Abstract
Twin studies document substantial heritability for successful abstinence from smoking. A genome-wide association study has identified markers whose allele frequencies differ with nominal p < 0.005 in nicotine-dependent clinical trial participants who were successful vs unsuccessful in abstaining from smoking; many of these results are also supported by data from two additional samples. More study is required to precisely determine the variance in quitting success that can be accounted for by the SNPs that are currently identified and to precisely classify individuals who may display varying degrees of genetic vs environmental effects into quitters or nonquitters. However, the data at hand do allow us to model the effects of genotypic stratification in smoking cessation trials. We identify relationships between the costs of identifying and genotyping prospective trial participants vs the costs of performing the clinical trials. We quantitate the increasing savings that result from genetically-stratified designs as recruiting/genotyping costs go down and trial costs increase. This model helps to define the circumstances in which genetically-stratified designs may enhance power and reduce costs for smoking cessation clinical trials.
Introduction
Nicotine dependence imposes health burdens on smokers and financial burdens on society. Better aids for smoking cessation and improved understanding of individual differences in the ability to successfully quit smoking are thus both important. One approach to enhancing studies aimed at improving smoking cessation would be to utilize emerging information from studies of the molecular genetics of successful smoking cessation. However, no prior work of which we are aware has modeled the potential efficacy of incorporating such molecular genetic information into clinical trials of aids for smoking cessation.
Twin studies indicate that success at achieving abstinence from smoking is substantially heritable 1–8 with about 0.5 genetic and 0.5 environmental influences. Clinical trials of aids to smoking cessation reveal that about 10% vs 20% of research volunteers who receive placebo vs active treatments remain abstinent at 1–2 month follow-up periods 9–11. We have recently reported 520,000 SNP genome-wide association studies of nicotine dependent individuals who successfully abstain from smoking for at least 6 weeks (“quitters”) vs those who do not achieve and/or maintain this abstinence (“nonquitters”). 2,321 of the 520,000 tested SNPs display allele frequencies that distinguish quitters from nonquitters with nominal p < 0.005 12. This initial study was not powered sufficiently to identify all of the SNP allelic frequencies that separate quitters from nonquitters. However, many of the positive SNPs from this study receive support from studies of two additional independent samples 13. These 2,321 SNP allelic frequencies thus provide one reasonable initial representation of the SNP allelic frequencies that will eventually be identified as reproducibly distinguishing quitters from nonquitters. It is important to note that replication in independent samples will be required to determine precisely how much of the variance in quitting success was actually accounted for by these SNPs, how much this model can predict about behavior of individuals with varying degrees of genetic effects and how well the model correctly classifies individuals into quitter versus non-quitters. However, we can thus use the allelic frequencies for the SNPs that we have identified to model the effects of genomic stratification on clinical trials of nicotine quit success, even though we anticipate that an overlapping but different set of SNPs will eventually provide the best ability to distinguish individuals who will be more vs less likely to succeed during attempts to quit smoking. We use additive models of combined genetic and environmental features to develop a composite index of the risks for participants in a clinical trial to experience success in smoking cessation. Additive models represent only one of a variety of ways in which genetic and environmental factors could combine to produce disease risk. Additive models do not provide separate terms for gene × gene (g × g) or gene × environment (g × e) interactions. Nevertheless, such models are mathematically straightforward. Additive models also fit well with the additive genetic values for heritability (a2) that come from twin studies 3. We thus use additive models as a first approximation that can be refined in subsequent studies as the precise nature and magnitude of g × g and g × e interactions becomes clearer. Since the total weight of environmental factors roughly equals the total weight of genetic factors, we re-label the numerical values from the allele frequency estimates to estimate environmental features that alter success in smoking cessation.
We thus report an initial model of the effects of genotype-based stratification on clinical trials for smoking cessation. This model uses simulated individuals who have each been assigned an additive risk of smoking cessation success based on 2321 genetic and 2321 environmental elements. While seven of the clusters of nominally-positive SNPs from different studies do display significant linkage disequilibrium that is not taken into account here, we do not directly consider the modest error that this linkage disequilibrium provides for such a first approximation model. We use threshold approaches to defining the individuals who will quit smoking with placebo vs active treatment. We use data from the costs and sizes of current clinical trials from our experience and those of pharmaceutical industry associates 14, 15.
Results
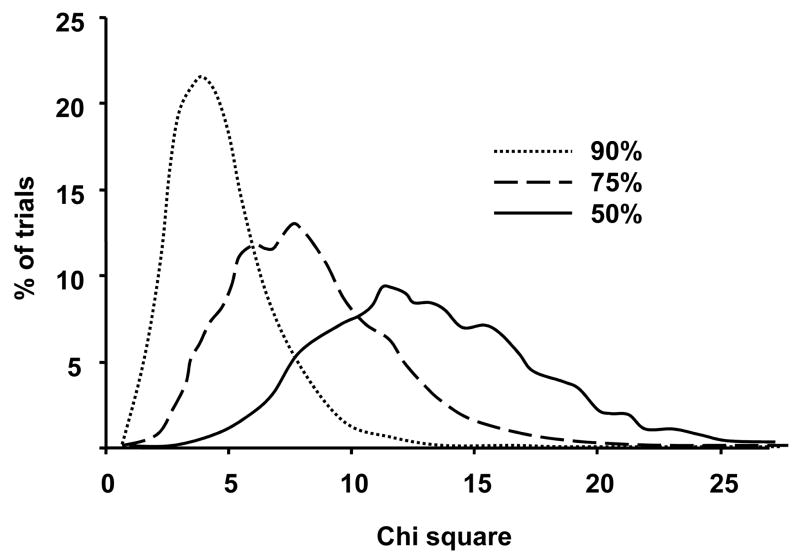
We first compare results from “random population” vs “genetically stratified” samples for small, medium and large clinical trials. For each of 100,000 Monte Carlo simulation trials of “random population” samples, 10% and 90% of the mock trial participants are sampled randomly from populations (n = 100,000 each) of “genetic quitters” and “genetic nonquitters”, respectively. For each of 100,000 Monte Carlo trials of “genetically stratified” samples, 50% and 50% of mock trial participants in placebo and treatment groups are sampled from “genetic quitters” and “genetic nonquitters”, respectively. χ2 values for the significance of the difference between 0.1 (“placebo”) and 0.2 (“treatment”) quit rates are compared for each trial (Fig 1).
Figure 1. Distributions of χ2 values for n = 200 clinical trials for smoking cessation success.
The distributions of the significance value for placebo (0.1 quit success) vs treatment (0.2 quit success) group effects in 100,000 simulation trials of subject groups selected in three ways are shown. Dotted line: Random assignment (90% genetic nonquitters). Dashed line: half-maximal genetic stratification (75% genetic nonquitters), Solid line: total genetic stratification (50% genetic nonquitters) (p < 0.05 threshold corresponds to χ2 >3.84)
For small trials with n = 40, power to detect a p < 0.05 effect is 0.14 with genetically-based stratification and 0.006 without stratification. For medium sized trials with n = 200, power is 0.99 and 0.34, respectively. For large studies with n = 2000, power is virtually 1 for either design.
We can also model effects of genetic stratification based on assays that provide only half of the maximal possible genetic information (“half-max”). For n = 200 studies, power is 0.91 for half-max and 0.34 for non-stratified samples. To achieve the 0.91 power that a half max stratified study achieves with n = 200, a non-stratified study would require n = 450.
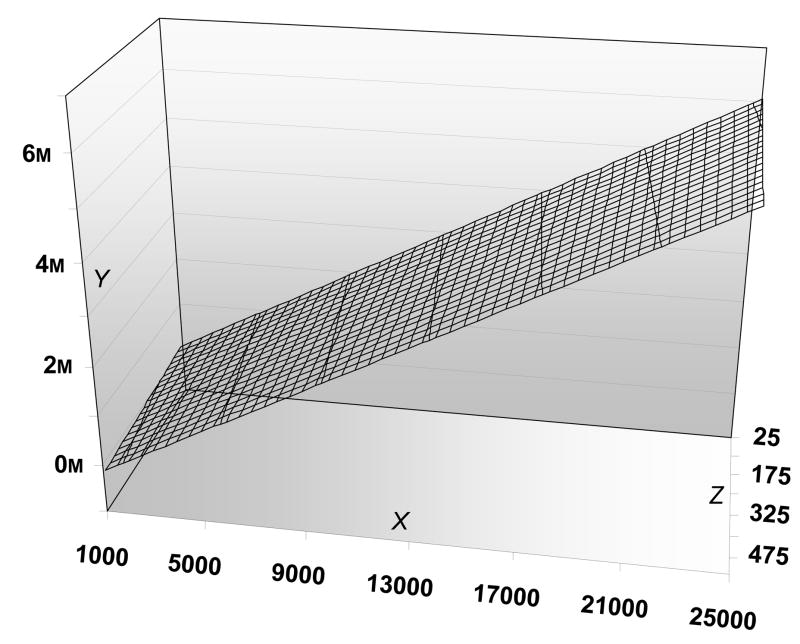
We can thus explore cost/benefit analyses for half max stratification. To identify 100 treatment and 100 placebo subjects for 0.9 power, “half-max” designs require recruiting and genotyping 1019 individuals. An nonstratified trial with the same 0.9 power would need to study 225 treatment and 225 placebo participants. In Fig 2, we assume $150/person genotyping costs and allow additional recruiting and trial costs to vary. When we calculate the net difference between stratified and nonstratified trials, the direct savings for trials with half max stratification rise from 4 to 15 million dollars as the costs/participant rise from the approximately $4000 characteristic of “academic” trials to the $25,000/participant costs characteristic of some pharmaceutically-sponsored trials (JR, unpublished results). As genotyping costs rise from $150 to $500/individual, the same overall relationships still hold (Figure 2, legend).
Figure 2. Costdifferences in clinical trials: half-max genetic stratification vs random assignment.
Cost differences (Y axis, in millions of US dollars) with 0.9 power to achieve p < 0.05 that are stratified by genotypes (half max genetic influences, n = 1019 genotyped, n= 200 participants) vs conventional design (p < 0.05, n = 450) as recruitment costs (Z axis, in US dollars) vary from $25 – $500 subject and trial costs (X axis, in US dollars) vary from $1,000 – $25,000/subject.
We assume genotyping costs of $150 for data on the main surface of the plot. If genotyping costs rise to $300/subject, and trial/recruiting costs are1000/25 or 25,000/500 savings from half-max genetic stratification are $ −69,925 and $5,659,800, respectively. If genotyping costs rise to $500/subject, and trial/recruiting costs are1000/25 or 25,000/500 savings from half-max genetic stratification are $ −273,725 and $5,456,000, respectively.
Discussion
Trials for smoking cessation are favorable places for quantitating the effects of genetic stratification on clinical trial design, power and costs for several reasons: 1) trials for smoking cessation often result in about 10 – 20% of participants achieving abstinence in placebo and actively treated groups 9–11, respectively; 2) costs of such trials are known; 3) twin study data provide good estimation of the magnitudes of additive genetic and environmental influences on smoking cessation success 5–8. Many of the other assumptions necessary for modeling smoking cessation success can also be supported by some data: 1) SNP allele frequencies that distinguish successful from unsuccessful quitters in an initial publication can be used as a surrogate for a future dataset that will contain all such allele frequencies and detailed information concerning linkage disequilibrium between them 12; 2) relabeled data from these SNP allelic frequencies can be used as a surrogate for the individual “environmental” elements that represent the 50% of environmental determinants of quitting success 6 and 3) data from twin studies that evaluate heritability of success in smoking cessation outside of clinical trials 6 can be used to estimate the genetic and environmental features of the likelihood of nicotine quit success that apply within clinical trials We can also use reasonable assumptions that include a threshold model to approximate smoking cessation in placebo and treatment groups.
It is important to note that the results of the current modeling do not distinguish between the benefits of genotypic stratification in a) reducing variance or b) enhancing treatment response magnitude; both effects are likely here. It is also important to note other novel and/or limiting features of the present work. 1) We are aware of no other example that uses such an approach to clinical trials focused on any other complex disorder, perhaps because there are few treatment responses for which both the genome wide association and the twin datasets, both required for the present analyses, are available. 2) The current approach uses each SNP as an independent marker and does not account for “functional” SNP status or linkage disequilibium between SNPs. The present approach does not provide terms for gene × gene or gene × environment interactions. While future models that take these issues into account could improve their predictive power, our identification of only a limited number of SNPs in strong linkage disequilbrium with each other suggests that an additive model may provide only limited error in the current setting. 3) Genotypes are associated, probabilistic risk factors; use of genotype profiles to enroll individuals in clinical studies might have ethical implications that may be greatest if alternative therapies are not available. 4) While selection of matched samples of “responsive” individuals for all arms of a clinical trial may decrease type II error due to lower variance in the dataset, drugs that might not be successful in general population samples might succeed in restricted responder subpopulations. Such results might mandate use of pretreatment genotyping to match individuals with treatments in clinical practice. 5) The twin data cited here provide estimates of heritability for smoking cessation among what are likely to be heterogeneous groups of smokers that include: self-quitters who might have been able to quit without treatment and smokers who require the benefits of the clinical trial in order to quit, perhaps in part due to co-morbid psychopathology 16. 6) Other alternative approaches that utilize stratification based on histories of psychopathologies such as major depression or of traumatic life events might add information to or even supplant parts of the molecular genetic analyses presented here. 7) The current paper uses variance estimates from the SNPs that we have identified from a published dataset for smoking cessation success. This paper models effects of arbitrarily-chosen sizes (eg 100% and 50% of genetic influences) for illustrative purposes. One goal of ongoing research in this area is to use the sets of SNPs that we have identified retrospectively in prospective studies of quitting success in independent samples. Such data will then allow us to make much better empirical estimates of the fraction of the actual variance in quitting success that can be actually accounted for by these sets of SNPs, allowing us to understand how well these particular SNPs correctly classify individuals into quitter vs non-quitter classes. Nevertheless, it is important to stress that we do not have current accurate information concerning the fraction of the variance in quitting success was actually accounted for by the 2311 SNPs modeled here, about how predictive the model would be with varying degrees of genetic effects, since we use the data from twin studies, and about how well the SNPs modeled here correctly classify individuals into quitter versus non-quitters, since the arguments are based on stochastic responses in groups of subjects, rather than on complete predictive knowledge of individiuals’ behaviors. 8) Application of these approaches assumes that we have information about SNPs whose allele frequencies help to distinguish successful from nonsuccessful quitters in the populations being studied. Issues relating to population stratification might require preliminary work when this approach is applied to trials of smoking cessation success in novel populations for which baseline allele frequency differences might render SNP markers less informative.
The number of SNPs used in the current model comes from a single study. Many of these results have obtained substantial support from data from two additional samples 13. Despite this encouraging data, genotypes at these 2,311 SNPs cannot presently be considered to provide reliable estimates of quitting success or to correctly classify all individuals into quitter and non-quitter groups. The general conclusions of the work presented in this paper are independent of the exact nature of the SNPs used for these analyses, and should be applicable for any set of genomic markers that captures the fractions of the genetic component of successful smoking cessation used for these modeling approaches.
Despite the cautions noted above, the results of the current simulation studies appear to justify careful consideration of use of genotypic stratification for medium sized trials (eg n = 200) that are characteristic of phase II drug testing. Such consideration should increase as increasing numbers of studies validate the roles of theses individual SNPs and haplotypes in predicting success in smoking cessation. Since phase II trials often provide the first clear-cut indication of human efficacy for a novel therapeutic agent, positive results from genetically- stratified n = 200 phase II studies could then allow focused genotyping in larger phase III studies. Positive results from phase III studies, in turn, could provide more accurate estimations of the likelihood that the novel treatment would work in quitters and in nonquitters from the general population.
Refinements in the present model are thus certain to take place as we obtain more data on more of the allelic variants that distinguish quitters from nonquitters. Even this initial model, however, underscores the likely benefits of applying molecular genetics to studies of smoking cessation. Such approaches could also inform design of pharmacological trials for other addictions and for other indications in which complex genetics would be likely to influence trial outcomes.
Materials and Methods
Data used and assumptions used to model effects of genotyping on clinical trial costs and power include: a) Data: 0.5 heritability for quit success from twin studies; Assumptions: 0.5 genetic influence on therapeutic trial outcomes; b) Data: little evidence for large gene × environment interaction terms in twin datasets; Assumption: additive genetic + environmental influences in our model; c) Data: allele frequencies from one “genome wide association” study of nicotine quit success; Assumption: this data can represent all studies; d) Data: power calculations document 0.45 –0.9 power to detect alleles that influence quit success from our initial genome scanning study; Assumptions: genotypes can readily assess 0.5 of genetic influences; e) Data: about 0.1 of smokers quit in placebo groups and about 0.2 in treatment groups; Assumption: set threshold for the combined genetic and environmental scores for each individual in each sample so that 0.1 of placebo and 0.2 of treated smokers are successful quitters; f) Data: the magnitude of environmentally derived influences on smoking quit success matches the magnitude of genetic influences from twin studies; Assumption: we can use the magnitude and variance of genetic components to approximate the variance of environmental features. Simulations assume that a threshold model for quit success and treatment effects provides a good first approximation of the underlying effects.
For this modeling, we use frequencies of allelic variants at 2311 SNPs that display the largest differences between nicotine dependent research volunteers with continuous 6 week abstinence from smoking after treatment with nicotine replacement and mecamylamine vs research volunteers who were not abstinent at this 6 week time point10. To construct the samples for simulation studies, we use the 2311 allele frequencies to first built computerized representations of two subpopulations: a) 100,000 “genetic quitters” and b) 100,000 “genetic nonquitters”, each assigned genotypes at the 2311 SNPs for which quitter and nonquitters differ at nominal p < 0.005 with randomly-assigned values with probabilities based on each SNP’s allele frequencies in successful and unsuccessful quitters, respectively. Each individual in both genetic quitter and genetic nonquitter samples also received a set of 2311 “environmental” factors” (relabeled from the 2311 genetic factors) that are assumed to contribute to the observed 0.9/0.1 nonsuccess/success ratio. The value for each of these environmental factors is thus randomly assigned from a group of relabeled values that come 0.9 from nonquitter allele frequencies and 0.1 from quitter allele frequencies.
To simulate a conventional study design, we use these mock individuals to build 100,000 different mock study samples. Each contains 0.1 “genetic quitters” and 0.9 “genetic nonquitters”. We assume that treatment effects are such that 0.1 of individuals quit with placebo and 0.2 quit with active treatment.
To simulate two genetically-stratified study designs, we use these mock individuals to build 100,000 different samples that are each 0.5 or 0.25 “genetic quitters” and 0.5 or 0.75 “genetic nonquitters”. The 0.75 vs 0.25 comparison assumes that we can assess ½ of the total genetics with assays likely to be available in the near future.
We seek χ2 values for the difference between treatment and placebo for each of these 100,000 mock study samples. We note the distribution of these χ2 values. The fraction of trials for which the 0.1 vs 0.2 difference reaches nominal significance is reported as the power.
To examine the effects of genotype-based stratification on the cost of clinical trials, we graph the results of the following formula, based on the assumption that we can capture ½ of the total genetic influences with genotyping that costs $150, $300 or $500/individual: Difference in costs based on conventional designs vs design with genotype-based stratification is equal to: [(Recruiting cost × 450) + (Trial cost × 450)] −[(Recruiting cost × 1019) + (Genotyping cost × 1019) + (Trial cost × 200)].
Acknowledgments
We acknowledge support by the NIH IRP (NIDA), DHSS, unrestricted support for studies of adult smoking cessation to the Duke Center for Nicotine and Smoking Cessation Research from Philip Morris USA, Inc and advice on the manuscript and statistical approaches from Dr Greg Samsa.
Contributor Information
George R Uhl, Molecular Neurobiology Branch, NIH-IRP, NIDA, Suite 3510, 333 Cassell Drive Baltimore, Maryland 21224.
Tomas Drgon, Molecular Neurobiology Branch, NIH-IRP, NIDA, Suite 3510, 333 Cassell Drive Baltimore, Maryland 21224.
Catherine Johnson, Molecular Neurobiology Branch, NIH-IRP, NIDA, Suite 3510, 333 Cassell Drive Baltimore, Maryland 21224.
Jed E Rose, Dept of Psychiatry and Behavioral Sciences and Center for Nicotine and Smoking Cessation Research, Duke University, Durham NC 27708.
References
- 1.Uhl GR, Elmer GI, Labuda MC, Pickens RW. Genetic influences in drug abuse. In: Gloom FE, Kupfer DJ, editors. Psychopharmacology: The Fourth Generation of Progress. Raven Press; New York: 1995. pp. 1793–2783. [Google Scholar]
- 2.Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, et al. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55(11):967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 3.Karkowski LM, Prescott CA, Kendler KS. Multivariate assessment of factors influencing illicit substance use in twins from female-female pairs. Am J Med Genet. 2000;96(5):665–670. [PubMed] [Google Scholar]
- 4.True WR, Heath AC, Scherrer JF, Xian H, Lin N, Eisen SA, et al. Interrelationship of genetic and environmental influences on conduct disorder and alcohol and marijuana dependence symptoms. Am J Med Genet. 1999;88(4):391–397. doi: 10.1002/(sici)1096-8628(19990820)88:4<391::aid-ajmg17>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 5.Xian H, Scherrer JF, Madden PA, Lyons MJ, Tsuang M, True WR, et al. The heritability of failed smoking cessation and nicotine withdrawal in twins who smoked and attempted to quit. Nicotine Tob Res. 2003;5(2):245–254. [PubMed] [Google Scholar]
- 6.Broms U, Silventoinen K, Madden PA, Heath AC, Kaprio J. Genetic architecture of smoking behavior: a study of Finnish adult twins. Twin Res Hum Genet. 2006;9(1):64–72. doi: 10.1375/183242706776403046. [DOI] [PubMed] [Google Scholar]
- 7.Carmelli D, Swan GE, Robinette D, Fabsitz R. Genetic influence on smoking--a study of male twins. N Engl J Med. 1992;327(12):829–833. doi: 10.1056/NEJM199209173271201. [DOI] [PubMed] [Google Scholar]
- 8.Morley KI, Lynskey MT, Madden PA, Treloar SA, Heath AC, Martin NG. Exploring the inter-relationship of smoking age-at-onset, cigarette consumption and smoking persistence: genes or environment? Psychol Med. 2007;37(9):1357–1367. doi: 10.1017/S0033291707000748. [DOI] [PubMed] [Google Scholar]
- 9.Tonnesen P, Norregaard J, Simonsen K, Sawe U. A double-blind trial of a 16-hour transdermal nicotine patch in smoking cessation. N Engl J Med. 1991;325(5):311–315. doi: 10.1056/NEJM199108013250503. [DOI] [PubMed] [Google Scholar]
- 10.Kenford SL, Fiore MC, Jorenby DE, Smith SS, Wetter D, Baker TB. Predicting smoking cessation. Who will quit with and without the nicotine patch. JAMA. 1994;271(8):589–594. doi: 10.1001/jama.271.8.589. [DOI] [PubMed] [Google Scholar]
- 11.Jones RL, Nguyen A, Man SF. Nicotine and cotinine replacement when nicotine nasal spray is used to quit smoking. Psychopharmacology (Berl) 1998;137(4):345–350. doi: 10.1007/s002130050629. [DOI] [PubMed] [Google Scholar]
- 12.Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE. Molecular genetics of nicotine dependence and abstinence: whole genome association using 520,000 SNPs. BMC Genetics. 2007;8:10. doi: 10.1186/1471-2156-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, Rose JE, et al. Molecular genetics of successful smoking cessation: convergent genome-wide association results. Arch Gen Psychiatry. 2008 doi: 10.1001/archpsyc.65.6.683. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornuz J, Gilbert A, Pinget C, McDonald P, Slama K, Salto E, et al. Cost-effectiveness of pharmacotherapies for nicotine dependence in primary care settings: a multinational comparison. Tob Control. 2006;15(3):152–159. doi: 10.1136/tc.2005.011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall SM, Lightwood JM, Humfleet GL, Bostrom A, Reus VI, Munoz R. Cost-effectiveness of bupropion, nortriptyline, and psychological intervention in smoking cessation. J Behav Health Serv Res. 2005;32(4):381–392. doi: 10.1007/BF02384199. [DOI] [PubMed] [Google Scholar]
- 16.Hughes JR, Giovino GA, Klevens RM, Fiore MC. Assessing the generalizability of smoking studies. Addiction. 1997;92(4):469–472. [PubMed] [Google Scholar]