Abstract
In eukaryotic mismatch repair (MMR), degradation of the error-containing strand initiates at nicks or gaps that can be up to a kilobase away from the mispair. These discontinuities may be the ends of Okazaki fragments or the 3′-termini of the leading strands during replication, whereas the termini of invading strands may fulfill this role during recombination. Here we show that, in extracts of human cells, MMR can initiate also at sites of ongoing base excision repair. Although unlikely under normal circumstances, this situation may arise in vivo during somatic hypermutation (SHM) and class switch recombination of Ig genes, where activation-induced cytidine deaminase (AID) generates multiple U/G mismatches in the variable or switch regions. Uracil should normally be excised by base excision repair (BER), but we show here that MMR proteins activated by a nearby mismatch interfere with uracil processing to generate long single-stranded gaps. We postulate that, in a subset of the repair events, filling-in of the MMR-generated gaps might be catalyzed by the error-prone polymerase-η, rather than by the high-fidelity polymerase-δ. Because polymerase-η has a propensity to misinsertions opposite adenine residues, the above mechanism would help explain why SHM affects not only C/G, but also A/T base pairs.
Keywords: antibody diversity, class switch recombination
SHM is initiated by activation-induced cytidine deaminase (AID) (1), which converts multiple cytidines to uridines in the vicinity of complementarity determining regions, as well as around the switch loci of the Ig genes (2–4). Rather than repairing these uracils by error-free base excision repair (BER), pathogen-activated B-cells process these lesions in a highly error-prone manner, such that essentially any type of mutation, transition or transversion, can arise at C/G sites. However, mutations at A/T pairs appear with similar frequency, and these cannot be assigned to error-prone BER. Genetic studies with knockout mice have implicated the mismatch repair genes Exo1, Msh2, and Msh6, as well as the gene encoding polymerase-η, in A/T mutagenesis. The latter enzyme has a propensity for misinsertions opposite A in the template strand (5), but it isn't known to participate in BER, and it is unclear how it might be involved in error-prone DNA synthesis at the Ig loci.
The involvement of the mismatch repair (MMR) system in SHM was also puzzling. First, Mlh1 and Pms2, which are essential for MMR, affected SHM to a substantially lesser extent than Msh2 and Msh6, which suggested that the hypermutation process did not involve canonical MMR. Second, there was no evidence implicating MMR proteins in the processing of uracil-containing lesions, even though the Msh2/Msh6 heterodimer (MutSα) is known to bind U/G mispairs (6). That is because eukaryotic MMR initiates at strand discontinuities, such as nicks or gaps that are distal to the mispair (7–9). No such termini are known to exist in the proximity of AID-generated uracils.
AID is believed to be targeted to its sites of action by transcription (10–12). A single deamination event in a transcription bubble or in transiently underwound DNA would generate a uracil residue in one of the strands, which would give rise to a U/G mispair once transcription had moved on and the DNA had reannealed. Under normal circumstances, this lesion should be addressed by BER, where the uracil is removed by one of four uracil DNA glycosylases (UNG2, TDG, MBD4, or SMUG1), giving rise to an abasic (apurinic or apyrimidinic; AP) site. Subsequent cleavage of the sugar-phosphate backbone by an AP-endonuclease provides an entry point for polymerase-β, which extends the 3′-terminus of the nick by inserting a dCMP residue. Concurrently, pol-β cleaves off the baseless sugar-phosphate residue at the 5′-end of the gap to leave a nick, which is sealed by DNA ligase III (3, 13). We wanted to test whether this canonical BER process takes place also in extracts of B-cells and whether we could find any evidence of the involvement of MMR in U/G processing as suggested by the genetic studies.
Results
Covalently Closed Substrates Containing a Single Mismatch Are Refractory to MMR.
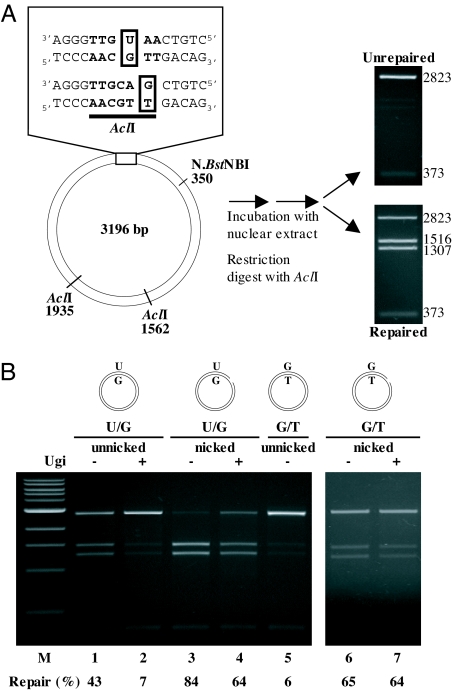
We constructed supercoiled phagemid substrates containing a single U/G or G/T mispair in the recognition sequence of the AclI endonuclease. The presence of these mispairs makes the DNA refractory to cleavage at this site, but correction of U/G to C/G, or of G/T to A/T, will regenerate the restriction site; digestion of the heteroduplex DNA with AclI will then give rise to two new fragments of 1516 and 1307 nucleotides (Fig. 1A). [Note that, in this work, mismatches are designated such that the first letter indicates the nucleotide in the complementary strand of the phagemid and the second letter denotes the nucleotide in the viral strand of the heteroduplex. In substrates containing more than one mismatch, the order in which they are listed indicates their respective positions. Thus, in the substrate G/T-U/G, the mispaired uracil is situated 3′ (downstream) from the mispaired guanine residue in the same (complementary) strand, whereas in the U/G-G/T heteroduplex, the uracil is 5′ (upstream) from the guanine, also in the complementary strand. Where nicks were introduced into the complementary strand, these were either 5′ or 3′ from the mispaired guanine of the G/T or the mispaired uracil of the U/G mispair in the AclI site, as specified in the text.] As shown in Fig. 1B, the U/G repair reaction was very efficient (lane 1). Inhibition of the two most active uracil glycosylases in extracts of human cells (14), TDG (by immunodepletion) and UNG2 (by the uracil glycosylase inhibitor; Ugi), made the substrate refractory to cleavage (lane 2), which confirmed that the U/G mispair was processed by BER in this extract. [Note that all extracts used in this study were depleted of TDG. In this way, uracil processing by BER could be inhibited simply by adding Ugi to the extracts (14).] The G/T mismatch in the supercoiled substrate was also not repaired (lane 5). This confirmed that circular heteroduplexes lacking nicks or gaps are refractory to MMR, because the latter strand discontinuities represent essential entry points for exonuclease 1 (EXO1), which catalyses the degradation of the discontinuous strand of the heteroduplex up to and approximately 150 nucleotides past the mispair (15). Correspondingly, the U/G and G/T substrates in the same substrates were very efficiently processed by MMR to C/G and A/T, respectively, when a nick was introduced approximately 350 nucleotides 5′ from the U or the G (lanes 4 and 6, respectively). That the G/T substrate was efficiently repaired even when BER was inhibited (lane 7) showed that neither UNG2 nor TDG were required for the nick-directed MMR process. This experiment also confirmed that the extracts contained no endonuclease capable of nicking the G/T or U/G substrates.
Fig. 1.
BER- and MMR-catalyzed repair of U/G and G/T mispairs in extracts of BL2 cells. (A) Schematic representation of the circular heteroduplex substrates carrying either a single U/G or G/T mispair in the recognition site of AclI endonuclease at nucleotide 44 or 46, respectively. The substrates were constructed by primer extension as described previously (14). The positions of the other AclI cleavage sites and of the N.BstNBI site, where a nick can be introduced selectively into the outer strand, are indicated. The numbering relates to the inner (viral) strand of the heteroduplex. The substrates were incubated with the cell extracts in the presence or absence of the UNG inhibitor Ugi, either covalently closed or nicked as shown above the respective figures. In the absence of repair, digestion of the phagemid DNA gave rise to fragments of 2,823 and 373 bp. Repair of the U/G mispair to C/G, or of the G/T to A/T regenerated the third AclI restriction site, such that the phagemid DNA was cut into 3 fragments of 1,516, 1,307, and 373 nucleotides. The efficiency of the repair reaction was estimated by ImageQuant from scans of ethidium bromide-stained agarose gels. (B) Repair of the U/G or G/T mismatches in covalently closed (lanes 1, 2, 5) or nicked (lanes 3, 4, 6, 7) heteroduplex substrates in extracts of BL2 cells. [Note that, in all repair experiments in this study, the extracts were immunodepleted of TDG, because this enzyme could be shown to process U/G mispairs with high efficiency in human cell extracts (24)]. MBD4 and SMUG1, the other known uracil glycosylases, are inactive under our reaction conditions, such that UNG2 is the only remaining uracil-processing enzyme in the TDG-depleted extracts. The U/G mispair in the unnicked substrate was efficiently processed by BER (lane 1), and this reaction could be inhibited by the addition of Ugi (lane 2). In a nicked substrate, the U/G mispair was addressed also by MMR (lane 3). Correspondingly, only the BER, and not the MMR reaction, was inhibited by Ugi (lane 4). The control experiments showed that the unnicked substrate G/T was refractory to MMR (lane 5), but was efficiently processed when a nick was introduced into the G strand by N.BstNBI (lane 6). Addition of Ugi to the reaction containing the nicked G/T substrate did not influence the efficiency of the repair process. The numbers under the lanes represent an average of at least 2 independent experiments. M, molecular size marker.
Partially-Processed U/G Mispairs Can Serve as Initiation Sites for MMR.
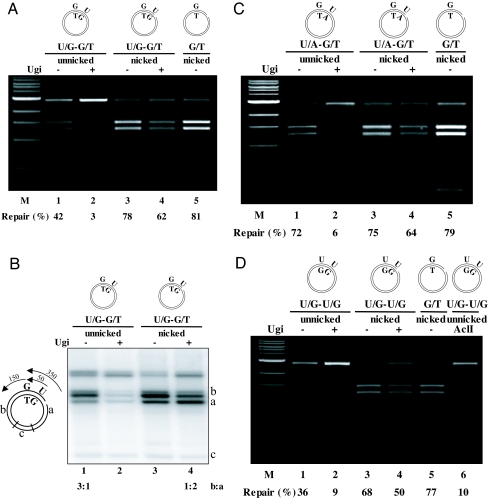
The above experiments confirmed that a single U/G mispair in a covalently closed circular substrate is not subject to MMR-dependent processing, despite being efficiently recognized by the mismatch recognition factor MutSα (a heterodimer of MSH2 and MSH6) (6), which is highly abundant in our extracts (data not shown) and generally in extracts of rapidly proliferating B-cells (16). However, available evidence suggests that AID catalyses several deaminations per molecule (17–19), which might give rise to several U/G mispairs in close proximity. We argued that in such a scenario, the picture might change. Upon mismatch recognition, MutSα undergoes an ATP-dependent conformational change, which converts it into a sliding clamp that causes it to leave the mismatch and diffuse along the DNA contour in search of a nick (20). On a covalently closed substrate containing a single U/G, the clamp would load at the mismatch, bind ATP, slide away, and fall off the DNA upon ATP hydrolysis, having failed to find a nick. In contrast, on a substrate containing more than one U/G, a MutSα sliding clamp loaded at one mispair might interfere with uracil processing by BER at nearby U/Gs and use such partially repaired sites to load EXO1. To test this hypothesis, we constructed a supercoiled U/G-G/T substrate with the uracil residue 54 bp 5′ from the guanine of the G/T mispair in the AclI restriction site. [A G/T rather than a U/G mispair in the AclI site was used in these experiments, because G/T is refractory to BER under our experimental conditions (Fig. 1B, lane 5), yet its processing efficiency by MMR is comparable to U/G (cf. lanes 4 and 6).] In the control reaction, the U/G-G/T substrate contained, in addition to the two mispairs, also a nick 296 nucleotides 5′ from the uracil. As shown in Fig. 2A, the G/T mismatch in the nicked G/T control substrate (lane 5) and in the nicked U/G-G/T substrate (lane 3) was corrected with high efficiency to A/T. The latter reaction was slightly inhibited by the addition of Ugi (lane 4), which was expected, as, upon BER inhibition, all repair events must commence 350 rather than 54 nucleotides away from the mispair. Importantly, however, the G/T mismatch in the unnicked U/G-G/T substrate was also corrected with appreciable efficiency (lane 1), and this repair could be almost totally inhibited by the addition of Ugi (lane 2). This shows that uracil processing in these extracts is not concerted and that BER intermediates are accessible to other enzymes, possibly because polymerase-β levels (and thus presumably also BER efficiency) are reduced in BL2 cells (21). In this case, MutSα loaded at the downstream G/T mismatch apparently loads EXO1 at the cleaved AP-site appearing after uracil excision to initiate a strand displacement reaction that results in the conversion of the G/T mispair to an A/T pair.
Fig. 2.
Intermediates of uracil processing can be used as MMR initiation sites in substrates containing a uracil residue upstream from a mispair. (A) A covalently closed circular substrate carrying a U/G mispair 54 nucleotides 5′ from a G/T mispair in the AclI site was repaired with high efficiency (lane 1), but this reaction could be inhibited by the addition of Ugi (lane 2). This shows that uracil processing by UNG2 and/or TDG was indispensable for this MMR-catalyzed process. Ugi inhibition could be overcome by the introduction of a nick into the substrate (lanes 3, 4). The nicked G/T substrate was used as a control (lane 5). M, molecular size marker. (B) Left panel shows the schematic representation of the radiolabeled MMR repair tracts starting either at the uracil residue or at the nick. AclI cuts the repaired phagemid into fragments a, b, and c. If the MMR repair tract starts at the uracil residue and continues approximately 150 nucleotides past the AclI site, fragment a should contain a stretch of 54 and fragment b of approximately 150 nucleotides labeled with [32P]dAMP. If the MMR repair tract starts at the nick and continues approximately 150 nucleotides past the AclI site, fragment a should contain a labeled stretch of 350 and fragment b of approximately 150 nucleotides. Right panel shows an autoradiograph of the repair reactions similar to lanes 1–4 in panel A above, carried out in the presence of [32P]dATP. (C) A uracil residue opposite adenine can function similarly to that in a U/G mispair. In this experiment, the G/T substrate contained a U/A base pair approximately 50 nucleotides 5′ from the G. Lanes as above. (D) In a covalently closed substrate containing two U/G mispairs, such as would arise through the repeated action of AID, the mispair in the AclI site (lane 1) was repaired to C/G with an efficiency similar to that observed in A above. The higher background in the Ugi-inhibited reaction (lane 2) is due to the fact that AclI partially cleaves DNA containing a U/G mispair in its recognition sequence, as shown in the digest of the untreated supercoiled substrate (lane 6). The numbers under the lanes represent an average of at least 2 independent experiments. M, molecular size marker.
MMR Events Commence at the Uracil Residue or at the Nick.
That the repair event in the supercoiled U/G-G/T substrate indeed initiated at the uracil is shown in Fig. 2B, in which the above assay was repeated using radioactive dATP. Because EXO1-catalyzed strand degradation is known to continue approximately 150 nucleotides past the mismatch (15), the repair tract initiating at the uracil residue should be approximately 200 nucleotides long. Thus, 1/4 of the radioactivity should be in the AclI fragment a and 3/4 in fragment b (see Fig. 2B Left), as was the case (lane 1). In contrast, the tract length in the nicked U/G-G/T substrate should be approximately 450 nucleotides, and thus 2/3 of the radioactivity should be in fragment a and 1/3 in fragment b, providing that only MMR was involved in the processing. Also this was the case (lane 4). When BER was active (lane 3), the band pattern resembled that seen with the supercoiled substrate, which indicated that most of the MMR events were starting at the uracil residue rather than at the nick.
Partially-Processed U/A Base Pairs Can also Serve as MMR Initiation Sites.
Having demonstrated that a uracil residue in a U/G mispair can act as a cryptic strand discontinuity and thus provide an entry point for MMR, we wanted to test whether uracils incorporated opposite adenines behave in a similar way. To this end, we constructed a U/A-G/T substrate with the uracil 54 nucleotides 5′ from the guanine in the G/T mismatch in the AclI site. As shown (Fig. 2C, lane 1), the U/A was used at least as efficiently as the U/G (Fig. 2C, lane 1) for MMR initiation.
MMR Interferes With BER also on a U/G-U/G Substrate.
Should AID generate several uracil residues, and thus several U/G mispairs, per molecule, as suggested by studies from the Goodman and Storb laboratories (17, 18), processing of the uracils by BER, or of the U/G mispairs by MMR, could theoretically begin at any one of the deaminated sites. Because of the short repair patch generated during BER, no interference between the processing of sites as close as approximately 10 nucleotides would be expected. In contrast, if both BER and MMR were involved, the outcome of the repair would depend on where the first strand break was introduced. In the above experiments, we used a combination of U/G and G/T mispairs to show that uracil processing provides an entry point for mismatch-activated strand displacement. To demonstrate the relevance of this finding to SHM, we constructed a substrate containing two U/G mismatches (Fig. 2D). Incubation of this supercoiled substrate with B-cell extracts resulted in an efficient repair of the U/G within the AclI site to a C/G (lane 1), which indicated that the MMR-catalyzed strand displacement initiated at the upstream uracil. Addition of Ugi inhibited the reaction (lane 2), which showed that the strand displacement process depended on a uracil glycosylase activity. When the U/G-U/G substrate contained also a nick 350 nucleotides upstream, the repair by MMR was more efficient (lane 3) and was only slightly inhibited by the addition of Ugi (lane 4), which showed that MMR is the predominant pathway in B-cell extracts acting on U/G mispairs in nicked substrates.
MutLα Is Dispensable for Uracil-Directed 5′ → 3′MMR.
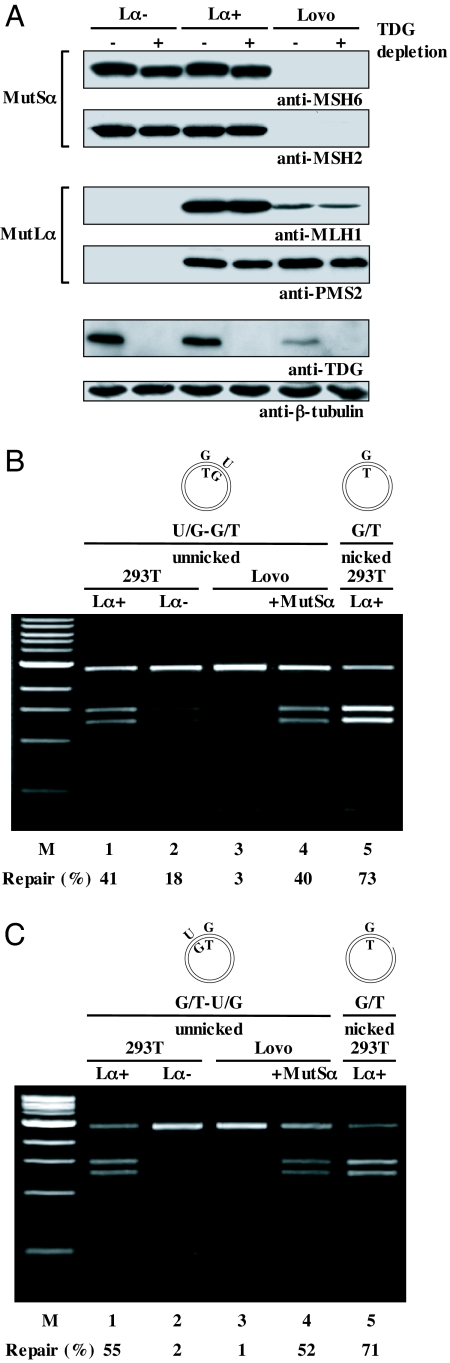
Mismatch repair is a bidirectional process, where the strand degradation reaction can take place in either 5′ → 3′ or 3′ → 5′ direction (8, 15). In our scenario, if mismatch- and ATP-activated MutSα encounters a nick 5′ from the mispaired guanine or uracil, it can load EXO1, which then catalyzes the degradation of the nicked strand in a 5′ → 3′ direction until the mispair had been corrected to A/T or C/G, respectively. Importantly, whereas this reaction is stimulated by the MLH1/PMS2 heterodimer MutLα, it is not absolutely dependent on it. In contrast, when the strand discrimination signal lies 3′ from the mispaired guanine or uracil, MutSα recruits MutLα, which then introduces several nicks into the nicked strand, both upstream and downstream from the mispair (22). These nicks are then used as excision initiation sites and the strand degradation proceeds in the 5′ → 3′ direction, as dictated by the polarity of EXO1. When the strand degradation begins at nicks situated 5′ from the mispair, the repair polymerase filling-in the EXO1-generated gap will replace the mispaired G or U with A or C, respectively. To test whether this factor requirement applies also to the uracil-directed MMR, we incubated the U/G-G/T substrate with extracts lacking either MutSα or MutLα. Extracts of 293T-Lα+ cells (23) express all 4 MMR proteins (Fig. 3A) and are MMR-proficient, as judged also by the > 70% efficiency of G/T → A/T repair in a 5′ nicked heteroduplex substrate (Fig. 3B, lane 5). In these extracts, the G/T → A/T repair in the supercoiled U/G-G/T substrate was approximately 40% (lane 1). Down-regulation of MLH1 expression in the 293T-Lα cells by doxycycline leads to a concurrent PMS2 degradation (Fig. 3A) and to a MMR-defect (23). In these 293T-Lα− extracts, the efficiency of G/T repair in the supercoiled U/G-G/T substrate was reduced to approximately 20% (lane 2). In contrast, the same substrate incubated with extracts of MutSα-deficient LoVo cells was fully refractory to AclI cleavage (lane 3), but repair efficiency could be restored by the addition of purified recombinant MutSα (lane 4), which shows that solely the lack of the latter factor was responsible for the MMR defect in the LoVo extracts.
Fig. 3.
Dependence of the uracil-initiated MMR reaction on MutSα and MutLα. (A) Western blot of extracts of human 293T-Lα and LoVo cells probed with MSH2, MSH6, MLH1, PMS2, and TDG antibodies. Although LoVo extracts lack the MutSα constituents MSH2 and MSH6, 293T-Lα+ extracts contain all 4 MMR proteins. However, when the cells are grown in the presence of doxycycline (293T-Lα−), MLH1 expression is turned off, and PMS2 is degraded in the absence of its cognate partner (23). β-Tubulin was used as the loading control. (B) The unnicked U/G-G/T substrate carrying a uracil residue 54 nucleotides 5′ from the G was processed with high efficiency in MMR-proficient extracts of 293T-Lα+ cells (lane 1). The G/T repair was less efficient in extracts of 293T-Lα− cells that do not express MutLα (lane 2). In contrast, extract of LoVo cells, which lack MutSα, were deficient in G/T repair (lane 3), unless supplemented with purified recombinant MutSα (lane 4). Lane 5 contains the nicked G/T control. (C) The unnicked G/T-U/G substrate carrying a uracil residue approximately 50 nucleotides 3′ from the G was processed with high efficiency in MMR-proficient extracts of 293T-Lα+ cells (lane 1). The G/T repair was detected neither in extracts of 293T-Lα− (lane 2) nor of LoVo (lane 3) cells. When the LoVo extracts were supplemented with purified recombinant MutSα, the substrate was processed efficiently (lane 4). Lane 5 contains the nicked G/T control. The numbers under the lanes represent an average of at least 2 independent experiments. M, molecular size marker.
Uracil-Directed 3′ → 5′ MMR Requires MutLα.
The above results show that a MutSα defect abolishes 5′ → 3′ repair, whereas MutLα deficiency only decreases its efficiency. This agrees with the finding from the Modrich laboratory, which demonstrated that MutLα increases the efficiency of 5′ → 3′ MMR, but that it is essential for the 3′ → 5′reaction (22). To test whether this applies also to the uracil-directed MMR, we constructed the G/T-U/G substrate, which contained the uracil residue 54 nucleotides 3′ from the mispaired G. Unlike in the U/G-G/T substrate, where approximately 20% 5′ → 3′ MMR was detected in the absence of MutLα (Fig. 3B, lane 2), the supercoiled G/T-U/G substrate, which requires 3′ → 5′ MMR, was refractory to repair in both 293T-Lα− (Fig. 3C, lane 2) and LoVo (lane 3) extracts. In contrast, the G/T-U/G and the control G/T substrate carrying a nick approximately 300 nucleotides 3′ from the mispaired guanine were repaired with similar efficiencies in the MMR-proficient 293T-Lα+ extracts (lanes 1 and 5) and in LoVo extracts supplemented with MutSα (lane 4). The absolute dependence of the MMR-dependent in vitro U/G repair on MutSα, but only a partial dependence on MutLα, corroborate the results of genetic studies, which showed that loss of Mlh1 affected SHM to a substantially lesser extent than mutations in Msh2 (3). Taken together, these results imply that the MutSα-dependent 5′ → 3′ process predominates during this phase of diversification of Ig genes.
Discussion
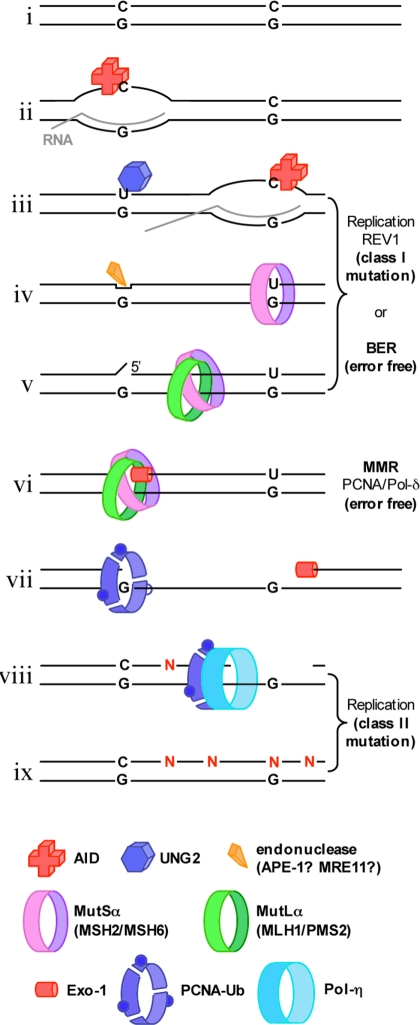
Based on our results, we propose that SHM could proceed as outlined in Fig. 4: AID is recruited to an Ig promoter (i), where it deaminates a cytosine residue in single-stranded DNA of a transcription bubble (ii). This gives rise to a U/G mispair in duplex DNA once the bubble has moved on (iii). This mispair may be detected by MutSα, but cannot be processed by MMR because it has no nicks in the vicinity. MutSα will therefore slide off and the uracil will be made available for processing by BER. If BER were to be interrupted immediately after the action of UNG2 or TDG, the uncleaved AP-site might persist until DNA replication, where its by-pass by REV1 would give rise to mutations at C/G sites (iii). This situation changes once a second cytosine deamination takes place in the moving transcription bubble (iv). Should a partially processed deamination site lie within approximately 1 kb (the maximum distance between a mismatch and a nick), the MutSα sliding clamp activated by the newly formed U/G (v) may interrupt the BER process at the distal uracil. If MutSα were to encounter an AP-site cleaved either by APE1 or MRE11, it might load EXO1 (vi), and the subsequent strand degradation would give rise to a single-stranded region spanning the distance between the first deamination site and approximately 150 nucleotides past the second one (vii). This gap would normally be filled in by the replicative polymerase-δ in an error-free manner. However, because the polymerase processivity factor PCNA in activated B-cells needs to be ubiquitylated (vii) in order for SHM to occur (25) and because ubiquitylated PCNA has a high affinity for the error-prone polymerase-η (26), it is possible that the ubiquitylated PCNA will recruit polymerase-η to a subset of the MMR repair patches (viii). Gap-filling by this error-prone enzyme (ix), which is known to introduce noncomplementary nucleotides (N) opposite Ts (27), would give rise to mutations at A/T base pairs (28).
Fig. 4.
Putative mechanism of somatic hypermutation. See text for details.
Our data help explain many of the enigmatic results obtained in genetic and biochemical experiments (2–4). It must be remembered, however, that the mutations observed in hypermutated B-cells arise through several distinct processes of DNA metabolism, which are at least in part redundant. Thus, the mechanism described in this study likely represents just one of several pathways leading to SHM, which coexists with others. None of these pathways need be particularly efficient, given that cells producing high-affinity antibodies are selected for. Importantly, the redundancy in SHM ensures that the loss of a single gene is unlikely to lead to immune deficiency. This helps ensure the survival of the organism on an evolutionary time scale.
Materials and Methods
Substrates, Nuclear Extracts, and in Vitro MMR Assays.
The detailed procedure has been described previously (14). Briefly, heteroduplex DNA substrates containing a G/T or a U/G mismatch within an AclI restriction site in the 46-bp polylinker of a pGEM13Zf(+) derivative were constructed by primer extension, using the mismatch-containing 50-mer oligonucleotides (U/G: 5′-GGC CGC GAT CTG ATC AGA TCC AGA CGT CTG TCA AUG TTG GGA AGC TTG AG-3′; G/T: 5′-GGC CGC GAT CTG ATC AGA TCC AGA CGT CTG TCG ACG TTG GGA AGC TTG AG-3′) as primers (uracil or mispaired residues are highlighted in bold) and the single-stranded phagemid DNA as template. To introduce a second U/G mismatch or U/A base pair in addition to the mismatch in the AclI restriction site, 89 (85)-mer oligonucleotides containing an additional uracil at the indicated site were used (U/G-U/G: 5′-CCA GTG AAT TGT AAT AUG AAC ACT ATA GGG CGA ATT GGC GGC CGC GAT CTG ATC AGA TCC AGA CGT CTG TCA AUG TTG GGA AGC TTG AG-3′; U/G-G/T: 5′-CCA GTG AAT TGT AAT AUG AAC ACT ATA GGG CGA ATT GGC GGC CGC GAT CTG ATC AGA TCC AGA CGT CTG TCG ACG TTG GGA AGC TTG AG-3′; U/A-G/T: 5′- CCA GTG AAT TGT AAU ACG AAC ACT ATA GGG CGA ATT GGC GGC CGC GAT CTG ATC AGA TCC AGA CGT CTG TCG ACG TTG GGA AGC TTG AG-3′; G/T-U/G: 5′-CCA GAC GTC TGT CG A CGT TGG GAA GCT TGA GTA TTC TAT AGT GTC ACC TAA ATA GCT TGG CGT AAT UAT GGT CAT AGC TGT TTC C-3′). Isolation of the desired supercoiled heteroduplex substrates and the MMR assays were carried out as described, using 100 ng (47.5 fmol) heteroduplex DNA substrate and 150 μg of nuclear extracts from BL2, 293T-Lα+ (MLH1+), 293T-Lα− (MLH1−) or Lovo cells in a total volume of 30 μL.
UDG Inhibition and TDG-Immunodepletion of Nuclear Extracts.
Protein A Dynabeads were washed twice with 30 mM Hepes-KOH, pH 7.5, 7 mM MgCl2. Anti-TDG antibody (1:10,000) was added and the beads were incubated for 2 h at 4 °C. They were then washed 3 times with the above buffer and stored at 4 °C. The extracts were immunodepleted of TDG by incubating with 6.3 μL antibody-preadsorbed Dynabeads for 30 min at 4 °C and subsequently used for in vitro MMR assays. Where indicated, UNG2 was inhibited by the addition of 3.6 μL UGI (7.2 units) per 150 μg nuclear extracts and incubation for 10 min at 37 °C.
Antibodies and Reagents.
The anti-TDG antibody (rabbit polyclonal) was a generous gift of Primo Schar. The UDG inhibitor UGI was obtained from New England Biolabs and the Protein A Dynabeads were obtained from Dynal Biotech, Invitrogen.
Acknowledgments.
The authors wish to express their gratitude to Myriam Marti for excellent technical assistance, to Barbara Schöpf for the generous gift of purified recombinant MutSα, to Alessandro Sartori for critical reading of the manuscript, and to Ruedi Aebersold and Hans Hengartner for helpful comments. The financial support of the Bonizzi-Theler Stiftung and the European Community (grant nos. LSHC-CT-2005–512113 and FP7-HEALTH-2007B-223545) to J.J., the Swiss National Science Foundation (grant no. 3100/068182.02/1) to D.C., J.J., and S.S., and of UBS AG to F.F. and J.J. is also gratefully acknowledged.
Footnotes
The authors declare no conflict of interest.
References
- 1.Honjo T, Nagaoka H, Shinkura R, Muramatsu M. AID to overcome the limitations of genomic information. Nat Immunol. 2005;6:655–661. doi: 10.1038/ni1218. [DOI] [PubMed] [Google Scholar]
- 2.Vallur AC, Yabuki M, Larson ED, Maizels N. AID in antibody perfection. Cell Mol Life Sci. 2007;64:555–565. doi: 10.1007/s00018-007-6434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 4.Longerich S, Basu U, Alt F, Storb U. AID in somatic hypermutation and class switch recombination. Curr Opin Immunol. 2006;18:164–174. doi: 10.1016/j.coi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 6.Wilson TM, et al. MSH2-MSH6 stimulates DNA polymerase eta, suggesting a role for A:T mutations in antibody genes. J Exp Med. 2005;201:637–645. doi: 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 8.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: Functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 9.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duquette ML, Pham P, Goodman MF, Maizels N. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005;24:5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- 11.Nambu Y, et al. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 12.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 13.Dianov GL, Sleeth KM, Dianova II, Allinson SL. Repair of abasic sites in DNA. Mutat Res. 2003;531:157–163. doi: 10.1016/j.mrfmmm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Fischer F, Baerenfaller K, Jiricny J. 5-Fluorouracil is efficiently removed from DNA by the base excision and mismatch repair systems. Gastroenterology. 2007;133:1858–1868. doi: 10.1053/j.gastro.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Fang WH, Modrich P. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J Biol Chem. 1993;268:11838–11844. [PubMed] [Google Scholar]
- 16.Marra G, et al. Expression of human MutS homolog 2 (hMSH2) protein in resting and proliferating cells. Oncogene. 1996;13:2189–2196. [PubMed] [Google Scholar]
- 17.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 18.Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc Natl Acad Sci USA. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen HM, Ratnam S, Storb U. Targeting of the activation-induced cytosine deaminase is strongly influenced by the sequence and structure of the targeted DNA. Mol Cell Biol. 2005;25:10815–10821. doi: 10.1128/MCB.25.24.10815-10821.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iaccarino I, Marra G, Dufner P, Jiricny J. Mutation in the magnesium binding site of hMSH6 disables the hMutSalpha sliding clamp from translocating along DNA. J Biol Chem. 2000;275:2080–2086. doi: 10.1074/jbc.275.3.2080. [DOI] [PubMed] [Google Scholar]
- 21.Poltoratsky V, Prasad R, Horton JK, Wilson SH. Down-regulation of DNA polymerase beta accompanies somatic hypermutation in human BL2 cell lines. DNA Repair (Amst) 2007;6:244–253. doi: 10.1016/j.dnarep.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 23.Cejka P, et al. Methylation-induced G(2)/M arrest requires a full complement of the mismatch repair protein hMLH1. EMBO J. 2003;22:2245–2254. doi: 10.1093/emboj/cdg216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neddermann P, Jiricny J. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc Natl Acad Sci USA. 1994;91:1642–1646. doi: 10.1073/pnas.91.5.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arakawa H, et al. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 2006;4:e366. doi: 10.1371/journal.pbio.0040366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 27.Lehmann AR, et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 28.Pham P, Zhang K, Goodman MF. Hypermutation at A/T sites during G. U mismatch repair in vitro by human B-cell lysates. J Biol Chem. 2008;283:31754–31764. doi: 10.1074/jbc.M805524200. [DOI] [PMC free article] [PubMed] [Google Scholar]