Abstract
The coronavirus spike protein (S) plays a key role in the early steps of viral infection, with the S1 domain responsible for receptor binding and the S2 domain mediating membrane fusion. In some cases, the S protein is proteolytically cleaved at the S1–S2 boundary. In the case of the severe acute respiratory syndrome coronavirus (SARS-CoV), it has been shown that virus entry requires the endosomal protease cathepsin L; however, it was also found that infection of SARS-CoV could be strongly induced by trypsin treatment. Overall, in terms of how cleavage might activate membrane fusion, proteolytic processing of the SARS-CoV S protein remains unclear. Here, we identify a proteolytic cleavage site within the SARS-CoV S2 domain (S2′, R797). Mutation of R797 specifically inhibited trypsin-dependent fusion in both cell–cell fusion and pseudovirion entry assays. We also introduced a furin cleavage site at both the S2′ cleavage site within S2 793-KPTKR-797 (S2′), as well as at the junction of S1 and S2. Introduction of a furin cleavage site at the S2′ position allowed trypsin-independent cell–cell fusion, which was strongly increased by the presence of a second furin cleavage site at the S1–S2 position. Taken together, these data suggest a novel priming mechanism for a viral fusion protein, with a critical proteolytic cleavage event on the SARS-CoV S protein at position 797 (S2′), acting in concert with the S1–S2 cleavage site to mediate membrane fusion and virus infectivity.
Keywords: membrane fusion, proteolytic processing, virus entry
Enveloped viruses access their host cells by a process of membrane fusion that is mediated by a specific fusion, or “spike” protein, encoded by the virus and embedded in the viral envelope (1, 2). Such proteins are currently grouped into 3 distinct structural classes, with the so-called class I fusion proteins typically primed for fusion activation by proteolytic cleavage (3, 4). Fusion activation can be activated by low pH, receptor binding, or a combination of the two, with the cleavage event typically occurring in the vicinity of the viral fusion peptide, which becomes exposed upon activation-dependent conformational changes of the spike protein and initiates the fusion reaction following its insertion into the host cell membrane (5). In several cases, such as highly pathogenic avian influenza virus, mutations in the cleavage site (monobasic vs. polybasic) and subsequent changes in the molecular basis of proteolytic cleavage by trypsin-like or furin-like enzymes have profound implications on virulence (6, 7). As such, understanding spike protein cleavage is fundamental to an understanding of viral pathogenesis.
The severe acute respiratory syndrome coronavirus (SARS-CoV) emerged in 2003 as a significant threat to human health, and coronaviruses still represent a leading source of novel viruses for emergence into the human population. The coronavirus spike protein (S) mediates both receptor binding (via the S1 domain) and membrane fusion (via the S2 domain) and shows many features of conventional class I fusion proteins, including the presence of distinct heptad repeats within the fusion domain (8). Coronaviruses exist in 3 distinct groups, and their spike proteins appear to differ considerably in their proteolytic activation (9). Whereas some coronaviruses, notably the group 3 avian infectious bronchitis virus (IBV), are efficiently cleaved at the boundary between the S1 and S2 domains, many other coronaviruses apparently remain uncleaved. Early reports analyzing heterologously expressed SARS-CoV spike protein indicated that most of the protein was not cleaved (10, 11), but there was some possibility of limited cleavage at the S1–S2 position (11). However, S1–S2 cleavage could be enhanced by expression of furin family enzymes (12).
Analysis of both SARS-CoV infection and transduction by SARS-CoV S-pseudotyped virions has indicated that the virus is sensitive to inhibitors of endosomal acidification (13–17), and it has been shown that SARS-CoV S-pseudotyped virions use the endosomal protease cathepsin L to infect cells (18, 19). These data suggested that SARS-CoV S required a novel, endocytic protease-primed cleavage event during virus entry, in contrast to the majority of class I viral fusion proteins that are primed during virus assembly or following release from the cell. However, in addition to cathepsin L, several other proteases have been shown to cleave SARS-CoV S. Early reports showed that S1–S2 cleavage was enhanced by exogenous trypsin (20), and it was subsequently shown that SARS-CoV infection is enhanced by the addition of exogenous proteases, such as trypsin, thermolysin, and elastase (14, 17), as well as by expression of factor Xa (21). These data suggested that an alternative, nonendosomal route of SARS-CoV entry exists. Notably, infection mediated by exogenous proteases was considered to be 100– to 1,000-fold more efficient than by the endosomal route (17). Based on cleavage patterns on SDS/PAGE gels, the predominant cleavage event mediated by the exogenous protease appears to be at the S1–S2 boundary, and subsequent biochemical analysis by N-terminal sequencing identified R667 (SLLR-S) as a site of trypsin cleavage (22). Similarly, exogenous cathepsin L can also cleave the S1–S2 junction at residue T678 (VAYT-M) (23).
Although cleavage of SARS-CoV S is readily achieved at the S1–S2 boundary, it is notable that mutation of the 2 basic residues in this region (R667 and K672) failed to affect trypsin-primed membrane fusion, despite blocking S1–S2 cleavage (24), suggesting that the proteolytic processing of SARS-CoV S that leads to an activation of infectivity may occur at a different site. Here, we investigate the proteolytic cleavage sites in SARS-CoV S and characterize a novel cleavage site in the S2 domain (S2′), which acts in concert with the S1–S2 cleavage site to mediate SARS-CoV S-mediated membrane fusion and virus infectivity.
Results
Role of the S1–S2 Boundary in Trypsin-Mediated Activation of SARS-CoV S Membrane Fusion.
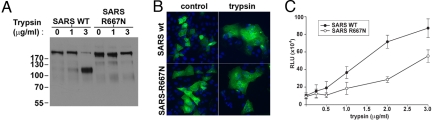
To investigate more precisely the role of proteolytic cleavage at the S1–S2 boundary on SARS-CoV S-mediated membrane fusion, we first mutated residue R667 to N to inhibit trypsin-mediated cleavage. As shown in Fig. 1A, wild-type SARS-CoV S was readily cleaved at the S1–S2 boundary by trypsin, whereas the R667N mutant showed no detectable cleavage at this position. We next monitored the fusion properties of the wild-type and mutant S protein in cell–cell fusion assays. Vero E6 cells expressing the wild-type SARS-CoV S or the R667 mutant were treated with trypsin and processed for immunofluorescence microscopy to visualize syncytia formation. Syncytia formation for the R667N mutant was the same as for the wild-type protein (Fig. 1B). These data are consistent with previous reports (24). To quantify the syncytia formation, we used a luciferase-based cell–cell fusion assay. BHK cells were cotransfected with the wild-type or mutant SARS-CoV S protein and with a plasmid encoding the luciferase gene driven by a T7 polymerase promoter. After 24 h, these cells were overlaid with VeroE6 cells transiently expressing the T7 polymerase. Cell–cell fusion was then induced by treating the cells with different doses of trypsin for 20 min. The amount of fusion increased with the dose of trypsin for the wild-type protein, reaching a plateau at 2 μg/mL trypsin (Fig. 1C). The mutant R667N was still able to cause cell–cell fusion; however, the fusion was not as efficient as that observed for the wild-type protein (maximum fusion for the mutant was ≈50% of the fusion of the wild-type protein). These data suggest that although the R667 at the S1–S2 boundary does have some role in cleavage activation of SARS-CoV S-mediated membrane fusion, other residues are likely to have more critical roles.
Fig. 1.
Role of the S1–S2 boundary in trypsin-mediated activation of SARS-CoV S membrane fusion. (A) BHK cells transiently expressing SARS-CoV S wild type (SARS wt) or the SARS-R667N mutant were treated with 1 μg/mL trypsin for 30 min or 1 h. Samples were analyzed by Western blot analysis, with the C-terminal part of the spike protein detected by using an anti-C9 antibody. (B) VeroE6 cells expressing SARS-CoV S wild type (SARS wt) or the SARS-R667N mutant were treated with 2 μg/mL trypsin for 20 min, then incubated for 40 min in absence of trypsin and analyzed by immunofluorescence microscopy using an anti-S monoclonal antibody (green), with nuclei counterstained with Hoechst 33548 (blue). (Magnification: 200×.) (C) BHK cells cotransfected with plasmids encoding the SARS-CoV S wild type (SARS wt) or SARS-R667N, as well as a plasmid encoding luciferase under the control of the T7 polymerase were overlaid with VeroE6 cells expressing the T7 polymerase. Cell–cell fusion was induced by treating the cells with increasing amount of trypsin for 20 min. At 5 h after fusion induction, the cells were lysed to monitor luciferase activity. Results are presented as relative light units (RLU). Error bars represent the standard error of the mean for 3 independent experiments.
Identification of Additional Cleavage Sites Within the Coronavirus S Protein.
To investigate the possibility that additional, possibly cryptic, cleavage sites exist within the coronavirus spike protein, we examined a comprehensive selection of spike proteins from coronaviruses in groups 1, 2, and 3 by using the ProP 1.0 server (www.cbs.dtu.dk/services/ProP/). This server predicts arginine and lysine propeptide cleavage sites in eukaryotic protein sequences by using an ensemble of neural networks. Based on the known importance of furin sites in viral fusion proteins (25), we used furin-specific prediction as the default. One coronavirus spike protein in particular showed a very strong prediction for furin cleavage within the S2 domain (Fig. S1). This spike protein, from the IBV strain Beaudette, belongs within coronavirus group 3 and is well-established to be cleaved at the S1–S2 boundary via a furin cleavage site (26). Most IBV strains [e.g., the prototype strain Massachusetts 41 (M41)] have a restricted tropism, being able to infect only primary chicken cells in tissue culture; however, the Beaudette strain presents an extended tropsim and is able to infect a wide range of cell lines (27). Alignment of the IBV spike protein shows that all database sequences of the Beaudette strain contain a second consensus furin cleavage site within S2, which is absent in all other IBV strains (Fig. S2). This furin cleavage site (-RRRR- or -RRKR-) is located between residues 687 and 690, and we named it S2′. To determine whether this second furin site is actually cleaved in IBV S, we analyzed the profile of the IBV Beaudette and M41 spike proteins by Western blot analysis, using an antibody specific for the S2 domain (28). Two cleavage products (65 kDa and 50 kDa) were clearly observed for the Beaudette strain, representing cleavage at S1–S2 as well as at S2′, whereas the M41 presented only a single cleaved product (65 kDa) due to cleavage at S1–S2. (Fig. S2). These data suggest that the second cleavage site can be present in the coronavirus S2 domain, which may have important properties for membrane fusion and virus entry.
IBV Beaudette was the only coronavirus for which the ProP server predicted a furin site clearly above threshold within S2. However, analysis of the SARS-CoV S protein (group 2b) revealed a relatively high cleavage potential by furin at the S2′ position, with very limited cleavage potential at the S1–S2 boundary (Fig. S1), indicating the possibility of a cryptic cleavage site at the S2′ position. The S2′ site of SARS-CoV S comprises a dibasic motif K796, R797 (Fig. S2) that could be such a cleavage site, and so we decided to investigate the role of these 2 basic residues in the membrane fusion and entry of SARS-CoV.
Introduction of a Furin Site at S2′ Induces SARS-CoV S-Mediated Membrane Fusion.
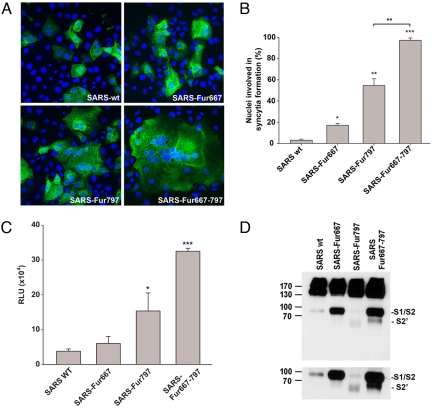
To examine the potential use of the SARS-CoV S1–S2 and S2′ positions as sites for proteolytic cleavage, we first introduced furin cleavage recognition sites at these locations by making the following mutations 664SLLRSTSQSI-SLLRRSRRSI671 (S1–S2) and 792LKPTKRSF-LKRTKRSF799 (S2′). Expression of these mutants in VeroE6 cells induced cell–cell fusion in the absence of any trypsin treatment to different extents (Fig. 2A). As described previously (29), introduction of the furin site at position 667 (furin667) enhanced cell–cell fusion to a modest extent (17% of nuclei in syncytia) compared with wild-type spike, where <5% of nuclei were in syncytia (Fig. 2B). In contrast, the mutant furin797 showed extensive syncytia formation, with ≈55% of cells in syncytia. We also created an additional mutant with furin recognition sites at both S1–S2 and at S2′ (furin667furin797). This mutant protein induced formation of very large syncytia, with >95% of cells in syncytia. To quantify the level of membrane fusion induced by these mutants, we used a luciferase-based cell–cell fusion assay to quantify the fusion activity of these mutants (Fig. 2C). Luciferase activity observed for the furin667 mutant was only slightly increased (1.3 ± 0.15-fold). Insertion of the furin site at the S2′ position (furin797) markedly increased the luciferase activity (4.3 fold ± 3.5-fold) but, as seen in Fig. 2B, this effect was dramatically increased (12.3 ± 3.6-fold) by addition of the furin cleavage site at S1–S2 (furin667furin797).
Fig. 2.
Introduction of a furin site at S2′ induces SARS-CoV S-mediated membrane fusion. (A) VeroE6 cells expressing SARS-CoV S wild type (SARS wt) or the mutants SARS-Fur667, SARS-Fur797, or SARS-Fur667-797 were analyzed by immunofluorescence microscopy using an anti-S monoclonal antibody (green), with nuclei counterstained with Hoechst 33548 (blue). (Magnification: 200×.) (B) VeroE6 cells expressing SARS-CoV S wild type (SARS wt) or furin mutants were analyzed by immunofluorescence microscopy, and the number of nuclei involved in syncytia formation was counted for each mutant. Error bars represent the standard error of the mean for 3 independent experiments. Data were analyzed by using a Student's t test. (C) BHK cells were cotransfected with SARS-CoV S wild type (SARS wt) or furin mutants and with a T7-driven luciferase plasmid and then were overlaid with VeroE6 cells expressing the T7 polymerase. Luciferase activity was then measured after 8 h. Results are presented as relative light units (RLU). Error bars represent the standard error of the mean for 3 independent experiments. Data were analyzed by using a Student's t test. (D) BHK cells expressing SARS-CoV S wild type (SARS wt) or furin mutants were analyzed by Western blot analysis, with the C-terminal part of the spike protein detected by using an anti-myc antibody. (Lower) A longer exposure time of a portion of the Western blot analysis to more clearly show the cleavage products for the mutant furin797 (F797).
We next analyzed the cleavage efficiency of these mutants in a cell surface biotinylation assay (Fig. 2D). The SARS-CoV spike protein was present on the cell surface mainly as an uncleaved protein, but a weak band corresponding to cleavage at S1–S2 (85–95 kDa) also was observed. In contrast, the SARS spike protein harboring a cleavage site at the S1–S2 boundary (furin667) was efficiently cleaved into S1 and S2 subunits. With a single furin site at the S2′ position (furin797), a cleaved product of (65–70 kDa) was clearly produced, although the cleavage product was only detected after longer exposure times. The presence of the 65- to 70-kDa product occurred along with a decrease in the amount of S2 (85–95 kDa). The mutant with the double furin cleavage site (furin667furin797) also showed a clear band corresponding to the cleavage at S2′, but this cleavage product was less abundant compared with the cleavage product produced at the S1–S2 boundary. Taken together, these data suggest that SARS-CoV can be proteolytically cleaved at the S2′ position to activate membrane fusion, and that cleavage at the S1–S2 boundary facilitates both cleavage at S2′ (797) and S2′-activated membrane fusion.
Mutation of R797 at SARS-CoV S2′ Inhibits Trypsin-Induced Membrane Fusion.
To determine the role of the dibasic residues at the SARS-CoV S2′ position in membrane fusion in more detail, we examined the effects of mutating residues K796 and R797 on trypsin-primed fusion. We generated different mutants: the single mutations K796A and R797N, as well as the double mutant K796A/R797N. We first monitored the cell surface expression of these mutants by cell surface biotinylation assay. Mutation of the dibasic residues reduced cell surface expression of the protein; however, cell surface expression could be rescued by incubating cells at 32 °C (Fig. S3). All of the mutants showed at least 62% of the expression of the SARS wild-type protein on the cell surface (Fig. S3). We then analyzed the fusion properties of these mutants. We first analyzed syncytia formation by immunofluorescence microscopy (Fig. 3A). Trypsin treatment of VeroE6 cells expressing the SARS-CoV wild-type protein induced robust syncytia formation, whereas syncytia formation was virtually undetectable for cells expressing the mutant K796A/R797N. Similar results were obtained for the cells expressing the R797N mutant. However, the mutant K796A still showed efficient trypsin-induced syncytia formation. To quantify this effect, we used a luciferase-based cell–cell fusion assay (Fig. 3B). In line with microscopy data, these results show that mutation of both basic residues K796A/R797N strongly inhibited trypsin-induced membrane fusion activated by SARS-CoV S (2.5% ± 1.1%). Analysis of the role of each residue independently shows that mutation of K796 only partially affected the fusion induced by trypsin (79.5% ± 4.9%), whereas mutation of R797 completely blocked membrane fusion induced by trypsin (2.5% ± 0.8%). As shown for Fig. 1, the mutation R667N (S1–S2) had limited fusion activity (48.8% ± 2.9%).
Fig. 3.
Mutation of R797 at SARS-CoV S2′ inhibits trypsin-induced membrane fusion. (A) VeroE6 cells expressing SARS-CoV S wild type (SARS wt) or the S2′ mutants were treated with 2 μg/mL trypsin for 20 min, then incubated for 40 min in the absence of trypsin and analyzed by immunofluorescence microscopy using an anti-S monoclonal antibody (green), with nuclei counterstained with Hoechst 33548 (blue). (Magnification: 200×.) (B) BHK cells, cotransfected with plasmids encoding the SARS-CoV S wild type (SARS wt) or mutants and a plasmid encoding luciferase under the control of the T7 polymerase, were overlaid with VeroE6 cells expressing the T7 polymerase. Cell–cell fusion was induced by treating the cells with 1 μg/mL trypsin for 20 min. At 5 h after fusion induction, the cells were lysed to monitor luciferase activity. Results are presented as relative light units (RLU). Error bars represent the standard error of the mean for 3 independent experiments. Data were analyzed by using an ANOVA test.
SARS-CoV S-Mediated Virus Entry Is Dependent on Cleavage at S2′, as Well as at the S1–S2 Boundary.
To analyze the role of the S1–S2 and S2′ cleavage sites in vivo, in the context of the viral particles we used a murine leukemia virus (MLV)-based pseudotyping system that produces particles that induce luciferase expression upon transduction of permissive cells (30). We produced SARS-CoV S-pseudotyped MLV particles containing the different mutants and analyzed trypsin-induced infection of cells. To increase the transduction efficiency in this system, we used BHK cells coexpressing the SARS-CoV receptor ACE2 (31), as well as DC-SIGN, which has been shown to increase the level of SARS-CoV infection (32). Cells were pretreated for 1 h in presence of NH4Cl to inhibit infection by the endosomal pathway, the pseudotyped virons were bound at the cell surface for 2 h in presence of NH4Cl, and fusion was induced by a short trypsin treatment at 37 °C in the presence of NH4Cl. The cells were then incubated for 30 min in presence of NH4Cl, and luciferase activities were measured after 48 h. The results show that mutation of the dibasic residues K796A, R797N completely inhibited trypsin-primed infection of the pseudoparticles, suggesting that these residues play a key role in SARS-CoV infection induced by trypsin (Fig. 4; see also Fig. S4). As expected, the effect of mutation K796A was weak (84.8% ± 14% of the wild-type level), but mutation of R797 induced a strong decrease of infection induced by trypsin (3.8% ± 2.3% of the wild-type level). Surprisingly, whereas mutation R667N decreased cell–cell fusion induced by trypsin by ≈50%, infection recovery induced by trypsin was strongly impaired (21.4% ± 10.6% of the wild type). This suggests that in the context of the virus infection, cleavage at the S1–S2 boundary may play a more important role than in cell–cell fusion. To further confirm the role of the S1–S2 cleavage in the context of trypsin-induced infection, we generated pseudoparticles containing the furin667 mutation and analyzed trypsin-induced infection of cells. The results show that the infection recovered by this mutant was substantially higher than infection recovered for the wild-type protein (216% ± 78%). We also assessed the effect of the R797N mutation on virus entry in the absence of endosomal acidification. This mutant showed no effect on transduction in the absence of NH4Cl and trypsin (Fig. S5), indicating that R797 is not a critical residue for the endosomal route of SARS-CoV entry, which is presumably primed by cathepsin L cleavage.
Fig. 4.
Role of S1/S2 and S2′ cleavage in virus entry mediated by SARS-CoV S. BHK cells coexpressing ACE2 and DC-SIGN were pretreated for 1 h with 25 mM NH4Cl. Virions pseudotyped with the SARS-CoV S wild type (SARS wt) or the different mutants were bound at 4 °C for 2 h in the presence of drug. Viral entry was induced by a 5-min treatment of the cells at 37 °C with trypsin. The results are expressed as the percentage of infection recovery obtained for the wild type. Error bars represent the standard error of the mean for 3 independent experiments. Data were analyzed by using a Student's t test.
Discussion
Triggering of membrane fusion by viral fusion proteins is often preceded by a critical proteolytic priming event. We show here a novel mechanism of priming for the SARS-CoV S protein, based on sequential proteolytic cleavage at 2 distinct sites. Class I viral fusion proteins typically undergo a single cleavage event that primes the fusion machine (1); however, the respiratory syncytial virus (RSV) F protein is known to be cleaved twice, with both cleavage events being necessary for full fusion activity in vivo (33, 34). In the case of RSV, however, the 2 events appear to occur independently, which is in contrast to the mechanism of SARS-CoV S fusion presented here, which suggests that cleavage is a sequential event—with cleavage at the S1/S2 boundary promoting subsequent cleavage at the S2′ position, which in turn leads to the triggering of membrane fusion.
Our original rationale for cleavage at the CoV S2′ position was based on the efficient furin-based cleavage at this location of the IBV spike protein. Our bioinformatic studies (Fig. S1) suggested that instead of the efficient cleavage occurring with IBV S, there was the possibility of a cryptic, poorly used cleavage site at the S2′ position of wild-type SARS-CoV S. Even with the introduction of a consensus furin site, only a relatively low fraction of the SARS-CoV S protein was cleaved in our assays, suggesting the site is less exposed in the 3-dimensional structure of SARS-CoV S compared with IBV. However, this small fraction of cleaved SARS-CoV S is sufficient to prime the fusion event. This raises the general suggestion that not all spike proteins on a coronavirus particle are necessarily cleaved identically; i.e., there is a heterogeneous population of cleaved and uncleaved spikes. It is also possible that the dibasic site at SARS-CoV S2′ is a recognition site for other members of the proprotein convertase family. Many cell types express these furin-related enzymes, which typically are less stringent in their requirements for basic residues, having the general consensus BxB, where x = 0, 2, or 4 residues (35–37). Indeed, the KR sequence at S2′ has previously been considered to a possible cleavage site for SARS-CoV S (12). Although S2′ cleavage is not readily apparent for wild-type S, the use of proprotein convertase family members by SARS-CoV remains a possibility and will be explored in future studies.
The experiments reported here explore SARS-CoV S-mediated membrane fusion by furin or trypsin, presumably at the cell surface or during virus assembly. However, SARS-CoV infection is known to be sensitive to inhibitors of endosomal acidification (13–15), via the action of the endosomal protease cathepsin L (18, 19). As expected, infection of cells by virus particles pseudotyped with the wild-type SARS-CoV spike was sensitive to inhibitors of endosomal acidification and cathepsin L. However, none of the point mutants described in this paper showed an effect on virus entry in the absence of endosomal acidification or cathepsin L inhibitors (Fig. S5). This may be due to the relatively indiscriminate recognition site of cathepsin L, compared with trypsin or furin (37)—leading to the suggestion that either cathepsin L is not involved in S2′ cleavage, or that cathepsin-based cleavage can still occur at satellite sites in the S2′ region. Based on the trypsin-mediated activation of membrane fusion shown here, our data suggest that R797 is an important site of S processing. However SARS-CoV S has primarily respiratory tropism, and so we suggest that in vivo, a trypsin-like serine protease localized to the respiratory tract may cleave SARS-CoV S, as is the case for influenza virus (38). Based on the situation with many other class I viral fusion proteins, it is possible that cleavage at S2′ exposes a viral fusion peptide. This will be explored in future experiments.
In the case of bovine respiratory syncytial virus F, the 2 furin-mediated cleavage events lead to the release of a 27-aa peptide, termed virokinin, that is a bioactive peptide of the tachykinin family and an important pathogenicity factor (39, 40). In the case of Ebola virus GP, cleavage by cathepsins B and L results in the shedding of an extensive part of the fusion protein, which is important for both attachment and fusion (41–44). It remains possible that the domain between S1–S2 and S2′ is released from the trimeric S ectodomain upon protease action.
One possibility for our mutants where furin sites were introduced at S1–S2 and/or S2′ is that we serendipitously created a basic heparin sulfate-binding site, and so we cannot rule out the possibility of cross-talk between heparin sulfate-binding and furin cleavage for SARS-CoV S, as previously suggested for other viruses (45, 46, 47).
While this work was in progress, Taguchi and coworkers (48) generated a SARS-CoV S construct with a furin site at the S2′ position (793KPTKR797 to 793KRRKR797) and have shown SARS-CoV S activation at the cell surface in a trypsin-independent manner. This work is in general agreement with the data presented here and highlights the importance of residue R797 in the context of SARS-CoV infection. Multiple sequence alignments show that this arginine residue is extremely well-conserved across all coronaviruses. This raises the possibility that cleavage in the S2′ region is a common mechanism for fusion activation across the Coronaviridae. In support of this is the finding that a neutralizing epitope for the anti-MHV antibody 5B19, which is known to block fusion activation, binds at the S2′ position of the MHV spike (49). It has also become apparent that cleavage at the S1/S2 boundary can occur in group 1 coronaviruses, which were previously thought to be uncleaved (9, 47). Thus, the mechanism of fusion activation by sequential cleavage at 2 distinct sites revealed here for SARS-CoV may be applicable to all coronaviruses. In the case of paramyxoviruses, insertion of the 2 furin cleavage sites of the RSV F protein into the Sendai virus F protein led to both enhanced cell–cell fusion and a decreased dependency on the HN attachment protein for activity (50). It will be interesting to explore the possibility that modulation of the 2 coronavirus spike cleavage sites might reduce the reliance on species-specific receptors and play a role in host range or viral pathogenesis.
Materials and Methods
Cell–Cell Fusion Assays.
For visualization of syncytia, VeroE6 cells were grown on coverslips in 24-well plates and transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After 24 h, cells were treated for 20 min in serum-free medium containing various concentrations of trypsin to induce fusion. Medium was replaced by medium containing 10% FBS, and the cells were incubated for 40 min, fixed with 3% paraformaldehyde, and processed for indirect immunofluorescence essentially as described previously (51) by using the monoclonal anti-SARS-CoV S antibody 341CD (National Institutes of Health Biodefense and Emerging Infections Research Resources Repository, Manassas, VA). Images were captured by using a Nikon E600 microscope with a 20× Plan Apo objective (numerical aperture, 0.75) and equipped with a Sensicam EM camera (Cooke Corp.), as well as IPLab software (Scanalytics).
For syncytia formation quantification of the mutants containing furin cleavages sites, cells were fixed without any protease treatment. The percentage of cells involved in syncytia formation was determined by counting the number of cells containing 2 or more nuclei, compared with the number of total nuclei in transfected cells. At least 150 cells were counted for each mutant, and the experiment was repeated 3 times.
For quantitative cell–cell fusion assays based on luciferase expression, BHK cells were grown in 24-well plates and cotransfected with wild-type or mutant SARS-CoV S-expressing plasmids, as well as a plasmid expressing luciferase under the control of a T7 promoter. VeroE6 cells grown in a 60-mm dish were also transfected with a plasmid encoding the T7 polymerase. After 24 h, transfected BHK cells were overlaid with transfected VeroE6 cells and cocultured for 3 h. Cells were then incubated for 20 min in serum-free medium containing various concentrations of trypsin to induce fusion. The medium was replaced with fresh medium containing 10% FBS, and the cells were incubated for 6 h before lysis. Luciferase activity was detected by using a Luciferase Assay Kit (Promega), and light emission was measured by using a Glomax 20/20 luminometer (Promega).
Production of Pseudotyped Virions.
Pseudotyped virions were produced essentially as described previously (30), with plasmids for pseudotyping kindly provided by Jean Dubuisson (Lille Pasteur Institute, Lille, France). Briefly, 293T cells were cotransfected with an MLV-based transfer vector encoding luciferase, an MLV Gag-Pol packaging construct, and an envelope glycoprotein-expressing vector (pcDNA3.1-SARS-CoV S) by using Exgen 500 (Fermentas), as recommended by the manufacturer. Cells were incubated at 32 °C for 72 h, and after harvesting the supernatants were filtered through 0.45-μm pore-sized membranes.
For transduction of pseudoparticles, BHK cells were first cotransfected with equal amounts of plasmid pcDNA3.1(-)/ACE2 (kindly provided by Michael Farzan, New England Primate Research Center, Southborough, MA) and pcDNA3-DC-SIGN (National Institutes of Health AIDS Research & Reference Reagent Program, Germantown, MD). Cells were preincubated for 1 h in presence of 25 mM NH4Cl at 37 °C to inhibit virus entry through the endosomal route, and then were transferred to ice. Pseudotyped virions were bound at 4 °C for 2 h in RPMI containing 0.2% BSA, 20 mM Hepes, and 25 mM NH4Cl. Cells were then warmed by addition of RPMI containing 3 μg/mL trypsin, 0.2% BSA, 20 mM Hepes, and 25 mM NH4Cl and were incubated for 5 min in a water bath at 37 °C. The cells were then incubated for 30 min in complete medium containing 25 mM, and the medium was replaced. Luciferase activity was detected by using a Luciferase Assay Kit (Promega) and light emission measured by using a Glomax 20/20 luminometer (Promega).
For additional information see SI Methods.
Supplementary Material
Acknowledgments.
We are indebted to the late Beverley Bauman for her encouragement and support during the early part of this work. We also thank Dr. Michael Farzan and Dr. Jean Dubuisson for providing plasmids, A. Damon Ferguson for technical assistance, Xiangjie Sun for helpful discussions, Ruth Collins for helpful advice, and Andrew Regan for critical reading of the manuscript. This work was supported in part by the Cornell University Nanobiotechnology Center (NBTC), an STC program of the National Science Foundation under agreement number ECS-987677, and by National Institutes of Health Grant R03 AI060946.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809524106/DCSupplemental.
References
- 1.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 4.Lamb RA, Jardetzky TS. Structural basis of viral invasion: Lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai AL, Li Y, Tamm LK. In: Protein–Lipid Interactions. Tamm LK, editor. Weinheim, Germany: Wiley-VCH; 2005. pp. 279–303. [Google Scholar]
- 6.Klenk HD, Matrosovich M, Stech J. In: Animal Viruses: Molecular Biology. Mettenleiter TC, Sobrino F, editors. Norfolk, UK: Caister Academic; 2008. pp. 253–303. [Google Scholar]
- 7.Klenk HD, Garten W. In: Cellular Receptors for Animal Viruses. Wimmer E, editor. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1994. pp. 241–280. [Google Scholar]
- 8.Wentworth DE, Holmes KV. In: Coronaviruses: Molecular and Cellular Biology. Thiel V, editor. Norfolk, UK: Caister Academic; 2007. pp. 3–31. [Google Scholar]
- 9.Bosch BJ, Rottier PJ. In: Nidoviruses. Perlman S, Gallagher T, Snijder EJ, editors. Washington, DC: ASM Press; 2008. pp. 157–178. [Google Scholar]
- 10.Song HC, et al. Synthesis and characterization of a native, oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J Virol. 2004;78:10328–10335. doi: 10.1128/JVI.78.19.10328-10335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao X, et al. The SARS-CoV S glycoprotein: Expression and functional characterization. Biochem Biophys Res Commun. 2003;312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergeron E, et al. Implication of proprotein convertases in the processing and spread of severe acute respiratory syndrome coronavirus. Biochem Biophys Res Commun. 2005;326:554–563. doi: 10.1016/j.bbrc.2004.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofmann H, et al. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J Virol. 2004;78:6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simmons G, et al. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci USA. 2004;101:4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang ZY, et al. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J Virol. 2004;78:5642–5650. doi: 10.1128/JVI.78.11.5642-5650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ujike M, et al. Heptad repeat-derived peptides block the protease-mediated direct entry from cell surface of SARS coronavirus but not entry via endosomal pathway. J Virol. 2007;82:588–592. doi: 10.1128/JVI.01697-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuyama S, et al. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc Natl Acad Sci USA. 2005;102:12543–12547. doi: 10.1073/pnas.0503203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang IC, et al. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J Biol Chem. 2005;10:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simmons G, et al. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao YX, et al. Cleavage and serum reactivity of the severe acute respiratory syndrome coronavirus spike protein. J Infect Dis. 2004;190:91–98. doi: 10.1086/421280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du L, et al. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochem Biophys Res Commun. 2007;359:174–179. doi: 10.1016/j.bbrc.2007.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F, et al. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J Virol. 2006;80:6794–6800. doi: 10.1128/JVI.02744-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosch BJ, Bartelink W, Rottier PJ. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J Virol. 2008;82:8887–8890. doi: 10.1128/JVI.00415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simmons G, Rennekamp AJ, Bates P. Proteolysis of SARS-associated coronavirus spike glycoprotein. Adv Exp Med Biol. 2006;581:235–240. doi: 10.1007/978-0-387-33012-9_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klenk HD, Garten W. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 1994;2:39–43. doi: 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 26.Cavanagh D, et al. Coronavirus IBV: Partial amino terminal sequencing of spike polypeptide S2 identifies the sequence Arg-Arg-Phe-Arg-Arg at the cleavage site of the spike precursor propolypeptide of IBV strains Beaudette and M41. Virus Res. 1986;4:133–143. doi: 10.1016/0168-1702(86)90037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. 2007;38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 28.Karaca K, Naqi S, Gelb J., Jr Production and characterization of monoclonal antibodies to three infectious bronchitis virus serotypes. Avian Dis. 1992;36:903–915. [PubMed] [Google Scholar]
- 29.Follis KE, York J, Nunberg JH. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology. 2006;350:358–369. doi: 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J Exp Med. 2003;197:633–642. [Google Scholar]
- 31.Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han DP, Lohani M, Cho MW. Specific asparagine-linked glycosylation sites are critical for DC-SIGN- and L-SIGN-mediated severe acute respiratory syndrome coronavirus entry. J Virol. 2007;81:12029–12039. doi: 10.1128/JVI.00315-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez-Reyes L, et al. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc Natl Acad Sci USA. 2001;98:9859–9864. doi: 10.1073/pnas.151098198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zimmer G, Budz L, Herrler G. Proteolytic activation of respiratory syncytial virus fusion protein. Cleavage at two furin consensus sequences. J Biol Chem. 2001;276:31642–31650. doi: 10.1074/jbc.M102633200. [DOI] [PubMed] [Google Scholar]
- 35.Seidah NG, et al. The activation and physiological functions of the proprotein convertases. Int J Biochem Cell Biol. 2008;40:1111–1125. doi: 10.1016/j.biocel.2008.01.030. [DOI] [PubMed] [Google Scholar]
- 36.Seidah NG, Day R, Marcinkiewicz M, Chretien M. Mammalian paired basic amino acid convertases of prohormones and proproteins. Ann NY Acad Sci. 1993;680:135–146. doi: 10.1111/j.1749-6632.1993.tb19680.x. [DOI] [PubMed] [Google Scholar]
- 37.Barrett AJ, Rawlings ND, Woessner JF. Handbook of Proteolytic Enzymes. London: Elsevier Academic; 2004. [Google Scholar]
- 38.Kido H, et al. Cellular proteinases trigger the infectivity of the influenza A and Sendai viruses. Mol Cells. 1999;9:235–244. [PubMed] [Google Scholar]
- 39.Valarcher JF, et al. Bovine respiratory syncytial virus lacking the virokinin or with a mutation in furin cleavage site RA(R/K)R109 induces less pulmonary inflammation without impeding the induction of protective immunity in calves. J Gen Virol. 2006;87:1659–1667. doi: 10.1099/vir.0.81755-0. [DOI] [PubMed] [Google Scholar]
- 40.Zimmer G, et al. Virokinin, a bioactive peptide of the tachykinin family, is released from the fusion protein of bovine respiratory syncytial virus. J Biol Chem. 2003;278:46854–46861. doi: 10.1074/jbc.M306949200. [DOI] [PubMed] [Google Scholar]
- 41.Lee JE, et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaletsky RL, Simmons G, Bates P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J Virol. 2007;81:13378–13384. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schornberg K, et al. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandran K, et al. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pasquato A, et al. Heparin enhances the furin cleavage of HIV-1 gp160 peptides. FEBS Lett. 2007;581:5807–5813. doi: 10.1016/j.febslet.2007.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Haan CA, et al. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J Virol. 2005;79:14451–14456. doi: 10.1128/JVI.79.22.14451-14456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Haan CA, et al. Cleavage of group 1 coronavirus spike proteins: How furin cleavage is traded off against heparan sulfate binding upon cell culture adaptation. J Virol. 2008;82:6078–6083. doi: 10.1128/JVI.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe R, et al. Entry from cell surface of SARS coronavirus with cleaved S protein as revealed by pseudotype virus bearing cleaved S protein. J Virol. 2008;82:11985–11991. doi: 10.1128/JVI.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taguchi F, Shimazaki YK. Functional analysis of an epitope in the S2 subunit of the murine coronavirus spike protein: Involvement in fusion activity. J Gen Virol. 2000;81:2867–2871. doi: 10.1099/0022-1317-81-12-2867. [DOI] [PubMed] [Google Scholar]
- 50.Rawling J, Garcia-Barreno B, Melero JA. Insertion of the two cleavage sites of the respiratory syncytial virus fusion protein in Sendai virus fusion protein leads to enhanced cell-cell fusion and a decreased dependency on the HN attachment protein for activity. J Virol. 2008;82:5986–5998. doi: 10.1128/JVI.00078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu VC, et al. The avian coronavirus infectious bronchitis virus undergoes direct low-pH-dependent fusion activation during entry into host cells. J Virol. 2006;80:3180–3188. doi: 10.1128/JVI.80.7.3180-3188.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.