Abstract
In development, timing is of the utmost importance, and the timing of developmental processes often changes as organisms evolve. In human evolution, developmental retardation, or neoteny, has been proposed as a possible mechanism that contributed to the rise of many human-specific features, including an increase in brain size and the emergence of human-specific cognitive traits. We analyzed mRNA expression in the prefrontal cortex of humans, chimpanzees, and rhesus macaques to determine whether human-specific neotenic changes are present at the gene expression level. We show that the brain transcriptome is dramatically remodeled during postnatal development and that developmental changes in the human brain are indeed delayed relative to other primates. This delay is not uniform across the human transcriptome but affects a specific subset of genes that play a potential role in neural development.
Keywords: human evolution, brain development, gene expression, heterochrony, chimpanzee
Humans differ from their closest living relatives, chimpanzees, in brain size and numerous cognitive traits (1–5). Humans also differ from chimpanzees in the timing of particular developmental landmarks. For example, female sexual maturity is reached between 8 and 9 years of age in chimpanzees and between 13 and 14 years in humans (6, 7). Studies of comparative primate morphology, some dating back to the 19th century, suggested that human ontogenesis proceeds at a slower rate than in other primates; consequently, adult humans retain features characteristic of juvenile primates. This type of heterochronic shift is known as neoteny (see ref. 6 and references therein). Neoteny has been ascribed a central role in human evolution (8); for example, as a possible explanation for the emergence of human-specific cognitive abilities through an extended period of high neuronal plasticity (4, 6, 9).
To date, human and chimpanzee ontogenesis have mainly been compared in terms of skeletal morphology. Results from these comparisons indicate that neoteny may indeed explain some human features, such as small jaws (10). They also show that neoteny is not a ubiquitous feature of the human phenotype (10–14). The reason for the large human brain size, for example, appears to be rapid early postnatal brain-growth rates rather than an extended brain-growth period in human infants (3). Meanwhile, the timing of human ontogenesis relative to that in other primates at the molecular and histological levels remains unexplored. For instance, it is unknown whether all genes expressed in the human brain show a consistent delay in expression timing relative to the chimpanzee or, alternatively, whether different structures or molecular networks are affected to different extents. More generally, how the transcriptome as a whole is affected by evolutionary shifts in developmental timing is an open question. Although studies in model organisms have previously documented how changes in gene expression timing during development can produce morphological and functional novelties (15, 16), this type of evolutionary change has not yet been investigated on a genome-wide scale.
More than 30 years ago, M. C. King and A. Wilson (17) proposed that identifying differences in the timing of gene expression during brain development between humans and apes would be crucial for understanding human evolution. Here we address this issue by analyzing genome-wide gene expression levels in human, chimpanzee, and macaque brains during postnatal development.
Results and Discussion
General Pattern of Expression Changes During Brain Development.
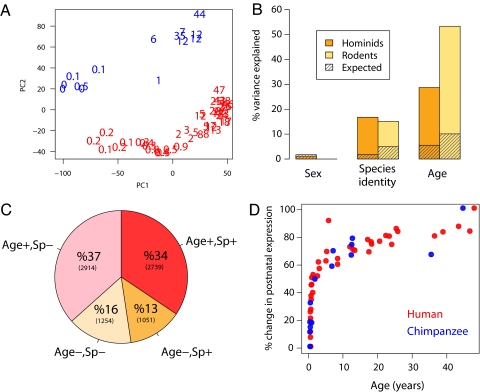
We analyzed gene expression levels in the dorsolateral prefrontal cortex (DLPFC) of 39 humans, 14 chimpanzees, and 9 rhesus macaques by using Affymetrix GeneChip Human Genome (GC HG)-U133 Plus 2.0 microarrays (see Materials and Methods and SI Appendix, Table S1). For both humans and chimpanzees, the individuals' age distributions cover the entire span of postnatal ontogenesis, with a particular focus on early life stages (Fig. 1A and SI Appendix, Fig. S1). In these individuals, we reliably detected and quantified the expression of 7,958 genes (Materials and Methods). Among these genes, we first analyzed the relative influence of 3 factors—age, sex, and species—on total expression variation among individuals. Quantitatively, age explains the largest part of the total expression variation at 29%, followed by species at 17% and sex at <2% (Fig. 1B). Although the effects of age and species are highly significant (permutation test, P < 0.001), the effect of sex is not (P = 0.54) (SI Appendix, Table S2). Thus, in our dataset, age has by far the greatest influence on expression levels. Consistently, we find that a striking 71% of the 7,958 genes expressed in the human brain change significantly during postnatal development [at a false discovery rate (FDR) of 10%] (Fig. 1C). Functionally, these genes are significantly enriched in a range of biological processes that include cell adhesion, synaptic transmission, and axonogenesis (permutation test for overall enrichment, P = 0.002) (SI Appendix, Table S3).
Fig. 1.
Expression variation during primate and rodent brain development. (A) The first 2 principle components of the human and chimpanzee DLPFC dataset. The numbers represent each individual's age in years. The first and second components explain 25% and 15% of the total variance and are significantly correlated with age (r = 0.86, P < 10−16) and species identity (r = 0.84, P < 10−16), respectively. Red, humans; blue, chimpanzees. (B) The mean proportion of the total variance explained by sex, species identity, and age across all expressed genes. The values for 39 humans and 14 chimpanzees (orange bars, left) are based on 7,958 genes. The values for rodents (yellow bars, right) are based on 8,362 genes measured in 18 individuals. The expected values are calculated as the median of 1,000 permutations of each factor. Note that the proportion of variance explained by sex does not exceed the random expectation in humans and chimpanzees, whereas in mice it is not estimated, because only males were used. (C) Proportions of age-related genes and genes showing significant expression differences between humans and chimpanzees in the DLPFC transcriptome. Age+/Age− represents genes showing/not showing a significant expression difference with age, and Sp+/Sp− represents genes showing/not showing a significant expression difference between species. The number of genes in each category is given in parentheses. (D) The percentage of global expression change relative to newborns. One-hundred percent change was designated as the difference between the youngest and oldest individuals (in humans or chimpanzees) in terms of the summary measure of global expression (see Materials and Methods). Each point represents an individual.
Next, we estimated when during human and chimpanzee brain development these expression changes take place. We find that in both species gene expression changes occur most rapidly during the first few years of life. Approximately 50% of the total expression change observed between newborns and 40-year-olds occurred within the first year of life (Fig. 1D). Furthermore, the overall trajectory of age-related expression changes in the chimpanzee brain, although based on fewer samples, closely resembles the human one (Fig. 1D). Notably, we also find similar trajectories of genome-wide expression changes with age in the brains of 2 species of mouse (Mus musculus and Mus spretus) from birth until adulthood (Materials and Methods and SI Appendix, Fig. S2). Furthermore, the trajectories of age-related changes also show a high correlation on the individual gene level, both between humans and chimpanzees (median Pearson r = 0.90) and between humans and mice (median r = 0.83) (SI Appendix, Table S4). Thus, the pattern of expression changes with age observed here is not particular to humans and chimpanzees and most likely reflects fundamental changes shared among mammals in the brain's molecular and histological organization during postnatal development.
Human–Chimpanzee Expression Differences.
We have found that age-related expression trajectories are generally conserved among humans, chimpanzees, and mice. Nevertheless, for individual genes the timing of expression changes may differ between species. To explore this issue, we tested each gene for expression differences between humans and chimpanzees by using multiple regression models. We find that 48% of age-related genes in humans are either expressed at significantly different levels or follow significantly different expression trajectories with age than in chimpanzees (at P < 0.0025, FDR = 10%) (Fig. 1C). Hence, despite the similarities between the expression profiles in human and chimpanzee brains described above, among all genes that change during prefrontal cortex development, approximately half change with age differently in the 2 species (Fig. 1C).
The proportion of genes classified as differently expressed between human and chimpanzee brains in this study is greater than previously reported (<10%) (18, 19) because of the larger sample size and different statistical criteria used here. Equalizing these factors, we find similar proportions of differently expressed genes (33% and 35%) as well as a significant overlap (50%, P < 10−10) between this and a published dataset (19).
A Test for Expression Heterochrony.
We next asked whether age-related expression differences between humans and chimpanzees reflect shifts in the timing of ontogenetic changes (i.e., heterochrony) between the 2 species. Heterochronic expression changes can be in the form of either delays (neoteny) or accelerations. Therefore, we designed a test to estimate both the direction and the amplitude of the shift in timing between expression profiles in the 2 species. In essence, this heterochrony test assumes that the species' ontogenetic trajectories are similar in shape (SI Appendix, Fig. S2) and then estimates whether introducing a difference in developmental timing significantly improves the fit between the 2 species' expression profiles (SI Appendix, Fig. S3).
Before applying this test to the human–chimpanzee comparison, we used it on a known developmental timing difference within the human brain: maturation of the human prefrontal cortex and of the caudate nucleus. The prefrontal cortex is among the last regions to mature during human brain ontogenesis (20, 21), but the caudate nucleus, a subcortical region of the brain, matures relatively early (20). To test whether we can detect a corresponding shift in gene expression timing, we compared gene expression patterns in the prefrontal cortex to those in the caudate nucleus in 13 human individuals aged 0–46 years old (Materials and Methods). We identified 2,979 genes as both age-related and differently expressed between the 2 brain regions. Applying the heterochrony test to these genes, we find that 2,261 of these genes show significant expression heterochrony between the 2 regions (at P < 0.05) and that for 58% of these genes the direction of the shift corresponds to slower maturation of the prefrontal cortex relative to the caudate nucleus (SI Appendix, Table S5). Thus, in line with anatomical observations, our test detects a substantial delay in postnatal development of the prefrontal cortex compared with the caudate nucleus on the gene expression level.
Testing the Neoteny Hypothesis of Human Evolution.
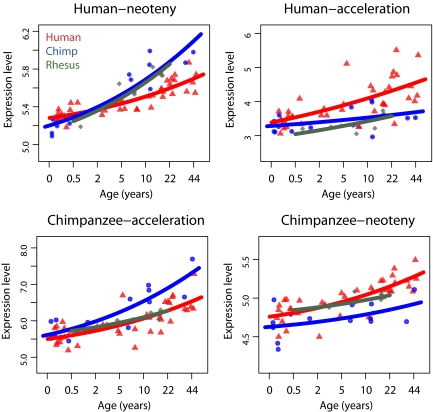
Next, to assess the differences in developmental timing between humans and other primates, we applied the heterochrony test to the human and chimpanzee expression data. To minimize possible sampling bias, we used a subset of 14 humans closely matching the 14 chimpanzees with respect to age and sex. Furthermore, we used the rhesus macaque as an outgroup to assign genes showing expression heterochrony to one of the following 4 phyloontogenetic categories: (i) human neoteny–expression changes occurred on the human lineage, and human expression corresponds to that in younger chimpanzees; (ii) human acceleration–expression changes occurred on the human lineage, and human expression corresponds to that in older chimpanzees; (iii) chimpanzee neoteny; and (iv) chimpanzee acceleration (Fig. 2).
Fig. 2.
Developmental shifts between humans and chimpanzees. The expression changes with age of 4 exemplary genes representing 4 phyloontogenetic patterns: human neoteny (EZH1), human acceleration (HER3), chimpanzee acceleration (ERBB2IP), and chimpanzee neoteny (MTMR2). The y axis shows normalized log2 expression levels. The x axis shows age in years. Each dot represents an individual; red, humans; blue, chimpanzees; dark green, rhesus macaques. The curves are fitted to the points using polynomial regression.
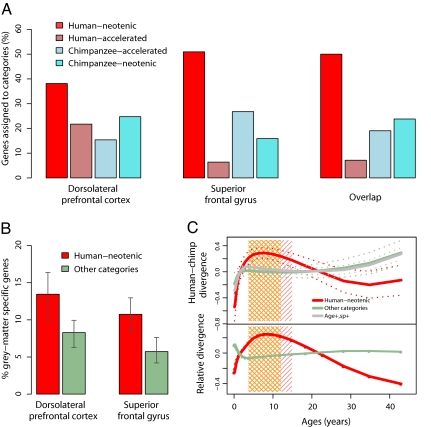
Among the 3,075 genes expressed in all 3 species, we could confidently assign 299 to one of these 4 categories. Of these 299 genes, approximately the same proportions (15 to 25%) fall into categories ii, iii, and iv. In contrast, we find approximately twice as many genes (38%) in category i, human neotenic genes (Fig. 3A). This enrichment is robust against the choice of P value cutoffs, models used to describe age-related changes of individual genes, and criteria used to define neoteny (Table 1 and SI Appendix, Table S6). Furthermore, we observe the same results by using the complete human dataset, rather than a subset of 14 individuals. In addition, we obtain qualitatively the same results regardless of our assumptions concerning the gestation-time difference between humans and chimpanzees (Table 1). By comparing humans with humans, we also estimate the FDR of the test as ≈10% (SI Appendix, Table S7).
Fig. 3.
Gene expression neoteny in the human brain. (A) The distribution of genes among phyloontogenetic categories in the 2 prefrontal cortex areas and their overlap. (B) The proportion of gray-matter-specific genes among human-neotenic genes (red) or among genes in the other 3 phyloontogenetic categories (green). The error bars indicate 95% confidence intervals estimated by bootstrapping across genes within a category 10,000 times. (C) (Upper) Changes in human–chimpanzee expression divergence with age. The solid lines indicate the mean normalized expression divergence between humans and chimpanzees across the age range for human-neotenic genes (red), genes in the other 3 phyloontogenetic categories (green), and all age-related and differentially expressed genes (Age+,sp+ genes, gray). The dotted lines indicate 95% confidence intervals estimated by bootstrapping across genes within a category 10,000 times. (Lower) Changes in human–chimpanzee divergence for human-neotenic genes (red) and for genes in the other 3 categories (green) relative to all age-related and differentially expressed genes. The shaded areas indicate the age range where the 95% bootstrap intervals of human-neotenic genes do not overlap with age-related and differentially expressed genes (pink) or with genes in the other 3 categories (orange). For Figs. 3B and 3C, as well as for the overlapping genes shown in 3A, we chose gene sets by using relaxed significance cutoffs (Table 1). Using other criteria yields the same principal results.
Table 1.
Expression heterogeneity test results
| Dataset | DLPFC |
SFG |
Overlap |
|||
|---|---|---|---|---|---|---|
| Numbers of humans vs. chimpanzees compared | 14 vs. 14 | 39 vs. 14 | 14 vs. 14 | 14 vs. 14 | 9 vs. 9 | 14 vs. 14, 9 vs. 9 |
| Assumed gestation-time difference in days | 0 | 0 | 20 | 51 | 0 | 0 |
| Number of expressed genes | 3,075 | 3,075 | 3,075 | 3,075 | 11,333 | 2,863 |
| Human-neotenic* | 114 | 154 | 121 | 131 | 234 | 6 |
| Human-accelerated* | 65 | 88 | 59 | 54 | 25 | 2 |
| Chimpanzee-accelerated* | 46 | 86 | 59 | 63 | 127 | 3 |
| Chimpanzee-neotenic* | 74 | 118 | 78 | 72 | 70 | 0 |
| P neoteny*† | 2 × 10−4 | 1 × 10−5 | 2 × 10−6 | 7 × 10−9 | 5 × 10−44 | 1 × 10−1 |
| P human specificity*‡ | 4 × 10−8 | 7 × 10−6 | 2 × 10−6 | 6 × 10−7 | 1 × 10−8 | 3 × 10−1 |
| Human-neotenic§ | 55 | 97 | 61 | 65 | 115 | 0 |
| Human-accelerated§ | 31 | 61 | 23 | 23 | 15 | 1 |
| Chimpanzee-accelerated§ | 24 | 45 | 21 | 25 | 60 | 2 |
| Chimpanzee-neotenic§ | 38 | 78 | 43 | 38 | 46 | 0 |
| P neoteny†§ | 6 × 10−3 | 3 × 10−3 | 2 × 10−5 | 4 × 10−6 | 1 × 10−20 | 1 |
| P human specificity‡§ | 3 × 10−4 | 8 × 10−6 | 6 × 10−6 | 1 × 10−5 | 2 × 10−5 | 1 |
| Human-neotenic¶ | 171 | 255 | 208 | 232 | 684 | 29 |
| Human-accelerated¶ | 100 | 138 | 107 | 97 | 111 | 3 |
| Chimpanzee-accelerated¶ | 81 | 148 | 97 | 110 | 403 | 9 |
| Chimpanzee-neotenic¶ | 121 | 193 | 129 | 122 | 242 | 12 |
| P neoteny†¶ | 1 × 10−5 | 2 × 10−9 | 7 × 10−9 | 3 × 10−14 | 9 × 10−102 | 1 × 10−6 |
| P human specificity‡¶ | 7 × 10−9 | 5 × 10−8 | 1 × 10−10 | 2 × 10−11 | 7 × 10−18 | 8 × 10−4 |
Shown are the numbers of genes assigned to phyloontogenetic categories in the DLPFC or SFG datasets under a range of assumptions and criteria and the overlap between these gene sets (also see SI Appendix, Table S6).
*FDR = 10% for age and species effects; P < 0.05 for heterochrony and lineage (1-sided) tests.
†The binomial test P value for neoteny with the alternative hypothesis that >50% of genes that are assigned to the human lineage show delayed development in humans relative to chimpanzees.
‡The binomial test P value for human specificity with the alternative hypothesis that >50% of genes that show delayed development in humans vs. chimpanzees are assigned to the human lineage.
§Stringent cutoffs: P < 0.01 in all 4 tests.
¶Relaxed cutoffs: P < 0.10 in all 4 tests.
To determine the generality and robustness of this result, we generated a second brain development dataset by using another prefrontal cortex region, the superior frontal gyrus (SFG), taking samples from 9 human, 9 chimpanzee, and 9 rhesus macaque individuals. All human and rhesus individuals differed from those used in the first dataset, and the experiment was conducted on a different microarray platform (Affymetrix Human Gene 1.0 ST). Applying the same analysis procedure to these data, we again find a significant excess of human neotenic genes compared with the other 3 phyloontogenetic categories (Fig. 3A and Table 1). The overlap between the human neotenic genes identified in the 2 brain regions is approximately twice as large as expected [Fisher's exact test, P < 0.01] (SI Appendix, Table S8). Thus, there is a reproducible, human-specific, neotenic shift in gene expression during postnatal maturation of the human prefrontal cortex, causing adult humans to resemble juvenile chimpanzees in their expression profiles (SI Appendix, Fig. S4).
This shift, however, affects a limited portion of the total cortical transcriptome (≈4%). By using simulations, we show that our test may fail to identify many real heterochronic changes (SI Appendix, Table S7), and therefore the actual percentage of genes affected by a human neotenic shift is likely to be higher. Still, based on the simulation results, we can confidently exclude the existence of a global neotenic shift affecting the entire transcriptome of the human frontal cortex (SI Appendix, Table S7). Thus, a parallel may be drawn between this result and previous morphological studies. Earlier formulations of the neoteny hypothesis of human evolution (e.g., that by L. Bolk) postulated that an ontogenetic shift affects human development in its entirety, including multiple organs and tissues (6). This notion was later modified and restricted the neotenic shift to brain growth (10, 11). Similarly, we find that the neotenic shift affects a limited group of genes expressed in the brain rather than the entire brain transcriptome. In a more general sense, this result suggests that mosaic evolution, as seen in brain structures among mammals (22), also applies to the evolution of gene expression patterns.
Properties of Human Neotenic Genes.
We analyzed the genes affected by the neotenic shift in the human prefrontal cortex with respect to their histological location, function, regulation, and expression timing. First, with respect to their histological location, we used published gene expression data from human gray and white matter (23) and found that, in both brain regions, human neotenic genes are significantly overrepresented among genes expressed specifically in gray matter (P = 0.06 and P = 0.0001 in DLPFC and SFG, respectively) but not among genes expressed in white matter (P > 0.6) (Fig. 3B and SI Appendix, Table S9).
Second, with respect to function, we find that in both brain regions human neotenic genes show a tendency to cluster in biological processes related to growth and development (SI Appendix, Tables S10 and S11). When each dataset was considered separately, this result was not significant (P > 0.1).
Third, with respect to expression timing, we asked whether the observed neotenic shift affects human postnatal ontogenesis in its entirety or is limited to a particular age interval. If the latter is true, human–chimpanzee expression divergence should increase at the corresponding age. We thus compared the extent of expression divergence across age among human neotenic genes to expression divergence among genes in the other 3 phyloontogenetic categories as well as of all age-related and differentially expressed genes. For all gene categories, human–chimpanzee expression divergence is relatively small after birth and subsequently increases. Human neotenic genes, however, diverge more rapidly than genes in the other categories during early adolescence, showing the largest difference to other groups ≈10 years of age (Fig. 3C). We reproducibly find the same result in the DLPFC dataset by using either all 39 or 14 selected human individuals. We find a similar increase in expression divergence around adolescence in the second dataset from the SFG (because of the limited sample size and wider age distribution in this dataset, we could not assess the timing and significance of this increase with confidence). Thus, at least in the DLPFC, the neotenic shift does not uniformly affect human life span but is particularly prominent in early adolescence.
Conclusion
By comparing the gene expression profiles in human, chimpanzee, and rhesus macaque prefrontal cortices throughout postnatal development, we have found that there is no uniform shift in the developmental timing between humans and other primates. We find instead a significant excess of genes showing neotenic expression in humans. This result is in line with the neoteny hypothesis of human evolution (6) and provides insight into the possible functional role of neoteny in human brain development. Specifically, we show that at least in one of the 2 cortical regions studied, the neotenic shift is most pronounced at the time when humans approach sexual maturity, a process known to be delayed in humans relative to chimpanzees or other primates (6, 24). Furthermore, the neotenic shift particularly affects a group of genes preferentially expressed in gray matter. Intriguingly, the timing of the shift also corresponds to a period of substantial cortical reorganization characterized by a decrease in gray-matter volume, which is thought to be related to synaptic elimination (21, 25, 26). The developmental pace of changes in gray-matter volume has been associated with the development of cognitive skills among humans (e.g., linguistic skills) (27) as well as with the development of disorders (e.g., attention-deficit/hyperactivity disorder) (28).
Although the precise causes and consequences of the human neotenic shift remain unknown, together these observations suggest that ontogenetic timing differences between the human and the chimpanzee prefrontal cortex transcriptomes may reflect differences in sexual and cognitive maturation between the 2 species. According to this logic, delayed gray-matter maturation in the human prefrontal cortex may extend the period of neuronal plasticity associated with active learning, thus providing humans with additional time to acquire knowledge and skills.
Materials and Methods
Sample Collection and Hybridization.
For the first primate dataset, we dissected postmortem DLPFC samples containing 90–95% gray matter from 39 humans (ranging in age from 0–47 years; 67% males), 13 chimpanzees (0–44 years; 64% males), and 9 rhesus macaques (1–18 years; 44% males), and caudate nucleus samples from 13 humans (0–46 years; 78% males) (SI Appendix, Table S1). RNA extracts from the dissections were used to generate labeled cRNA and hybridized to Affymetrix GC HG-U133 Plus 2.0 arrays according to the standard protocol. Among the 3 species, human samples have more detailed sample information. Testing the effects of various sample characteristics—such as sex, RNA quality, postmortem delay, prolonged agonal state, and brain tissue pH—on gene expression, we found that none systematically covaried with age (SI Appendix, Tables S12 and S13). Furthermore, although human and chimpanzee samples showed a slight but significant difference in RNA quality, accounting for this difference by using linear regression did not affect the results (SI Appendix, Table S6).
The second primate dataset was generated by using SFG samples from 9 humans (ranging in age from 0–66 years), 9 chimpanzees (0–44 years), and 9 rhesus macaques (0–28 years) and hybridized to Affymetrix GeneChip Human Gene 1.0 ST arrays. All humans and all but one rhesus macaque individual were males and differed from the individuals used in the first dataset. For chimpanzee samples, 5 of the 9 individuals were males, and there was no significant bias in the distribution of male and female samples with age. Furthermore, there was no significant difference in RNA quality between species or within each species with age (F test, P > 0.3). All samples were dissected by the same person (Z.Y.) and contained gray and white matter at an approximate ratio of 2:1.
For the mouse experiment, RNA from whole-brain samples of 9 M. musculus and 9 M. spretus individuals (0, 14, or 56 days old) were hybridized to Affymetrix MG-430 2.0 GeneChip arrays. Because mice reach sexual maturation at ≈42 days of age, the relative ages of the mice used here (0, 14, and 56 days) approximate the age distributions of the primates analyzed.
To analyze chimpanzee and rhesus macaque expression profiles in an unbiased way, in both primate datasets we masked the array probes that did not match the analyzed species' genomes perfectly and uniquely by using the corresponding reference genomes (SI Appendix, Table S14). Similarly, for mice we identified MG-430 2.0 probes that did not match the M. spretus genome by using a statistical algorithm that detects discordant expression patterns among probes within a probe set (SI Appendix, “Supporting Materials and Methods”); these probes were masked.
For the GC HG-U133 Plus2.0 and MG-430 2.0 arrays, to group probes into probe sets, we used published chip definition files based on Ensembl genes (29). Expression levels were summarized, log-transformed, and normalized by using the “rma” function in the R Bioconductor “affy” package (30). For the HuGene 1.0 arrays we used the “rma” algorithm adjusted to this novel array type to compute gene expression values and Affymetrix annotation files to map transcript clusters to Ensembl genes (SI Appendix, “Supporting Materials and Methods”).
In all experiments, probe sets with <8 probes remaining after the masking procedure were excluded from the analysis. Genes with detected expression levels above the background level in one third of the individuals in either species were considered expressed. Using different expression detection criteria did not affect our results (SI Appendix, Table S6). All original data files from the microarray experiments have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) with the accession numbers GSE11512 (GC HG-U133 Plus2.0 experiments) and GSE11528 (MG-430 2.0 experiments) (GSE15163 for the HuGene 1.0 ST experiments). The Chip Definition Files and R code used in the analysis can be found at www.picb.ac.cn/Comparative/data.html.
Analyses of Age and Species Effects.
For analyzing age-related changes, we used base-2, log-transformed ages (log-age) starting from the inferred conception date. Log-transformed ages are frequently used to analyze developmental phenotypes with parametric models (e.g., 31–33). In our dataset, this transformation yields a more uniform distribution of errors across ages than the linear age scale, making it more suitable for statistical analysis (34). Nonetheless, an analysis based on the linear age scale results in the same principal findings (SI Appendix, Table S6). Another factor related to age assignment is the difference in gestation time between species. Although humans are known to have a longer gestation time than chimpanzees, the exact extent of this difference is not certain and may vary among individuals. Therefore, to avoid any bias that can be caused by incorrect assumptions concerning the gestation-time difference, we assigned the same gestation time (280 days) to humans and chimpanzees in the main analysis. Such an assignment is conservative, because it ignores a shorter gestation period in chimpanzee and biases our results against finding human-specific neoteny. Using different gestation times for humans and chimpanzees did not affect our findings, including the lower percentage of detected neotenic shifts than would have been expected from a global shift (Table 1).
For calculating the proportion of variance explained by age, species identity, or sex among all expressed genes, we used cubic regression models with log-age and linear regression models with species identity or with sex, respectively. We used the multidimensional scaling algorithm “isoMDS” in the R “MASS” package (35) to calculate a 1D summary measure of global expression shown in Fig. 1D. We tested the age-related expression changes per gene by using polynomial regression models with log-age and employing the F test (SI Appendix, Table S15). For each gene, the regression model was chosen from all possible linear-to-cubic models by applying the adjusted r2 criterion (36). We tested human–chimpanzee expression differences per gene by using multiple regression models with log-age and species as factors and used the F test to assess significance (SI Appendix, Table S16). In both the age and species tests, FDRs were computed by 1,000 random permutations of age or species assignments and fixed at 10%. Differently expressed genes between species were assigned to the human lineage if the distances among the 9 rhesus expression values and the human expression age curve were significantly greater than the distances to the chimpanzee curve (1-sided Wilcoxon test, P < 0.05) and vice versa for genes assigned to the chimpanzee lineage (SI Appendix, Table S17). Using multiple regression models for lineage assignment gives the same results (SI Appendix, Table S6).
Testing Expression Heterochrony.
We identified and quantified differences in expression timing between humans and chimpanzees (or between the prefrontal cortex and the caudate nucleus) by testing whether a linear age transformation (i.e., an age shift) significantly minimizes the difference between the expression age trajectories of the 2 groups (see Fig. S3). To find the optimal age shift for each gene, we used a nonlinear least squares algorithm (NL2SOL) (“nls” function in the R “stats” package). The significance of the age transformation was assessed by using the F test (P < 0.05). We also tested whether the age transformation explains human–chimpanzee differences (or prefrontal cortex–caudate nucleus differences) at least as efficiently as a constant expression-level difference between the two groups (SI Appendix, “Supporting Materials and Methods”). We estimated the false-positive and false-negative rates by comparing humans with humans and by simulating transcriptome-wide age shifts on the orders of 10–30% and 70%, respectively (SI Appendix, Table S7).
Note that current literature distinguishes among 3 types of heterochrony: a difference in the onset of development, a difference in the rate of development, and a difference in the duration of development (13, 37). In this context, our test evaluates either a shift in the onset of development or a difference in developmental rates without distinguishing between the two, a result of our limited sample size (SI Appendix, “Supporting Materials and Methods”). Because of this limitation, we use the term “neoteny” to collectively describe all detectable situations in which adult human expression levels resemble expression levels of younger chimpanzees (SI Appendix, Fig. S4).
Characterization of Human-Neotenic Genes.
We compared human-neotenic genes to genes identified in the other 3 ontogenetic categories in terms of (i) their enrichment in gene ontology groups under the biological process taxonomy (38) by using the FUNC tool (39), which applies a correction for multiple testing; (ii) their enrichment in genes expressed in specific brain structures; and (iii) changes in divergence between humans and chimpanzees across age, calculated as the absolute distance between the human and chimpanzee expression age curves. In point ii, we identified 1,155 and 578 genes with gray- and white-matter-specific expression, defined as a 3-fold difference in expression between these structures by using published microarray data from human cortical gray and white matter (23) (SI Appendix, “Supporting Materials and Methods”). For all tests, we used phyloontogenetic gene sets defined based on FDRs as well as gene sets defined based on relaxed significance cutoffs (at P < 0.1) (see Table 1).
Supplementary Material
Acknowledgments.
We thank A. Fisher, H. R. Zielke, R. Vigorito, T. Arendt, W. Enard, and J. Visagie for assistance with experiments and analysis; Prof. R. Martin and the Anthropological Institute of the University of Zürich for sharing samples; S. Leigh and G. Bartzokis for sharing data and results; C. Green, R. E. Green, and 2 anonymous reviewers for suggestions on the manuscript; and T. Grossmann, H. B. Fraser, R. Mundy, J. Good, E. Herrmann, P. Ledoux, K. Strimmer, the Gene Expression and Theoretical Biology Groups in Leipzig, and the Comparative Biology Group in Shanghai for helpful discussions. This work was supported by Grant 2007CB947004 from the National Basic Research Program of the People's Republic of China, Chinese Academy of Sciences Knowledge Innovation Program KSCX2-YW-R-09, the Max Planck Society, and Bundesministerum für Bildung und Forschung Grant PPO-S25T11.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (GEO accession nos. GSE11528, GSE11512, and GSE15163).
This article contains supporting information online at www.pnas.org/cgi/content/full/0900544106/DCSupplemental.
References
- 1.Smith JM, Szathmary E. The Major Transitions in Evolution. New York: Oxford Univ Press; 1998. [Google Scholar]
- 2.Carroll SB. Genetics and the making of Homo sapiens. Nature. 2003;422:849–857. doi: 10.1038/nature01495. [DOI] [PubMed] [Google Scholar]
- 3.Leigh S. Brain growth, life history, and cognition in primate and human evolution. Am J Primatol. 2004;62:139–164. doi: 10.1002/ajp.20012. [DOI] [PubMed] [Google Scholar]
- 4.Langer J. The heterochronic evolution of primate cognitive development. Biol Theory. 2006;1:41–43. [Google Scholar]
- 5.Herrmann E, Call J, Hernandez-Lloreda MV, Hare B, Tomasello M. Humans have evolved specialized skills of social cognition: The cultural intelligence hypothesis. Science. 2007;317:1360–1366. doi: 10.1126/science.1146282. [DOI] [PubMed] [Google Scholar]
- 6.Gould S. J. Ontogeny and Phylogeny. Cambridge, MA: Harvard Univ Press; 1977. [Google Scholar]
- 7.de Magalhães JP. [Accessed May 2008];AnAge Database. 2006 build 9. Available at http://genomics.senescence.info/species.
- 8.Montagu MFA. Time, morphology, and neoteny in the evolution of man. Am Anthropol. 1955;57:13–27. [Google Scholar]
- 9.Johnson MH. Functional brain development in humans. Nat Rev Neurosci. 2001;2:475–483. doi: 10.1038/35081509. [DOI] [PubMed] [Google Scholar]
- 10.Penin X, Berge C, Baylac M. Ontogenetic study of the skull in modern humans and the common chimpanzees: Neotenic hypothesis reconsidered with a tridimensional procrustes analysis. Am J Phys Anthropol. 2002;118:50–62. doi: 10.1002/ajpa.10044. [DOI] [PubMed] [Google Scholar]
- 11.Shea BT. Heterochrony in human evolution: The case for neoteny reconsidered. Am J Phys Anthropol. 1989;32:69–101. [Google Scholar]
- 12.McNamara KJ. Shapes of Time. Baltimore: John Hopkins Univ Press; 1997. [Google Scholar]
- 13.Rice SH. In: Human Evolution Through Developmental Change. Minugh-Purvis N, McNamara KJ, editors. Baltimore: Johns Hopkins Univ Press; 2002. [Google Scholar]
- 14.Mitteroecker P, Gunz P, Bernhard M, Schaefer K, Bookstein FL. Comparison of cranial ontogenetic trajectories among great apes and humans. J Hum Evol. 2004;46:679–698. doi: 10.1016/j.jhevol.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Zakany J, Gerard M, Favier B, Duboule D. Deletion of a HoxD enhancer induces transcriptional heterochrony leading to transposition of the sacrum. EMBO J. 1997;16:4393–4402. doi: 10.1093/emboj/16.14.4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Kerr JQ, Min G. Molecular heterochrony in the early development of Drosophila. Proc Natl Acad Sci USA. 2000;97:212–216. doi: 10.1073/pnas.97.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 18.Caceres M, et al. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci USA. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khaitovich P, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- 20.Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- 21.Toga AW, Thompson PM, Sowell ER. Mapping brain maturation. Trends Neurosci. 2006;29:148–159. doi: 10.1016/j.tins.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton RA, Harvey PH. Mosaic evolution of brain structure in mammals. Nature. 2000;405:1055–1058. doi: 10.1038/35016580. [DOI] [PubMed] [Google Scholar]
- 23.Erraji-Benchekroun L, et al. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry. 2005;57:549–558. doi: 10.1016/j.biopsych.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 24.Hill K, et al. Mortality rates among wild chimpanzees. J Hum Evol. 2001;40:437–450. doi: 10.1006/jhev.2001.0469. [DOI] [PubMed] [Google Scholar]
- 25.Giedd JN, et al. Brain development during childhood and adolescence: A longitudinal MRI study. Nat Neurosci. 1999;2:861–863. doi: 10.1038/13158. [DOI] [PubMed] [Google Scholar]
- 26.Glantz LA, Gilmore JH, Hamer RM, Lieberman JA, Jarskog LF. Synaptophysin and postsynaptic density protein 95 in the human prefrontal cortex from mid-gestation into early adulthood. Neurosci. 2007;149:582–591. doi: 10.1016/j.neuroscience.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu LH, et al. Normal developmental changes in inferior frontal gray matter are associated with improvement in phonological processing: A longitudinal MRI analysis. Cereb Cortex. 2007;17:1092–1099. doi: 10.1093/cercor/bhl019. [DOI] [PubMed] [Google Scholar]
- 28.Shaw P, et al. Attention-deficit/hyperactivity disorder is characterized by a delay in cortical maturation. Proc Natl Acad Sci USA. 2007;104:19649–19654. doi: 10.1073/pnas.0707741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of genechip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 31.Clancy B, Darlington RB, Finlay BL. Translating developmental time across mammalian species. Neuroscience. 2001;105:7–17. doi: 10.1016/s0306-4522(01)00171-3. [DOI] [PubMed] [Google Scholar]
- 32.Lu L, Airey DC, Williams RW. Complex trait analysis of the hippocampus: Mapping and biometric analysis of two novel gene loci with specific effects on hippocampal structure in mice. J Neurosci. 2001;21:3503–3514. doi: 10.1523/JNEUROSCI.21-10-03503.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shupe JM, Kristan DM, Austad SN, Stenkamp DL. The eye of the laboratory mouse remains anatomically adapted for natural conditions. Brain Behav Evol. 2006;67:39–52. doi: 10.1159/000088857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sokal RR, Rohlf FJ. Biometry. New York: Freeman; 1995. [Google Scholar]
- 35.Venables WN, Ripley BD. Modern Applied Statistics with S. New York: Springer; 2002. [Google Scholar]
- 36.Faraway J. [Accessed May 2008];Practical Regression and ANOVA Using R. 2002 Available at: http://cran.r-project.org/doc/contrib/Faraway-PRA.pdf.
- 37.Alberch P, Gould SJ, Oster GF, Wake DB. Size and shape in ontogeny and phylogeny. Paleobiology. 1979;5:296–317. [Google Scholar]
- 38.Ashburner M, et al. Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prüfer K, et al. FUNC: A package for detecting significant associations between gene sets and ontological annotations. BMC Bioinformatics. 2007;8:41. doi: 10.1186/1471-2105-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.