DOI] [PubMed] [Google Scholar]
- 15.Bordenstein SR, Wernegreen JJ. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol Biol Evol. 2004;21:1981–1991. doi: 10.1093/molbev/msh211. [DOI] [PubMed] [Google Scholar]
- 16.Gavotte L, et al. Diversity, distribution and specificity of WO phage infection in Wolbachia of four insect species. Insect Mol Biol. 2004;13:147–153. doi: 10.1111/j.0962-1075.2004.00471.x. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann AA, Turelli M, Simmons GM. Unidirectional incompatibility between populations of Drosophila simulans. Evolution. 1986;40:692–701. doi: 10.1111/j.1558-5646.1986.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 18.Weeks AR, Turelli M, Harcombe WR, Reynolds KT, Hoffmann AA. From parasite to mutualist: rapid evolution of Wolbachia in natural populations of Drosophila. PLoS Biol. 2007;5:e114. doi: 10.1371/journal.pbio.0050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stouthamer R, Breeuwert JA, Luck RF, Werren JH. Molecular identification of microorganisms associated with parthenogenesis. Nature. 1993;361:66–68. doi: 10.1038/361066a0. [DOI] [PubMed] [Google Scholar]
- 20.Ogata H, Suhre K, Claverie JM. Discovery of protein-coding palindromic repeats in Wolbachia. Trends Microbiol. 2005;13:253–255. doi: 10.1016/j.tim.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Fuxelius HH, Darby AC, Cho NH, Andersson SG. Visualization of pseudogenes in intracellular bacteria reveals the different tracks to gene destruction. Genome Biol. 2008;9:R42. doi: 10.1186/gb-2008-9-2-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jolley KA, Wilson DJ, Kriz P, McVean G, Maiden MC. The influence of mutation, recombination, population history, and selection on patterns of genetic diversity in Neisseria meningitidis. Mol Biol Evol. 2005;22:562–569. doi: 10.1093/molbev/msi041. [DOI] [PubMed] [Google Scholar]
- 23.Martin DP, Williamson C, Posada D. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics. 2005;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- 24.Jiggins FM. The rate of recombination in Wolbachia bacteria. Mol Biol Evol. 2002;19:1640–1643. doi: 10.1093/oxfordjournals.molbev.a004228. [DOI] [PubMed] [Google Scholar]
- 25.Tamas I, et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 26.Cho NH, et al. The Orientia tsutsugamushi genome reveals massive proliferation of conjugative type IV secretion system and host-cell interaction genes. Proc Natl Acad Sci USA. 2007;104:7981–7986. doi: 10.1073/pnas.0611553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darby AC, Cho NH, Fuxelius HH, Westberg J, Andersson SG. Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet. 2007;23:511–520. doi: 10.1016/j.tig.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 28.Salzberg SL, et al. Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biol. 2005;6:R23. doi: 10.1186/gb-2005-6-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iturbe-Ormaetxe I, Riegler M, O'Neill SL. New names for old strains? Wolbachia wSim is actually wRi. Genome Biol. 2005;6:401. doi: 10.1186/gb-2005-6-7-401. author reply 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salzberg SL, et al. Correction: Serendipitous discovery of Wolbachia genomes in multiple Drosophila species. Genome Biol. 2005;6:402. doi: 10.1186/gb-2005-6-7-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kondo N, Nikoh N, Ijichi N, Shimada M, Fukatsu T. Genome fragment of Wolbachia endosymbiont transferred to X chromosome of host insect. Proc Natl Acad Sci USA. 2002;99:14280–14285. doi: 10.1073/pnas.222228199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fenn K, et al. Phylogenetic relationships of the Wolbachia of nematodes and arthropods. PLoS Pathog. 2006;2:e94. doi: 10.1371/journal.ppat.0020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hotopp JC, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 34.Nikoh N, et al. Wolbachia genome integrated in an insect chromosome: evolution and fate of laterally transferred endosymbiont genes. Genome Res. 2008;18:272–280. doi: 10.1101/gr.7144908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baldo L, Werren JH. Revisiting Wolbachia supergroup typing based on WSP: spurious lineages and discordance with MLST. Curr Microbiol. 2007;55:81–87. doi: 10.1007/s00284-007-0055-8. [DOI] [PubMed] [Google Scholar]
- 36.Berg OG, Kurland CG. Evolution of microbial genomes: sequence acquisition and loss. Mol Biol Evol. 2002;19:2265–2276. doi: 10.1093/oxfordjournals.molbev.a004050. [DOI] [PubMed] [Google Scholar]
- 37.Begun DJ, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sackton TB, et al. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39:1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- 39.Andersson B, Wentland MA, Ricafrente JY, Liu W, Gibbs RA. A “double adaptor” method for improved shotgun library construction. Anal Biochem. 1996;236:107–113. doi: 10.1006/abio.1996.0138. [DOI] [PubMed] [Google Scholar]
- 40.Sun LV, et al. Determination of Wolbachia genome size by pulsed-field gel electrophoresis. J Bacteriol. 2001;183:2219–2225. doi: 10.1128/JB.183.7.2219-2225.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 42.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 43.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 44.Salzberg SL, Delcher AL, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Badger JH, Olsen GJ. CRITICA: coding region identification tool invoking comparative analysis. Mol Biol Evol. 1999;16:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- 46.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 48.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 50.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 52.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.