Abstract
Background
The functional polymorphism that explains the established association of the androgen receptor (AR) with androgenetic alopecia (AGA) remains unidentified, but Copy Number Variation (CNV) might be relevant. CNV involves changes in copy number of large segments of DNA, leading to the altered dosage of gene regulators or genes themselves. Two recent reports indicate regions of CNV in and around AR, and these have not been studied in relation to AGA. The aim of this preliminary case-control study was to determine if AR CNV is associated with AGA, with the hypothesis that CNV is the functional AR variant contributing to this condition.
Methodology/Principal Findings
Multiplex Ligation-dependent Probe Amplification was used to screen for CNV in five AR exons and a conserved, non-coding region upstream of AR in 85 men carefully selected as cases and controls for maximal phenotypic contrast. There was no evidence of CNV in AR in any of the cases or controls, and thus no evidence of significant association between AGA and AR CNV.
Conclusions/Significance
The results suggest this form of genomic variation at the AR locus is unlikely to predispose to AGA.
Introduction
The importance of the gene encoding the androgen receptor (AR; NCBI NM_000044) in the development of androgenetic alopecia (AGA; OMIM %300710) is now well established. Our initial observation [1] of genetic association of AR with AGA has been replicated in three independent studies [2]–[4]. Although association with several single nucleotide polymorphisms (SNPs) has been demonstrated, none have proven causative. We have shown that known AR coding sequence polymorphisms such as triplet repeats are not associated with AGA [5], and mutation screening has identified no other relevant coding sequence mutations. Other forms of variation that might affect gene expression should now be considered.
Copy Number Variation (CNV) has been recognised recently as a common form of genomic variation [6]–[8] in which DNA segments (>1 kb) are altered in copy number compared to a reference genome. Recent reports have suggested that up to 12% of the genome is subject to CNV, accounting for at least 1% of genetic variation [6], [9].
CNV has been reported in the region of AR [6], [8] and SNPs can be co-inherited in linkage disequilibrium (LD) with CNV [6]. As CNV might result in variable copies of genetic regulatory regions or copies of the gene itself, such variation might underpin the tissue- and developmental stage-specific abnormalities in expression of AR seen in men predisposed to AGA.
We hypothesised that AR CNV could be associated with AGA. Therefore, our approach was to first search for evidence of CNV at the AR locus and then, if present, to test for association of CNV with AGA.
Materials and Methods
Subjects
The study was approved by the Ethics Review Committee of the Alfred Hospital Melbourne, and informed consent was obtained from each participant. Subjects were drawn from the population-based Victorian Family Heart Study cohort [10]. AGA phenotypes were gathered by way of direct assessment by trained observers and validated questionnaires [11]. Cases were 34 men aged 20–30 years (mean 27 years) with cosmetically significant baldness (Hamilton-Norwood types III vertex to VII, median type IV), and controls were 51 men aged 54–68 years (mean 56 years) with no indication of baldness (Hamilton-Norwood type I).
MLPA analysis
Peripheral blood was collected from each subject, and DNA was extracted using standard phenol-chloroform methods. Multiplex Ligation-Dependent Probe Analysis (MLPA) was performed to detect the presence of CNV around AR. Six probes were selected to test AR exons and one upstream non-coding region for the presence of CNV (probes 1 – 6; Table 1 and Figure 1). Two further probes (7 and 8) in unlinked autosomal loci were used as reference point controls (Table 1). Peak heights were analysed for the eight probes for each DNA sample, as outlined in White et al [12]. Briefly, peak heights for each probe were standardized to control peaks and the median ratio for each probe normalised to 1.0 [12]. We set threshold values for duplication and deletions of 1.5 and 0.5 respectively [13]. All samples were analysed at least twice.
Table 1. Sequences and genomic locations of the probes.
| Probe | Genomic Location | Sequence |
| 1 | ChrX: 66,481,078–66,481,135 | 5′ GGGTTCCCTAAGGGTTGGAGACTTAAGGGTACATAATAATGGGCAGTGG 3′ |
| 5′ ATTATGTGGACACCACAATTTGGAAGGGTCTAGATTGGATCTTGCTGGC 3′ | ||
| 2 | ChrX: 66,681,149–66,681,198 | 5′ GGGTTCCCTAAGGGTTGGAGCTTCAGCACTGCAGCCACGACCC 3′ |
| 5′ GCCTGGTTAGGCTGCACGCGGAGAGATCTAGATTGGATCTTGCTGGC 3′ | ||
| 3 | ChrX: 66,822,629–66,822,690 | 5′ GGGTTCCCTAAGGGTTGGACCGAAGGAAAAATTGTCCATCTTGTCGTCT 3′ |
| 5′ TCGGAAATGTTATGAAGCAGGGATGACTCTGGTCTAGATTGGATCTTGCTGGC 3′ | ||
| 4 | ChrX: 66,854,045–66,854,113 | 5′ GGGTTCCCTAAGGGTTGGAGCTTCCGCAACTTACACGTGGACGACCAGATGGCT 3′ |
| 5′ GTCATTCAGTACTCCTGGATGGGGCTCATGGTGTCTAGATTGGATCTTGCTGGC 3′ | ||
| 5 | ChrX: 66,858,400–66,858,453 | 5′ GGGTTCCCTAAGGGTTGGAGTACCGCATGCACAAGTCCCGGATGTA 3′ |
| 5′ CAGCCAGTGTGTCCGAATGAGGCACCTTCTAGATTGGATCTTGCTGGC 3′ | ||
| 6 | ChrX: 66,860,242–66,860,342 | 5′ GGGTTCCCTAAGGGTTGGACGAGAGAGCTGCATCAGTTCACTTTTGACCTGCTAATCAAG 3′ |
| 5′ TCACACATGGTGAGCGTGGACTTTCCGGAAATGATGGCAGAGATCTAGATTGGATCTTGCTGGC 3′ | ||
| 7 | Chr16: 3,772,843–3,772,914 | 5′ GGGTTCCCTAAGGGTTGGACCAGCTAGTGGAATTCAAAACACAATTGGTTCTGTTGGCACA 3′ |
| 5′ GGGCAACAGAATGCCACTTCTTTAAGTAACTCTAGATTGGATCTTGCTGGC 3′ | ||
| 8 | Chr22: 39,857,407–39,857,482 | 5′ GGGTTCCCTAAGGGTTGGACCAACCTAAGCACTGTTAGTCAGATTGATCCCAGCTCCAT 3′ |
| 5′ GAAAGAGCCTATGCAGCTCTTGGACTACCCTATCATCTAGATTGGATCTTGCTGGC 3′ |
Listed are the AR exonic and non-coding region probes (1–6), and control probes (7 and 8). Probes 7 and 8 were used as controls to normalize data and were designed in regions with no known CNV. Genomic location data based on human reference sequence (NCBI Build 36.1).
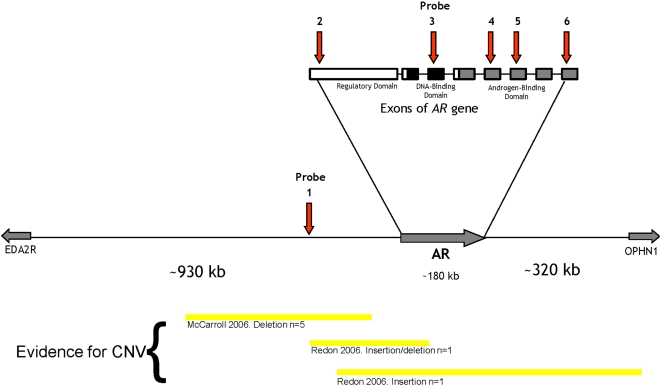
Figure 1. Location of the probes in relation to the androgen receptor.
Diagrammatic representation of part of chromosome × showing the androgen receptor (AR) and the upstream (EDA2R) and downstream (OPHN1) genes. The relative locations of the upstream region (probe 1) and the five probes within the exons of AR are represented by the arrows. The AR gene exons are represented as boxes, as is the location of prior evidence of copy number variation (CNV) in this region.
Results
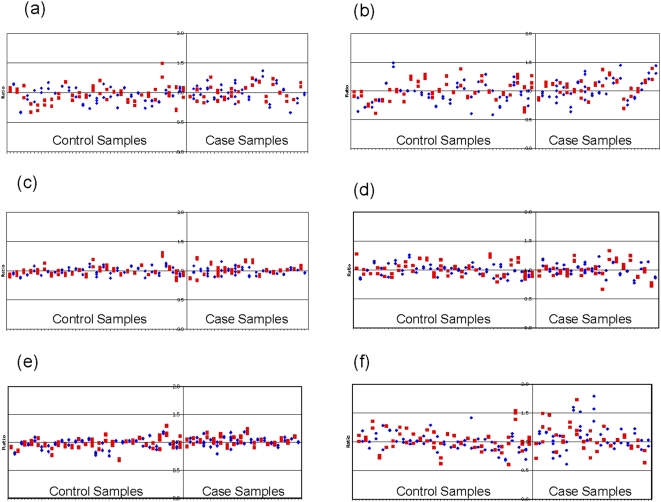
Figure 2 shows data for each of the six AR test probes in all case-control subjects. The standard deviation for each of the six AR test probes was within 15%, except for probes 2 and 6 which were slightly higher, possibly suggesting less reliable performance of the probes. Nevertheless, across the remaining samples, good coverage of the AR region was achieved by the higher performing probes, and the results provide no evidence of CNV in this region, either as deletion or duplication, in cases or controls.
Figure 2. (a-f): Normalised ratios for both case and control samples across the six test probes.
Each plot includes at least two replicates per sample represented by the same symbol, with the symbols alternating between each sample. Samples were normalised using two control probes (see Table 1). Normal range 0.5–1.5.
Discussion
This study examined CNV in and around AR and their potential relationship with AGA. CNV may contribute to differences in gene expression through duplications or deletions of genes or gene regulatory elements [14], and may in part account for genetic predisposition to common complex conditions such as AGA. However, we found no evidence of CNV in the regions of the AR that we examined. Insofar as our original hypothesis, we could therefore find no evidence of association of CNV with AGA.
Although data generated by probes 2 and 6 showed relatively high standard deviations, casting doubt on their reliability, the AR gene was well covered by the remaining probes. The lack of evidence for CNV at this locus supports the findings of a very recent study by McCarroll et al which has demonstrated a lack of CNV in the AR coding regions [15].
This study examined a locus on chromosome × in males, meaning that if changes in CNV dosages were detected, they would be expected to be at either a 0 (deletion) or 2 (duplication) level. Figure 2 demonstrates that the results for some samples showed normalised ratios of around 0.5 or 1.5, consistent with possible mosaicism within the cells of the blood samples from which the DNA was extracted. CNV mosaicism has been demonstrated in blood samples in monozygotic twins [16], and it is known that CNV may be variable between cell types within the same individual. Although the results presented here indicate no evidence of AR CNV in DNA derived from lymphocytes, we cannot rule out CNV around the AR in DNA contained in the cells of the hair follicles themselves.
Our preliminary investigation did not thoroughly investigate the downstream non-coding flanking region and other more distant sites from AR. However, we focussed on a region that we have already shown to be strongly associated with AGA and it would be likely that causative CNV, if they exist, would be in the region examined. Sample sizes used were relatively small, however power is augmented by the fact that association of AGA with AR is well-established and that the cases and controls were selected to represent high phenotypic contrast.
In the absence of evidence of AR CNV involvement in AGA, other forms of non-coding AR sequence variation, and also epigenetic mechanisms at this locus, should be considered in clarifying the role of AR as a genetic risk locus for AGA.
Acknowledgments
We thank Ms Margaret Stebbing, Professor John Hopper, Professor Graham Giles, the general practitioners and research nurses for their contributions to recruitment of VFHS study participants and Ms Angela Lamantia for DNA extraction. We also thank Dr Katrina Bell and Ms Lavinia Gordon for helpful suggestions regarding data presentation.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: JE: NHMRC Australia Capacity Building Grant in Population Health. SW: NHMRC Fellowship (Australia). www.nhmrc.gov.au. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ellis JA, Stebbing M, Harrap SB. Polymorphism of the androgen receptor gene is associated with male pattern baldness. J Invest Dermatol. 2001;116:452–455. doi: 10.1046/j.1523-1747.2001.01261.x. [DOI] [PubMed] [Google Scholar]
- 2.Hayes VM, Severi G, Eggleton SA, Padilla EJ, Southey MC, et al. The E211 G>A androgen receptor polymorphism is associated with a decreased risk of metastatic prostate cancer and androgenetic alopecia. Cancer Epidemiol Biomarkers Prev. 2005;14:993–996. doi: 10.1158/1055-9965.EPI-04-0778. [DOI] [PubMed] [Google Scholar]
- 3.Hillmer AM, Hanneken S, Ritzmann S, Becker T, Freudenberg J, et al. Genetic variation in the human androgen receptor gene is the major determinant of common early-onset androgenetic alopecia. Am J Hum Genet. 2005;77:140–148. doi: 10.1086/431425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy-Nissenbaum E, Bar-Natan M, Frydman M, Pras E. Confirmation of the association between male pattern baldness and the androgen receptor gene. Eur J Dermatol. 2005;15:339–340. [PubMed] [Google Scholar]
- 5.Ellis JA, Scurrah KJ, Cobb JE, Zaloumis SG, Duncan AE, et al. Baldness and the androgen receptor: the AR polyglycine repeat polymorphism does not confer susceptibility to androgenetic alopecia. Hum Genet. 2007;121:451–457. doi: 10.1007/s00439-006-0317-8. [DOI] [PubMed] [Google Scholar]
- 6.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 8.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 9.Beckmann JS, Estivill X, Antonarakis SE. Copy number variants and genetic traits: closer to the resolution of phenotypic to genotypic variability. Nat Rev Genet. 2007;8:639–646. doi: 10.1038/nrg2149. [DOI] [PubMed] [Google Scholar]
- 10.Harrap SB, Stebbing M, Hopper JL, Hoang HN, Giles GG. Familial patterns of covariation for cardiovascular risk factors in adults: The Victorian Family Heart Study. Am J Epidemiol. 2000;152:704–715. doi: 10.1093/aje/152.8.704. [DOI] [PubMed] [Google Scholar]
- 11.Taylor R, Matassa J, Leavy JE, Fritschi L. Validity of self reported male balding patterns in epidemiological studies. BMC Public Health. 2004;4:60. doi: 10.1186/1471-2458-4-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White SJ, Vink GR, Kriek M, Wuyts W, Schouten J, et al. Two-color multiplex ligation-dependent probe amplification: detecting genomic rearrangements in hereditary multiple exostoses. Hum Mutat. 2004;24:86–92. doi: 10.1002/humu.20054. [DOI] [PubMed] [Google Scholar]
- 13.Lalic T, Vossen RH, Coffa J, Schouten JP, Guc-Scekic M, et al. Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet. 2005;13:1231–1234. doi: 10.1038/sj.ejhg.5201465. [DOI] [PubMed] [Google Scholar]
- 14.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 16.Bruder CE, Piotrowski A, Gijsbers AA, Andersson R, Erickson S, et al. Phenotypically concordant and discordant monozygotic twins display different DNA copy-number-variation profiles. Am J Hum Genet. 2008;82:763–771. doi: 10.1016/j.ajhg.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]