Abstract
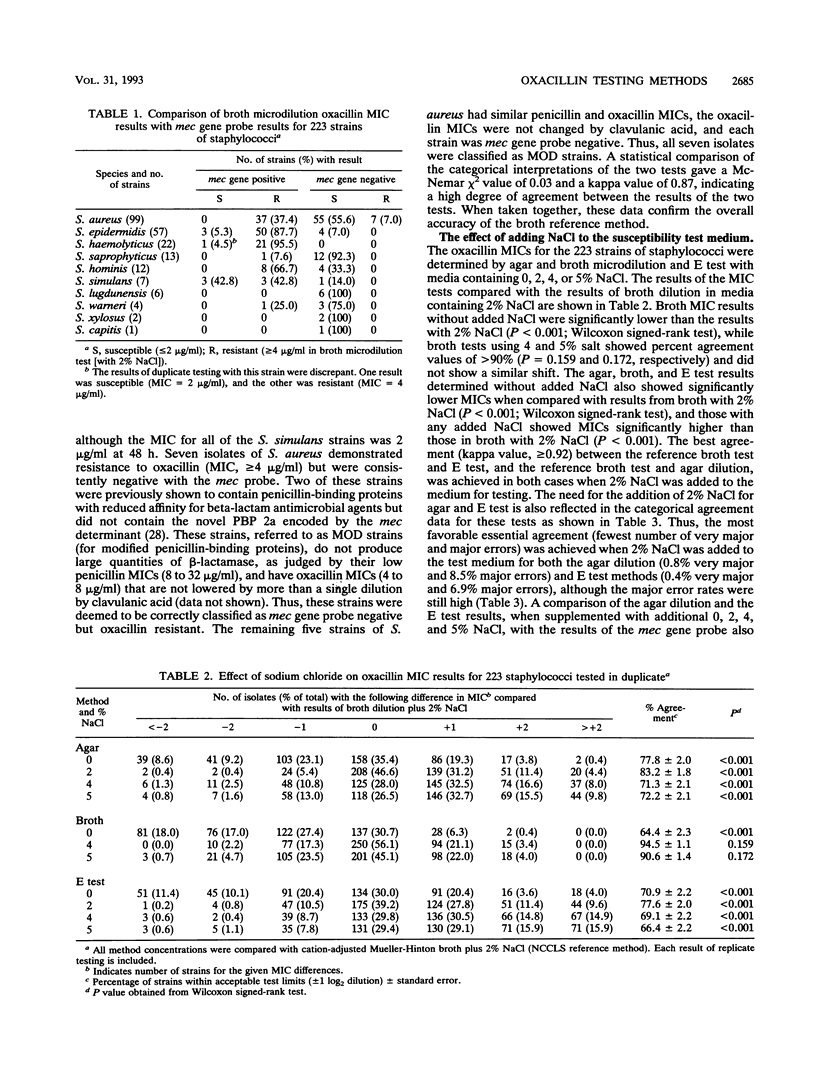
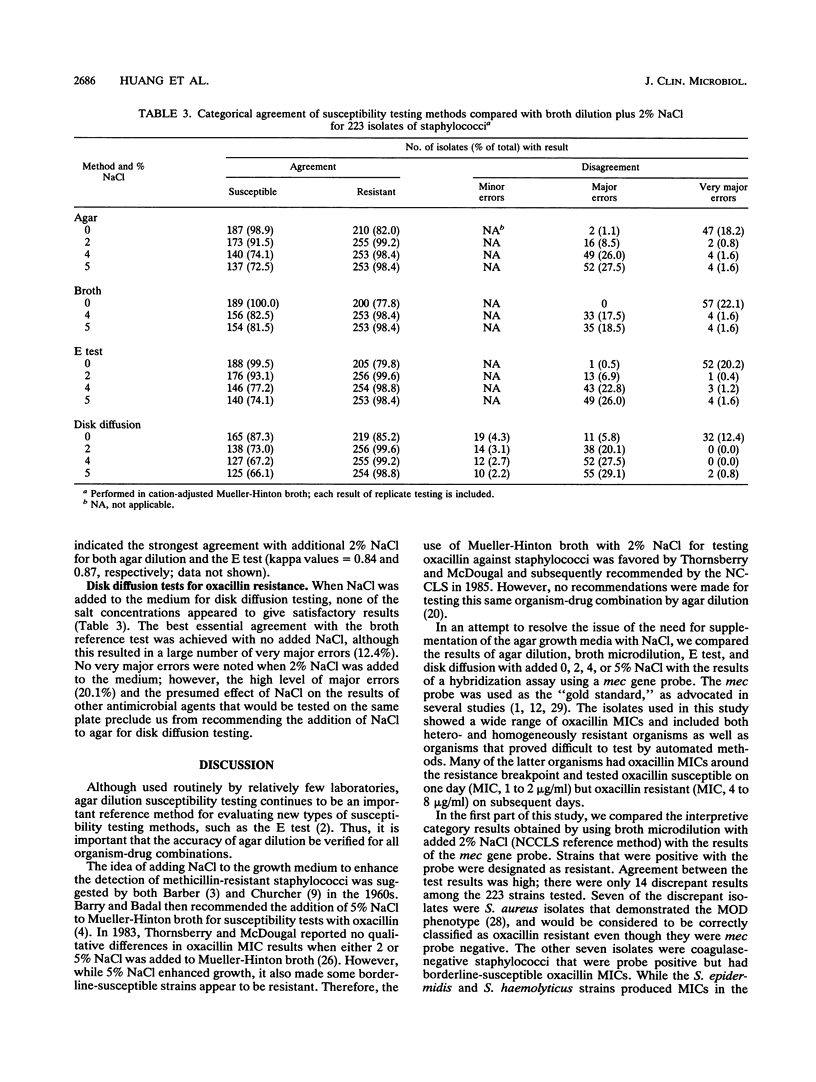
The need to add NaCl to agar media to ensure accuracy of results when testing staphylococci with oxacillin was investigated. The results of four antimicrobial susceptibility testing methods (agar and broth dilution, E test, and disk diffusion) in which the growth medium contained 0, 2, 4, or 5% NaCl were compared with the results of a hybridization assay using a mec gene probe. We tested 223 strains of staphylococci, 128 of which were mec gene positive. A total of 7 of the 128 positive strains were coagulase-negative staphylococci with 24-h oxacillin MICs of < or = 2 micrograms/ml. Ninety-five isolates were mec gene negative, including seven strains of Staphylococcus aureus with oxacillin MICs of > or = 4 micrograms/ml. The oxacillin MICs for mec gene-positive, oxacillin-resistant strains of staphylococci increased two- to fourfold with the addition of NaCl to the test medium, while the MICs for mec gene-negative strains did not change in the presence of added salt. Very major error rates for the agar dilution and E test methods in the absence of salt ranged from 18.2 to 20.2%. Major error rates for mec gene-negative S. aureus isolates were > 17% for all test methods when 4 or 5% NaCl was added to the test medium. The addition of 2% NaCl to Mueller-Hinton agar for testing of oxacillin resulted in very major error rates of < 1% for the agar dilution and E test methods although the major error rates for the two methods with added NaCl were 8.5 and 6.9%, respectively. The disk diffusion test did not perform well in this study, showing essential error rates of > or = 18.3%. We recommended the addition of 2% NaC1 to Mueller-Hinton agar when testing staphylococci with oxacillin by either the agar dilution or E test method. NaC1 should not be added for the disk diffusion test.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Archer G. L., Pennell E. Detection of methicillin resistance in staphylococci by using a DNA probe. Antimicrob Agents Chemother. 1990 Sep;34(9):1720–1724. doi: 10.1128/aac.34.9.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARBER M. NATURALLY OCCURING METHICILLIN-RESISTANT STAPHYLOCOCCI. J Gen Microbiol. 1964 May;35:183–190. doi: 10.1099/00221287-35-2-183. [DOI] [PubMed] [Google Scholar]
- Baker C. N., Stocker S. A., Culver D. H., Thornsberry C. Comparison of the E Test to agar dilution, broth microdilution, and agar diffusion susceptibility testing techniques by using a special challenge set of bacteria. J Clin Microbiol. 1991 Mar;29(3):533–538. doi: 10.1128/jcm.29.3.533-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry A. L., Badal R. E. Reliability of the microdilution technic for detection of methicillin-resistant strains of staphylococcus aureus. Am J Clin Pathol. 1977 May;67(5):489–495. doi: 10.1093/ajcp/67.5.489. [DOI] [PubMed] [Google Scholar]
- Barry A. L., Jones R. N. Reliability of high-content disks and modified broth dilution tests for detecting staphylococcal resistance to the penicillinase-resistant penicillins. J Clin Microbiol. 1987 Oct;25(10):1897–1901. doi: 10.1128/jcm.25.10.1897-1901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck W. D., Berger-Bächi B., Kayser F. H. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J Bacteriol. 1986 Feb;165(2):373–378. doi: 10.1128/jb.165.2.373-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers H. F., Archer G., Matsuhashi M. Low-level methicillin resistance in strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1989 Apr;33(4):424–428. doi: 10.1128/aac.33.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers H. F. Methicillin-resistant staphylococci. Clin Microbiol Rev. 1988 Apr;1(2):173–186. doi: 10.1128/cmr.1.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churcher G. M. A screening test for the detection of methicillin-resistant staphylococci. J Clin Pathol. 1968 Mar;21(2):213–217. doi: 10.1136/jcp.21.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerberding J. L., Miick C., Liu H. H., Chambers H. F. Comparison of conventional susceptibility tests with direct detection of penicillin-binding protein 2a in borderline oxacillin-resistant strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1991 Dec;35(12):2574–2579. doi: 10.1128/aac.35.12.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering R. V., Ruff E. A. Comparative analysis of conjugative plasmids mediating gentamicin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1983 Sep;24(3):450–452. doi: 10.1128/aac.24.3.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1986 Jan;29(1):85–92. doi: 10.1128/aac.29.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. B., Baker C. N., Banerjee S., Tenover F. C. Accuracy of the E test for determining antimicrobial susceptibilities of staphylococci, enterococci, Campylobacter jejuni, and gram-negative bacteria resistant to antimicrobial agents. J Clin Microbiol. 1992 Dec;30(12):3243–3248. doi: 10.1128/jcm.30.12.3243-3248.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Buescher G., Lewis N., Snyder S., Jungkind D. Detection of borderline oxacillin-resistant Staphylococcus aureus and differentiation from methicillin-resistant strains. Eur J Clin Microbiol Infect Dis. 1990 Oct;9(10):717–724. doi: 10.1007/BF02184683. [DOI] [PubMed] [Google Scholar]
- Maple P. A., Hamilton-Miller J. M., Brumfitt W. World-wide antibiotic resistance in methicillin-resistant Staphylococcus aureus. Lancet. 1989 Mar 11;1(8637):537–540. doi: 10.1016/s0140-6736(89)90076-7. [DOI] [PubMed] [Google Scholar]
- McDougal L. K., Thornsberry C. The role of beta-lactamase in staphylococcal resistance to penicillinase-resistant penicillins and cephalosporins. J Clin Microbiol. 1986 May;23(5):832–839. doi: 10.1128/jcm.23.5.832-839.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Minamide W., Wada K., Nakamura E., Teraoka H., Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991 Oct;29(10):2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngui-Yen J. H., Bryce E. A., Porter C., Smith J. A. Evaluation of the E test by using selected gram-positive bacteria. J Clin Microbiol. 1992 Aug;30(8):2150–2152. doi: 10.1128/jcm.30.8.2150-2152.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesch W., Strässle A., Berger-Bächi B., O'Hara D., Reynolds P., Kayser F. H. Cloning and expression of methicillin resistance from Staphylococcus epidermidis in Staphylococcus carnosus. Antimicrob Agents Chemother. 1988 Oct;32(10):1494–1499. doi: 10.1128/aac.32.10.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry C., Caruthers J. Q., Baker C. N. Effect of temperature on the in vitro susceptibility of Staphylococcus aureus to penicillinase-resistant penicillins. Antimicrob Agents Chemother. 1973 Sep;4(3):263–269. doi: 10.1128/aac.4.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry C., McDougal L. K. Successful use of broth microdilution in susceptibility tests for methicillin-resistant (heteroresistant) staphylococci. J Clin Microbiol. 1983 Nov;18(5):1084–1091. doi: 10.1128/jcm.18.5.1084-1091.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokue Y., Shoji S., Satoh K., Watanabe A., Motomiya M. Comparison of a polymerase chain reaction assay and a conventional microbiologic method for detection of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1992 Jan;36(1):6–9. doi: 10.1128/aac.36.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A., Drugeon H. B., de Lencastre H. M., Jabes D., McDougall L., Bille J. New mechanism for methicillin resistance in Staphylococcus aureus: clinical isolates that lack the PBP 2a gene and contain normal penicillin-binding proteins with modified penicillin-binding capacity. Antimicrob Agents Chemother. 1989 Nov;33(11):1869–1874. doi: 10.1128/aac.33.11.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A., Nachman S., Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1991 Jan;35(1):124–129. doi: 10.1128/aac.35.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukata K., Nonoguchi R., Song M. D., Matsuhashi M., Konno M. Homology of mecA gene in methicillin-resistant Staphylococcus haemolyticus and Staphylococcus simulans to that of Staphylococcus aureus. Antimicrob Agents Chemother. 1990 Jan;34(1):170–172. doi: 10.1128/aac.34.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre H., Sá Figueiredo A. M., Urban C., Rahal J., Tomasz A. Multiple mechanisms of methicillin resistance and improved methods for detection in clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 1991 Apr;35(4):632–639. doi: 10.1128/aac.35.4.632. [DOI] [PMC free article] [PubMed] [Google Scholar]


