Abstract
Background
The Eda-A1-Edar signaling pathway is involved in the development of organs with an ectodermal origin, including teeth. In mouse, mutants are known for both the ligand, Eda-A1 (Tabby), and the receptor, Edar (Downless). The adult dentitions of these two mutants have classically been considered to be similar. However, previous studies mentioned differences in embryonic dental development between Eda Ta and Edar dl-J mutants. A detailed study of tooth morphology in mutants bearing losses of functions of these two genes thus appears necessary to test the pattern variability induced by the developmental modifications.
Methodology/Principal Findings
3D-reconstructions of the cheek teeth have been performed at the ESRF (Grenoble, France) by X-ray synchrotron microtomography to assess dental morphology. The morphological variability observed in Eda Ta and Edar dl-J mutants have then been compared in detail. Despite patchy similarities, our detailed work on cheek teeth in Eda Ta and Edar dl-J mice show that all dental morphotypes defined in Edar dl-J mice resolutely differ from those of Eda Ta mice. This study reveals that losses of function of Eda and Edar have distinct impacts on the tooth size and morphology, contrary to what has previously been thought.
Conclusion/Signifiance
The results indicate that unknown mechanisms of the Eda pathway are implicated in tooth morphogenesis. Three hypotheses could explain our results; an unexpected role of the Xedar pathway (which is influenced by the Eda gene product but not that of Edar), a more complex connection than has been appreciated between Edar and another protein, or a ligand-independent activity for Edar. Further work is necessary to test these hypotheses and improve our understanding of the mechanisms of development.
Introduction
Genes of the Eda-A1/Edar signaling pathway are involved in the development of organs with an ectodermal origin, such as hair, glands, teeth [1]–[3] and palatal rugae [4]. The Eda gene, carried by the X chromosome, encodes the ligand (Eda-A1), and the Edar gene encodes its receptor (Edar). Eda Ta (Tabby) and Edar dl-J (Downless) mutant mice bear loss-of-function mutations for the Eda (ectodysplasinA) and the Edar (ectodysplasinA-receptor) genes, respectively [5], [6]. Consistent with their operation in a linear signalling pathway, the ectodermal organs of these mutants display similar gross phenotypes. However, differences between Eda and Edar mutants in the histological structure of the submandibular salivary gland have been reported [7]. Concerning dental morphology, only the dentition of Eda mutants has been deeply investigated. These studies revealed a high morphological diversity of the cheek dentition, characterized by modifications in the number of teeth and in the number and arrangement of cusps for homozygous and heterozygous mice [1], [8]–[13]. In contrast, no study has described the Edar dl-J dental phenotype, which is usually supposed to display the same dental defects as in Eda Ta mice [14]. However, differences between these mutants have been detected in the enamel knots, which are transient signalling centres that define the cusp pattern of the mature tooth [15]. The enamel knots of Eda Ta embryonic teeth are simply smaller in than those of WT mice [16], while Edar dl-J mutant molars have a structure termed the ‘enamel rope’ which is composed of enamel knot cells that are extended across the tooth primordium due to a failure of cell condensation [17]. This may indicate that Eda and Edar losses of function are likely to have different consequences on mature dental morphology. This prompted us to study the cheek dentition in Edar dl-J mutant mice to determine the extent to which Eda Ta and Edar dl-J phenotypes are similar and to discuss the implications of their putative differences and similarities.
Materials and Methods
Downlessj and Tabby mice
The Edar dl-J mice (FVB background) have been bred at the PBES of IFR 128 (Lyon). These mice carry a G to A transition mutation causing a glutamate to lysine substitution in the death domain of the Edar protein (E379K) [6]. Old studies on Edar mutant mice used either Edar dl [1], [9] or Edar sleek [3] mice. Edar dl-J mice have been used in more recent studies comparing the dental development and functional morphology of Eda and Edar mutant mice [17]. Comparison of Edar dl-J and Edar sleek dental morphology showed no differences [17]. Homozygous Eda and Edar mutant mice were identified according to external morphological criteria, such as the bald spot behind ears. Heterozygous Eda (female) mice were identified morphologically by the distinctive striping of the coat that gave these mutants their original name, ‘Tabby’. Heterozygous and wild-type Edar dl-J specimens exhibit similar external traits and were genotyped through PCR amplification of a 306 bp fragment covering the point mutation (primers: 5′ GTCTCAGCCCCACCGAGTTG and 3′ GTGGGGAGGCAGGTGGTACA), followed by sequencing. The Edar dl-J sample was composed of 20 heterozygous (Edar dl-J/+), 47 homozygous (Edar dl-J) and 5 control (WT) specimens. The Tabby sample used in comparison overlaps the sample studied by Kristenova et al. [10]. They carry the Eda Ta null allele of the Eda gene, carried by the X-chromosome. Eda Ta mice are on a mixed background (C57Bl6J+CBA), the sample included 60 heterozygous females (Eda Ta/+), 23 homozygous (Eda Ta/Ta) or hemizygous females (Eda Ta/0, which have a single X chromosome and display the same phenotype as homozygous [10]), 57 hemizygous males (Eda Ta/Y) and 40 WT mice. Mice were killed by cervical dislocation. The experimental protocol was designed in compliance with recommendations of the EEC (86/609/CEE) for the care and use of laboratory animals.
The uncertain homology of teeth between WT and mutant mice led us to adopt a nomenclature using Tx and Tx where T and x respectively symbolize the tooth and its rank within the row (e.g. T1 for the first upper cheek tooth, T1 for the first lower cheek tooth).
3D-data acquisition
Cheek teeth were examined using a Leica MZ16 stereomicroscope. Morphotypes have been defined for lower and upper tooth rows on the basis of the number and arrangement of cusps. Occlusal surface areas of cheek teeth were measured from digitized pictures using Optimas software. It has been demonstrated that X-ray synchrotron microtomography brings very high quality results for accurate imaging of small teeth [18]. Thus, tooth rows of a representative panel which covers the totality of observed morphologies were imaged using X-ray-synchrotron microtomography at the European Synchrotron Radiation Facility (ESRF, Grenoble, France). 3D-renderings were performed using VGStudiomax software.
Statistical tests
Statistical tests have been performed to compare tooth size. The Kruskal-Wallis analysis of variance was used to verify the significance of observed differences between Eda Ta and Edar dl-J mice. This is a non-parametric method, which tests equality of population medians among groups. The non-parametric Wilcoxon rank-test is used to indicate which groups are statistically different from the others.
Results
Wild-type mice
WT specimens from Eda Ta and Edar dl-J breeding colonies all display the normal dental morphology, without anomalies, and measurements show similar tooth size (ANOVA test at p = 0.05 threshold value). Thus, molars from WT from both genetic backgrounds were included in a single WT sample.
Heterozygous Eda Ta/+ and Edar dl-J/+ mice
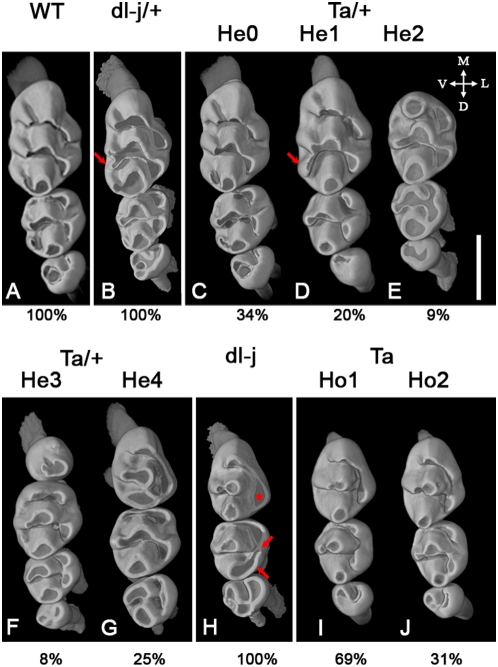
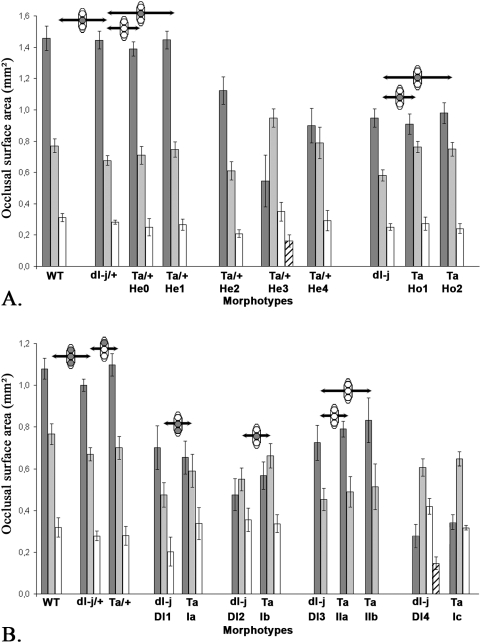
As Eda is located on the X chromosome while Edar is on an autosome, morphological differences linked to the X-inactivation process were predictable. Accordingly, in both the upper and lower cheek dentitions, heterozygous Edar dl-J/+ mice display dental morphologies very close to WT specimens (Fig. 1A–B, Fig. 2A–B) whereas Eda Ta/+ mice dentitions exhibit highly variable patterns (Fig. 1C–G, Fig. 2I–K). Edar dl-J/+ upper tooth rows differ from the WT morphology by the occurrence of a supplementary centro-vestibular cusp at the T1 (arrow on Fig. 1B). This pattern approaches the morphotype He1 of Eda Ta/+ mice (arrow on Fig. 1D) [13]. The size of the upper teeth only differs between WT, Eda Ta He1 and Edar dl-J by a smaller T2 in Edar dl-J (Fig. 3A). Upper tooth size is similar for Edar dl-J/+ and Eda Ta/+ He0, defined as the Tabby heterozygous morphotype exhibiting the wild-type morphology (presented Fig. 1C).
Figure 1. Upper tooth rows; wild-type morphology and morphotypes defined among EdaTa and Edardl-J mutant mice.
A: WT morphology, B: Edardl-j/+ morphology; C: morphotype Ta He0; D: morphotype Ta He1; E: morphotype Ta He2; F: morphotype Ta He3; G: morphotype Ta He4; H: Edardl-j morphology; I: morphotype EdaTa Ho1; J: morphotype EdaTa Ho2. The proportions indicated below the morphotypes are the occurrence frequency of the morphotypes. Images are obtained using X-ray synchrotron microtomography. Tooth orientation: M: mesial, D: distal, V: vestibular, and L: lingual. Scale bar: 1 mm. The structures indicated by arrows and asterisk are discussed in the main text.
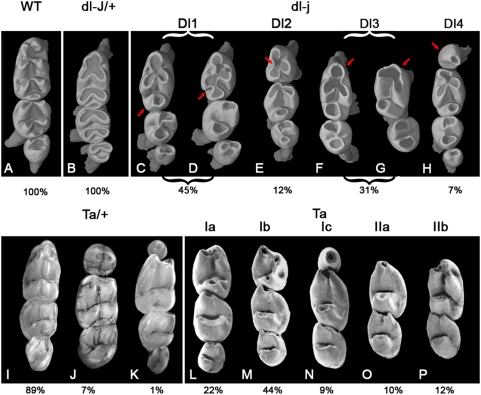
Figure 2. Lower tooth rows; wild-type morphology and morphotypes defined among EdaTa and Edardl-J mutant mice.
A: WT morphology; B: Edardl-J/+ morphology; C–D: Edardl-j morphotype Dl1, E: Edardl-j morphotype Dl2, F–G: Edardl-j morphotype Dl3, H: Edardl-j morphotype Dl4, I–K: EdaTa morphotypes, L: EdaTa morphotype Ia, M: EdaTa morphotype Ib, N: EdaTa morphotype Ic, O: EdaTa morphotype IIa, P: EdaTa morphotype IIb. The proportions indicated below the morphotypes are the occurrence frequency of the morphotypes. Images of Edar dl-J mice are obtained using X-ray synchrotron microtomography. Images of Eda Ta mice are obtained by photography and are taken from Kristenova et al. [10] and Peterkova et al. [30]. Same orientation as in Fig. 1. The structures indicated by arrows are discussed in the main text.
Figure 3. Size of the cheek teeth of Wild-type, Downless and Tabby heterozygous (+/−) and homozygous (−/−) mice.
A. Upper cheek teeth. Dark grey: T1, light grey: T2, white: T3, and hatched: T4. B. Lower cheek teeth. Dark grey: T1, light grey: T2, white: T3, and hatched: T4. Black bars indicate the standard-deviation of the mean. WT: Wild-type mice, dl-J/+: Edardl-J/+, Ta/+: EdaTa/+, dl-J: Edardl-J, Ta: EdaTa. The arrows indicate the statistical size comparison between morphologically close morphotypes, the three circles on each arrow represent from top to bottom the first, second and third cheek teeth, a grey-filled circle indicates a statistically significant difference in size between the two considered teeth according to a Wilcoxon rank-test at p = 0.05 threshold value, a white circle indicates the absence of statistical difference. Morphotypes that appear to be obviously different on the plot were found to have statistically significant differences.
Eda Ta/+ mice sometimes exhibit a small mesial lower tooth (Fig. 2J–K) which is never found in Edar dl-J/+ mice. Moreover, Edar dl-J/+ lower teeth are all smaller than those of the WT. They also differ from the Eda Ta/+ major morphotype by a smaller T1 (Fig. 3B). The almost normal morphology of upper and lower cheek dentitions of Edar dl-J/+ is thus far from the morphological diversity observed in Eda Ta/+.
Homozygous Eda Ta and Edar dl-J mice
Upper and lower cheek teeth clearly differ between Eda Ta and Edar dl-J mice in terms of morphology and size. Edar dl-J upper dentition is characterised by: (i) a T1 with a single lingual cusp (asterisk in Fig. 1H) linked by a crest to the mesial-most cusp, (ii) a T2 with a lingual interconnection between mesial and distal cusps (arrows in Fig 1H). T1 of Edar dl-J are similar to those of Eda Ta Ho1 (Fig. 1H–I). T1 of Edar dl-J and Eda Ta Ho1 statistically have the same size (Fig. 3A). However, T2 are statistically smaller in Edar dl-J (Fig. 3A) and the lingual ridge of Edar dl-J T2 does not occur in Eda Ta T2. The morphological differences between Eda Ta and Edar dl-J T2 can be explained by their crown size differences referring to the ‘patterning cascade mode of cusp development’ [19] in which the signaling centre succession, and consequently the number and position of cusps, is linked to the crown size.
The lower cheek dentition of Edar dl-J is more variable than the upper. Four morphotypes can be defined (Fig. 2C–G), specimens can exhibit two different morphotypes on left and right sides, none of the four morphotypes occurs in EdaTa dentition. (i) The first morphotype, Dl1 (for Downless1), includes 45% of the studied material. It is characterized by a three-toothed row and a four-cusp T1. The three mesial cusps form a three-leaf clover shape, while the fourth cusp is either isolated in the distal part of the tooth (85% of Dl1 tooth rows, arrowed in Fig. 2C) or connected to the others by a longitudinal crest (15% of Dl1 tooth rows, arrowed in Fig. 2D). No Eda Ta morphotype is similar to this Dl1 morphotype. (ii) The Dl2 morphotype (12% of examined tooth rows) also exhibits three cheek teeth, but the T1 only displays three cusps. As in Dl1 morphotype, the cusps form a three-leaf clover shape connected in the centre of the occlusal surface (arrow in Fig. 2E) but the distal cusp is missing. Except for cusp connections, the Dl2 morphotype approaches the EdaTa Ib morphotype (Fig. 2M), but differs by the size of the T2 (Fig. 3B). (iii) The Dl3 morphotype (31% of tooth rows) is characterized by rows with two lower cheek teeth (Fig. 2F–G). The T1 of the Dl3 morphotype resembles this of the Dl1 morphotype (Fig. 2C,F). However, the three mesial cusps display highly variable size and position. The most mesial element varies from a large rounded cusp (arrowed in Fig. 2F) to a highly reduced, almost absent, element (arrow in Fig. 2G). This latter morphology is alike the Eda Ta IIb morphotype (Fig. 2P). The Dl3 morphotype displays the same tooth size as Eda Ta IIa and IIb morphotypes (Fig. 3B). (iv) The Dl4 morphotype (7% of tooth rows) is characterized by a small T1 (arrow in Fig. 2H), 57% of the Dl4 tooth rows encompass three teeth, while 43% have a tiny T4. The T2 encompasses four cusps, three mesial interconnected and an isolated distal one. The presence of the reduced T1 is reminiscent of the Eda Ta Ic morphotype (Fig. 2N). However, the morphology of the T1 is highly different as it encompasses a higher number of cusps than that of the Eda Ta Ic morphotype, which moreover never exhibits 4 teeth.
Discussion
X-inactivation might explain differences between heterozygous Eda Ta/+ and Edar dl-J/+ mice
The many observed differences, and the higher variability of tooth rows, between Eda Ta/+ and Edar dl-J/+ mice can be explained by the fact that the Eda gene is X-linked and that Edar is carried by an autosome. Due to the X-inactivation effect in females [20], [21], EdaTa/+ mice are mosaics of cells with expression of a wild-type or a null Eda gene. This might induce a strong and random variability in the amount of Ectodysplasin-A1 protein available for dental development, explaining the higher morphological variability recorded in Eda Ta/+. This phenomenon does not occur in the Edar dl-J teeth since the gene is located on an autosome (chromosome 10).
Differences between homozygous Eda Ta and Edar dl-J mice imply other mechanisms
The same general trends in the reduction of the cusp number and tooth size are observed in both Eda Ta and Edar dl-J mutants. However, none of the Eda Ta and Eda dl-J tooth rows are identical. The two mutants are supposed to display total losses of function of the ligand and the receptor of the Eda pathway, respectively. According to Tucker et al. [17], Eda mice have small enamel knots while Edar dl-J mice have disorganised “enamel ropes”. As enamel knots are orchestrating the final mineralized crown pattern, differences of size and morphology, differences in enamel knots are likely to explain differences of size and morphology between Eda Ta and Edar dl-J dentitions.
Various hypotheses could explain these differences: (i) They could be linked to epistatic differences between the two different backgrounds of Eda Ta and Edar dl-J mice (respectively mixed C57Bl6J+CBA and FVB). However we consider this possibility to be unlikely to be the only explanation. Indeed, suppression of Eda action in embryonic CD-1 mice by addition of soluble forms of Edar to tooth explants results in the same phenotype as seen in Eda Ta mice on NMRI and mixed CBAT6T6xNMRI backgrounds but differs from the Edar dl-J on CD-1 background [17]. This indicates that differences observed between enamel knots in Eda Ta and Edar dl-J mice are not due to a difference in background but rather to an intrinsic difference between loss of Eda function and alteration of Edar function in Edar dl-J. (ii) Eda codes for two proteins, Eda-A1, which binds to Edar, and Eda-A2, which binds to Xedar [22]. The Xedar pathway is thus lost in Eda Ta mice but still present in Edar dl-J mice. An effect on tooth development of the loss of the Xedar pathway in the case of Eda loss of function and not in the Edar one might explain some of the differences that we report. Though a study of Xedar-null mice indicated no requirement for this gene in the normal development of ectodermal organs [23], dominant negative and constitutively active forms of this protein have been shown to have effects similar to those of Edar in developing chicken skin [24] and a compensatory action of Xedar that is revealed only upon loss of Edar function can not be ruled out. (iii) The Edar dl-J allele may not be a null mutation. Indeed, the Edar dl-J mutation causes a substitution in the death domain of the Edar protein, which connects to the Edar adapter with death domain, Edaradd [25]. However the intracellular domain also contains two sites for direct binding of TRAF proteins that are conserved among vertebrates [26]. These sites may allow Edaradd-independent signaling, even if, to date, no molecular study indicates utilisation of such an alternative pathway by Edar. Alternatively, the Edar dl-J mutants EdarE379K protein may retain some interaction with Edaradd allowing transmission of a weak signal through this pathway. Assays in cultured cells have given conflicting results on the nature of the signal output of EdarE379K. Yan et al. [22] reported that EdarE379K fails to activate NF-B, while Kumar et al. [27] found that the mutant protein retains a significant capacity to stimulate this pathway. Thus this hypothesis remains an attractive possibility that has to be experimentally tested. A complete loss of function of the gene may, in that case, reveal morphological differences with the present Downless genotype used in this study (iv) A ligand-independent activation mechanism for Edar could also explain the morphological differences observed. Indeed, the finding that elevation of Edar expression can rescue the hair phenotype of Eda Ta mutant embryos [28] suggests that Edar does undergo ligand-independent signaling, at least when expressed at sufficiently high levels. Alternatively, some transmembrane receptors are dependence receptors, such as the Netrin-1 receptor which displays ligand-independent activities [29]. Under this hypothesis, the absence of Eda that leads to a pathway containing an “active” unliganded Edar will lead to a different phenotype than the loss of Edar. This hypothesis could be tested by crossing Edar dl-J with Eda Ta mice. Such a cross should result in an Edar dl-J type phenotype. Another test could be a cross of Tabby mice with a loss-of-function mutant of the Edaradd gene that should also give rise to an Edar loss-of-function phenotype.
To sum up, loss of function of Eda or Edar leads to different dental morphologies, presumably linked to the differences in their enamel knot morphologies. These differences indicate unknown mechanisms of the Eda pathway involved in tooth morphogenesis. These unknown mechanisms we hypothesize to be based on an unexpected role of the Xedar pathway in tooth development, a more complex connection than thought between Edar and Edaradd, or on a ligand-independent activity for Edar.
Acknowledgments
We thank R. Peterkova and P. Kristenova for the loan of EdaTa specimens. Thanks also to J. Baruchel and ID 19 staff of the ESRF. We also acknowledge J.-J. Jaeger, P. Vignaud, G. Florent and C. Noël for their help and two anonymous reviewers for their comments.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study has been supported by the ANR (‘Quenottes’ program). Other supports have been provided by the regions Poitou-Charentes and Rhone-Alpes, the Fondation pour la Recherche Medicale, and the Fondation Singer-Polignac. SP holds a fellowship from CNRS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sofaer JA. Aspects of the tabby-crinckled-downless syndrome I. The development of Tabby teeth. Journal of Embryology and Experimental Morphology. 1969;22(2):181–205. [PubMed] [Google Scholar]
- 2.Grüneberg H. The Tabby syndrome in the mouse. Proceedings of the Royal Society of London Biological Sciences. 1971;179:139–156. doi: 10.1098/rspb.1971.0086. [DOI] [PubMed] [Google Scholar]
- 3.Sofaer JA. The teeth of the sleek mouse. Archives of Oral Biology. 1977;22:299–301. doi: 10.1016/0003-9969(77)90117-0. [DOI] [PubMed] [Google Scholar]
- 4.Charles C, Pantalacci S, Peterkova R, Peterka M, Laudet V, et al. Disruption of the palatal rugae pattern in Tabby (eda) mutant mice. European Journal of Oral Sciences. 2007;114:1–8. doi: 10.1111/j.1600-0722.2007.00482.x. [DOI] [PubMed] [Google Scholar]
- 5.Kere J, Srivastava AK, Montonen O, Zonana J, Thomas N, et al. X−linked anhidrotic (hypohidrotic) ectodermal dysplasia is caused by mutation in a novel transmembrane protein. Nature genetics. 1996;13(4):409–416. doi: 10.1038/ng0895-409. [DOI] [PubMed] [Google Scholar]
- 6.Headon DJ, Overbeek PA. Involvement of a novel Tnf receptor homologue in hair follicle induction. Nature genetics. 1999;22:370–374. doi: 10.1038/11943. [DOI] [PubMed] [Google Scholar]
- 7.Jaskoll T, Zhou Y-M, Trump G, Melnick M. Ectodysplasin receptor-mediated signaling is essential for embryonic submandibular salivary gland development. The Anatomical Record. 2003;271A:322–331. doi: 10.1002/ar.a.10045. [DOI] [PubMed] [Google Scholar]
- 8.Grüneberg H. The molars of the tabby mouse, and a test of the ‘single-active X-chromosome’ hypothesis. Journal of Embryology and Experimental Morphology. 1966;15(2):223–244. [PubMed] [Google Scholar]
- 9.Sofaer JA. Aspects of the tabby-crinckled-downless syndrome II. Observations on the reaction to changes of genetic background. Journal of Embryology and Experimental Morphology. 1969;22(2):207–227. [PubMed] [Google Scholar]
- 10.Kristenova P, Peterka M, Lisi S, Gendrault J-L, Lesot H, et al. Different morphotypes of functional dentition in the lower molar region of tabby (EDA) mice. Orthodontics and Craniofacial Research. 2002;5:205–214. doi: 10.1034/j.1600-0544.2002.02225.x. [DOI] [PubMed] [Google Scholar]
- 11.Mustonen T, Pispa J, Mikkola ML, Pummila M, Kangas AT, et al. Stimulation of ectodermal organ development by Ectodysplasin-A1. Developmental Biology. 2003;259:123–136. doi: 10.1016/s0012-1606(03)00157-x. [DOI] [PubMed] [Google Scholar]
- 12.Peterková R, Lesot H, Viriot L, Peterka M. The supernumerary cheek tooth in tabby/EDA mice - a reminiscence of the premolar in mouse ancestors. Archives of Oral Biology. 2005;50:219–225. doi: 10.1016/j.archoralbio.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 13.Charles C, Pantalacci S, Peterková R, Tafforeau P, Laudet V, et al. Effect of eda loss of function on upper jugal tooth morphology. The Anatomical Record. 2009;292:299–308. doi: 10.1002/ar.20804. [DOI] [PubMed] [Google Scholar]
- 14.Courtney J-M, Blackburn J, Sharpe PT. The Ectodysplasin and NFkB signalling pathways in odontogenesis. Archive of Oral Biology. 2005;50:159–163. doi: 10.1016/j.archoralbio.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 15.Vaahtokari A, Aberg T, Jernvall J, KerWen S, Thesleff I. The enamel knot as a signaling center in the developing mouse tooth. Mechanisms of Development. 1996;54:39–43. doi: 10.1016/0925-4773(95)00459-9. [DOI] [PubMed] [Google Scholar]
- 16.Pispa J, Jung H-S, Jernvall J, Kettunen P, Mustonen T, et al. Cusp Patterning Defect in Tabby Mouse Teeth and Its Partial Rescue by FGF. Developmental Biology. 1999;216:521–534. doi: 10.1006/dbio.1999.9514. [DOI] [PubMed] [Google Scholar]
- 17.Tucker AS, Headon DJ, Schneider P, Ferguson B, Overbeek P, et al. Edar/Eda interactions regulate enamel knot formation in tooth morphogenesis. Development. 2000;127:4691–4700. doi: 10.1242/dev.127.21.4691. [DOI] [PubMed] [Google Scholar]
- 18.Tafforeau P, Boistel R, Boller E, Bravin A, Brunet M, et al. Applications of X-ray synchrotron microtomography for non-destructive 3D studies of paleontological specimens. Applied Physics A: Materials Science & Processing. 2006;83:195–202. [Google Scholar]
- 19.Jernvall J. Linking development with generation of novelty in mammalian teeth. Proceedings of the National Academy of Sciences. 2000;97:2641–2645. doi: 10.1073/pnas.050586297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lyon M. Gene Action in the X-chromosome of the Mouse (Mus musculus L.). Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 21.Plath K, Mlynarczyk-Evans S, Nusinow D, Panning B. XIST RNA and the Mechanism of X chromosome inactivation. Annual Review of Genetics. 2002;36:233–278. doi: 10.1146/annurev.genet.36.042902.092433. [DOI] [PubMed] [Google Scholar]
- 22.Yan M, Wang L-C, Hymowitz SG, Schilbach S, Lee J, et al. Two-Amino Acid Molecular Switch in an Epithelial Morphogen That Regulates Binding to Two Distinct Receptors. Science. 2000;290:523–527. doi: 10.1126/science.290.5491.523. [DOI] [PubMed] [Google Scholar]
- 23.Newton K, French DM, Yan M, Frantz GD, Dixit VM. Myodegeneration in EDA-A2 Transgenic Mice Is Prevented by XEDAR Deficiency. Molecular and Cellular Biology. 2004;24:1608–1613. doi: 10.1128/MCB.24.4.1608-1613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drew CF, Lin CM, Jiang TX, Blunt G, Mou C, et al. The Edar subfamily in feather placode formation. Developmental Biology. 2007;305:232–245. doi: 10.1016/j.ydbio.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Headon DJ, Emmal S, Ferguson B, Tucker A, Justice M, et al. Gene defect in ectodermal dysplasia implicates a death domain adapter in development. Nature. 2001;914:913–916. doi: 10.1038/414913a. [DOI] [PubMed] [Google Scholar]
- 26.Pantalacci S, Chaumot A, Benoit G, Sadier A, Delsuc F, et al. Conserved features and evolutionary shifts of the EDA signaling pathway involved in vertebrate skin appendage development. Molecular Biology and Evolution. 2008;25:912–928. doi: 10.1093/molbev/msn038. [DOI] [PubMed] [Google Scholar]
- 27.Kumar A, Eby M, Sinha S, Jasmin A, Chaudhary P. The Ectodermal Dysplasia Receptor Activates the Nuclear Factor-kB, JNK, and Cell Death Pathways and Binds to Ectodysplasin A. Journal of Biological Chemistry. 2001;276:2668–2677. doi: 10.1074/jbc.M008356200. [DOI] [PubMed] [Google Scholar]
- 28.Mou C, Jackson B, Schneider P, Overbeek PA, Headon DJ. Generation of the primary hair follicle pattern. Proceedings of the National Academy of Sciences. 2006;103:9075–9080. doi: 10.1073/pnas.0600825103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehlen P, Rabizadeh S, Snipas S, Assa-Munt N, Salvesen G, et al. The DCC gene product induces apoptosis by amechanism requiring receptor proteolysis. Nature. 1998;395:801–804. doi: 10.1038/27441. [DOI] [PubMed] [Google Scholar]
- 30.Peterkova R, Lesot H, Peterka M. Phylogenetic memory of developing mammalian dentition. Journal of Experimental Zoology part B Molecular and Developmental Evolution. 2006;306B:1–17. doi: 10.1002/jez.b.21093. [DOI] [PubMed] [Google Scholar]