Abstract
HdeA has been shown to prevent acid-induced aggregation of proteins. With a mass of only 9.7 kDa, HdeA is one of the smallest chaperones known. Unlike other molecular chaperones, which are typically complex, multimeric ATP-dependent machines, HdeA is known to undergo an acid-induced dimer to monomer transition and functions at low pH as a disordered monomer without the need for energy factors. Thus, HdeA must possess features that allow it to bind substrates and regulate substrate affinity in a small and energy-independent package. To understand better how HdeA accomplishes this, we studied the conformational changes that accompany a shift to low pH and substrate binding. We find that the acid-induced partial unfolding and monomerization that lead to HdeA activation occur very rapidly (k >3.5 s−1). Activation exposes the hydrophobic dimer interface, which we found to be critical for substrate binding. We show by intramolecular FRET that the partially unfolded character of active HdeA allows the chaperone to adopt different conformations as required for the recognition and high-affinity binding of different substrate proteins. These efficient adaptations help to explain how a very small protein is rapidly activated and can bind a broad range of substrate proteins in a purely pH-regulated manner.
Keywords: HdeA, periplasm, posttranslational regulation
Bacteria possess the ability to respond to a changing environment by inducing transcriptional stress responses that defend against stress conditions. Although these stress responses are highly effective once fully operational, there is a significant time lag between the moment when the stress is first sensed and when defense and repair proteins are expressed (1). This lag creates a window during which cellular damage can occur. In contrast, posttranslational regulation of stress-related proteins eliminates this time lag and thus provides a much more rapid response to stress conditions. Two well-characterized examples of stress-related proteins that are activated at the posttranslational level are Hsp33 and the small heat shock protein Hsp26. Hsp33 is a redox-regulated chaperone specifically activated by oxidative stress conditions that lead to protein unfolding (2). Hsp26 is directly activated in response to elevated temperatures (3).
The major advantage of posttranslational regulation is the speed with which it occurs. However, it also wastes energy to produce and maintain inactive proteins. Thus, posttranslational regulation appears to be limited to potentially fatal stress conditions, which arise frequently and require an instantaneous response in order for the organism to survive. One such condition is acid stress, which is encountered by bacteria immediately after their ingestion when they enter the low-pH environment (pH 1–3) of the mammalian stomach (4). This poses a significant challenge to bacterial survival (5). In response, enteric bacteria have developed efficient protective systems that help them to survive the extreme pH of the mammalian stomach. Two of these systems are the highly effective glutamate- and arginine-dependent decarboxylases, which maintain a near neutral pH in the cytoplasm even during acid stress (6, 7). The pH of the periplasm, in contrast, equilibrates rapidly with the extracellular pH because of the porous nature of the outer membrane, which allows free diffusion of molecules smaller than 600 Da (8). Acidic pH conditions are known to cause protein unfolding (9), and this has been shown to cause periplasmic proteins to aggregate (10, 11). To combat acid-induced protein unfolding and aggregation in the periplasm, many bacteria express the small protein HdeA, which specifically senses low pH conditions (pH <3) where it becomes active as a chaperone (10, 12). This protein belongs to a class of stress-specific chaperones that are activated by the very stress conditions that they combat.
At neutral pH, HdeA exists as an inactive dimer. Once exposed to low pH conditions, however, HdeA has been shown to dissociate into chaperone-active monomers (10, 12). The kinetics of activation have not yet been studied, although this activation appears to be crucial for the protection of bacteria against low pH conditions; indeed, hdeA deletion strains of Escherichia coli and Shigella are extremely acid-sensitive (12, 13). Although the structure of the inactive dimeric HdeA from E. coli has been solved by X-ray crystallography (12), limited information is available on the structure, regulation, and substrate-binding activity of the active monomeric form of HdeA. Based on the structure of inactive HdeA, it has been proposed that the hydrophobic dimer interface may contribute to substrate binding in active HdeA monomers (12), although to date there is limited experimental evidence to support this. HdeA is known to unfold extensively at pH <3, and this has been proposed to transform HdeA into a globally disordered amphiphilic molecule, which possesses a hydrophobic substrate-binding surface and a highly charged region that aids in solubility of the chaperone–substrate complex (10). Disruption of this amphiphilic character, either by substituting charged amino acids for hydrophobic residues (V52K/V58K) or by deleting the highly charged N-terminal region of the protein, inhibits chaperone activity in vitro and acid resistance in vivo (14). The V52K/V58K mutant was shown to exhibit decreased substrate interactions (14). V52 resides at the periphery of the dimer interface of HdeA, whereas V58 is buried in the hydrophobic core of the monomer and is not near the dimer interface. Because a double mutant was used for this study, it is not possible to ascertain the relative contributions of each residue, and therefore the contribution of the hydrophobic dimer interface to substrate binding remains unknown. Moreover, it remains unclear how a “globally disordered” HdeA can recognize substrate proteins and what role structural disorder may play in chaperone function.
Here, we demonstrate that low pH very rapidly converts HdeA into its monomeric, chaperone-active conformation that, in contrast to previous reports, is only partially unfolded. This pH-induced dissociation exposes the hydrophobic residues of the HdeA dimer interface, which then serve as sites for unfolded substrate proteins to bind to the active monomer. Our intramolecular Förster resonance energy transfer (FRET) data suggest that the partially unfolded conformational ensemble of active HdeA allows the chaperone adaptively to bind substrates such that the conformation of HdeA can be molded to accommodate the structure of the substrate protein. To our knowledge, HdeA is the first molecular chaperone that utilizes its dimer interface for substrate binding and whose conformational flexibility plays such a critical role in the molecular recognition of different substrate proteins.
Results
Rapid Activation Makes HdeA Ideally Suited to Protect Proteins Against Rapid pH-Induced Aggregation.
HdeA has been shown to be an inactive dimer at neutral pH, but it becomes monomeric and chaperone-active at pH <3 (10, 12). Its activation has been proposed to involve transformation into a globally disordered conformation at low pH, which exposes hydrophobic surfaces that are likely to be involved in substrate binding (10). By using a combination of circular dichroism (CD) and fluorescence measurements, we found that HdeA is only partially unfolded by acid treatment, and it retains significant secondary structure but little tertiary structure in its active form [supporting information (SI) Fig. S1]. The C18–C66 disulfide bond in HdeA is critical for maintaining this residual structure, and reduction of this disulfide leads to a completely unfolded protein with no chaperone activity (Fig. S1).
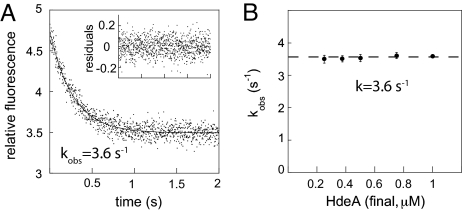
To determine how quickly HdeA undergoes conformational changes that convert a fully folded yet inactive protein into a partially unfolded highly active chaperone, we used stopped-flow fluorescence measurements. We made use of the pH-induced changes in HdeA tryptophan fluorescence, which include a change in the emission maximum (λmax), and a substantial decrease in fluorescence intensity (Fig. S1B). This fluorescence decrease can be used as a quantitative measure to assess the rate of pH-induced conformational changes in HdeA. As shown in Fig. 1A, we found that the decrease in HdeA fluorescence upon acid-induced partial unfolding occurs very rapidly (kobs = 3.6 s−1) and followed kinetics that were well described by a single exponential function. Variation of the HdeA concentration did not alter the rate of the reaction (Fig. 1B), indicating that we were monitoring intramolecular conformational rearrangements of HdeA.
Fig. 1.
HdeA rapidly changes conformation upon exposure to low pH. (A) Representative stopped-flow kinetic trace showing changes in intrinsic tryptophan fluorescence of 1 μM HdeA upon a rapid shift from pH 7 to pH 2.2. (Inset) Residuals for a single exponential fit. (B) Plot of kobs over a range of HdeA concentrations. The dashed line denotes the average value of 3.6 s−1.
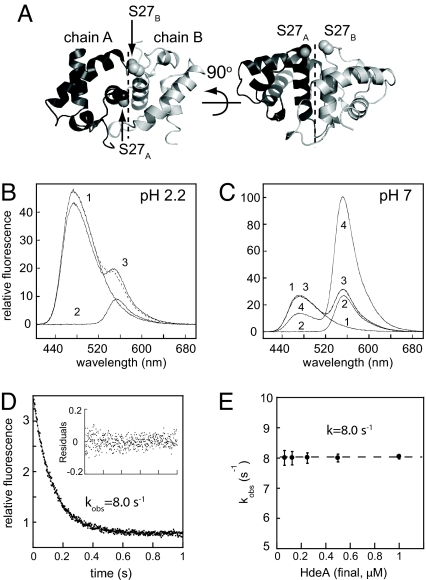
Two tryptophan residues (W16 and W82) are responsible for the intrinsic fluorescence of HdeA, and neither residue makes contact with an adjacent monomer at the dimer interface (12). Changes in intrinsic HdeA fluorescence are therefore most likely reporting on conformational changes within each monomer rather than changes in the HdeA oligomeric state (i.e., monomer ↔ dimer transitions). To monitor the oligomeric state of HdeA, we engineered the mutant HdeA(C27), which introduces a unique reactive cysteine near the dimer interface (Fig. 2A). The α-carbons of S27 in adjacent monomers are only 8.5 Å apart. This distance should result in a robust FRET signal when appropriate labels are attached at this position in the dimer and should allow us to monitor the rate of monomerization by following the acid-induced decrease in FRET. The dyes that we selected for these experiments are bimane and Alexa Fluor 532 (AF532), which are ideal for this purpose because their fluorescence is relatively insensitive to pH variations, and they exhibit substantial spectral overlap. Bimane served as the donor (exmax 390 nm, emmax 475 nm) and AF532 as the acceptor (exmax 532 nm, emmax 550 nm).
Fig. 2.
The oligomeric state of HdeA can be monitored by FRET. (A) Ribbon representation of the HdeA dimer [Protein Data Bank (PDB) ID code 1DJ8; chains A and B from reference 12] showing the position of S27, which was mutated to cysteine to provide a fluorescence-labeling site. (B) Fluorescence emission spectra of 0.5 μM bimane-labeled HdeA(C27) (trace 1), AF532-labeled HdeA(C27) (trace 2), or a 1:1 mixture of both (trace 3) recorded at pH 2.2 (λex = 390 nm). The sum of spectra 1 and 2 are shown as a dashed line. (C) Fluorescence emission spectra of 0.5 μM bimane-labeled HdeA(C27) (trace 1), AF532-labeled HdeA(C27) (trace 2), or a 1:1 mixture of both recorded at pH 7 (trace 3), or a 1:1 mixture of both mixed at pH 2.2 and then neutralized (trace 4). The sum of spectra 1 and 2 are shown as a dashed line, which fits nearly perfectly to trace 3 and is therefore not clearly visible. (D) Kinetics of pH-induced dimer dissociation as measured by a decrease in intermolecular FRET. Bimane- and AF532-labeled HdeA(C27) dimers were prepared by incubating 1:1 mixtures at low pH and subsequently diluting to a final concentration of 0.5 μM in pH 7 buffer. Dissociation of the dimers was then induced by rapidly shifting back to low pH by using a stopped-flow device. A representative trace is shown. (Inset) Residuals for a single exponential fit. (E) Plot of kobs over a range of HdeA concentrations. The dashed line denotes the average value of 8.0 s−1.
When we mixed equimolar amounts of bimane-labeled HdeA(C27) and AF532-labeled HdeA(C27) at either pH 2.2 (Fig. 2B) or pH 7.0 (Fig. 2C), we saw little evidence of energy transfer. The sum of the spectra (dashed trace) of the two individually labeled proteins (traces 1 and 2) was very similar to the spectrum of the mixture (trace 3). This result confirmed that at low pH HdeA is predominantly monomeric and does not interact with other monomers, whereas at neutral pH HdeA dimers are tightly associated and do not readily exchange subunits, consistent with the published Kd of ≈200 nM at pH 7 (12). When we incubated bimane- and AF532-labeled HdeA(C27) at a 1:1 ratio at pH 2.2 and then neutralized the mixture, however, we observed a significant decrease in bimane emission coupled with a large increase in AF532 emission (Fig. 2C, trace 4). This substantial degree of energy transfer from bimane to AF532 clearly indicates dimer formation upon neutralization. Addition of excess unlabeled HdeA decreased the FRET signal very slowly, with a half-time of ≈120 min, indicating that the mixed labeled HdeApH7.0 dimers are very stable.
These constructs provided us with a spectroscopic handle for monitoring the rate of HdeA pH-induced dissociation. We prepared mixed dimers of HdeA(C27)-bimane and HdeA(C27)-AF532, which exhibit high FRET efficiency, and subsequently monitored the decrease in FRET upon a rapid shift from pH 7.0 to 2.2. As was the case for the conformational changes monitored by tryptophan fluorescence, we found that pH-induced dissociation follows single-exponential kinetics with a rate of 8.0 s−1. This rate constant is also independent of the HdeA concentration (Fig. 2 D and E) but is somewhat faster than the rate determined for pH-induced conformational changes (Fig. 1). This slight discrepancy may be the result of the presence of the fluorescent probes near the dimer interface, which may increase the rate of dissociation.
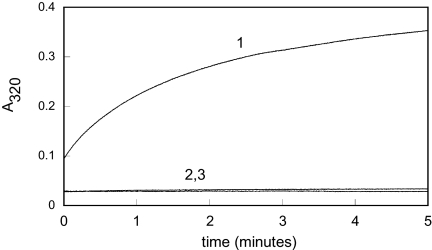
To assess whether the rapid conformational changes and monomerization correspond to the rate of chaperone activation, we compared the chaperone function of HdeA after a short-term (≈2 s) or long-term exposure (10 min) to low pH. We found that upon dilution into low pH buffer, chemically denatured rhodanese undergoes substantial aggregation (initial absorbance ≈0.1; Fig. 3, trace 1) within the time it takes to mix the components and begin the measurement (≈10 s). Importantly, this initial aggregation was suppressed in the presence of fully activated HdeA (initial absorbance ≈0.03; Fig. 3, trace 2). This significant difference in the initial absorbance values allowed us to determine whether a 2-s exposure of HdeA to pH 2.2 is sufficient to activate HdeA fully and to prevent the fast initial aggregation of rhodanese. No difference was observed between HdeA exposed to low pH for 2 s (Fig. 3, trace 3) or 10 min (Fig. 3, trace 2). In both cases, the initial absorbance was found to be near zero, indicating that no significant aggregation of rhodanese occurred even during the first few seconds after mixing, which indicates fast formation of HdeA–substrate complexes. These results clearly demonstrate that the rapid pH-induced unfolding and monomerization (as shown in Figs. 1 and 2) fully activate the HdeA chaperone function. This rapid activation makes HdeA ideally suited to bind proteins as they themselves unfold in response to acid, thereby preventing their aggregation.
Fig. 3.
Rapid conformational changes reflect the rate of chaperone activation. Rhodanese (4 μM) aggregation was monitored by apparent absorbance at 320 nm caused by light scatter in the absence (trace 1) or presence of 4 μM HdeA that was either exposed to low pH for 10 min (trace 2) or 2 s (trace 3) before mixing with Gdn-denatured rhodanese. Assays were performed in 100 mM potassium phosphate, 150 mM NaCl (pH 2.2) at 37 °C.
HdeA Hydrophobic Dimer Interface Serves a Second Role: Substrate Binding.
The data presented thus far suggest that conformational changes, monomerization, and functional activation of HdeA are tightly coupled and in all likelihood occur simultaneously. Further evidence for this correlation came from pH titration experiments in which we lowered the pH in a stepwise manner from neutral to pH 2 while monitoring the ability of HdeA to bind the hydrophobic dye, 4,4′-bis(1-anilinonaphthalene 8-sulfonate) (bis-ANS) and using FRET in a parallel experiment to monitor the oligomeric state of HdeA. We found that the midpoints of the pH titration for both bis-ANS binding and monomerization are very similar (Fig. S2), indicating that monomerization and the exposure of hydrophobic surfaces that are likely to be involved in substrate binding occur in parallel.
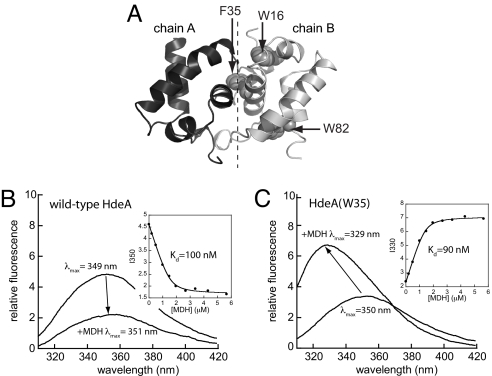
As has been noted in ref. 12, the surface of dimeric HdeA is highly charged, whereas the buried dimer interface is largely hydrophobic (see also Fig. S2). Because most molecular chaperones bind unfolded substrate proteins by hydrophobic interactions, this observation, in combination with our kinetic data, raises the intriguing possibility that the hydrophobic interface that promotes dimerization at neutral pH might also contribute to substrate binding at low pH. To test this hypothesis experimentally, we substituted the two intrinsic tryptophan residues of HdeA with nonfluorescent residues (W16F and W82L) and introduced a unique tryptophan residue at position 35, which makes extensive contacts with the adjacent HdeA monomer at the dimer interface (Fig. 4A; for nomenclature used for HdeA mutants, see Table S1). This tryptophan residue serves as a probe for direct monitoring of the solvent accessibility at this position in the absence and presence of bound substrate. To monitor the effect of substrate binding on HdeA tryptophan fluorescence, we used the tryptophan-free protein malate dehydrogenase (MDH), which we found to be an excellent model substrate for HdeA (Fig. S3).
Fig. 4.
Hydrophobic amino acids at the dimer interface of inactive HdeA contribute to substrate binding in active HdeA monomers. (A) Backbone representation of the HdeA dimer (PDB ID code 1DJ8; chains A and B from ref. 12) indicating positions of the two intrinsic tryptophan residues (W16 and W82), and F35, which was used to introduce a unique tryptophan residue at the dimer interface. Emission spectra (λex = 295 nm) of 1 μM wild-type HdeA (B) or HdeA(W35) (C) were recorded in the absence or presence of 2 μM MDH. Spectra were recorded in 100 mM potassium phosphate, 150 mM NaCl (pH 2.2) at 37 °C. (Insets) Peak intensity versus MDH concentration, which were fit to determine apparent Kd values (see see SI Materials and Methods).
Binding of MDH to wild-type HdeA resulted in only a small (2 nm) red shift of the emission maximum (Fig. 4B). This result indicates that the degree of solvent exposure of the two naturally occurring tryptophan residues, which do not make contacts at the dimer interface, does not significantly change upon substrate binding, implying that these residues are not directly involved in substrate binding. Plotting the fluorescence intensity versus MDH concentration enabled us to determine that HdeA and MDH form a tight complex (Kd = 100 nM; Fig. 4B Inset). Single deletions of either W16 or W82 revealed that the modest quenching effect of MDH is caused almost entirely by interactions with W16 (Fig. S4).
In contrast to wild-type HdeA, HdeA(W35), which contains a unique tryptophan residue at the dimer interface, showed a 21-nm blue shift in λmax upon MDH binding (Fig. 4C). These results strongly suggest that W35, which is maximally solvent-exposed in monomeric HdeApH2.2 (λmax = 350 nm), becomes buried and shielded from solvent upon MDH binding (λmax = 329 nm). Further support that W35 becomes buried upon MDH binding comes from potassium iodide (KI) quenching experiments. Iodide is a strong collisional quencher of tryptophan fluorescence, and KI quenching efficiency correlates well with tryptophan accessibility (15). We found that KI effectively quenched the fluorescence of wild-type HdeA, regardless of whether MDH was bound. In contrast, KI quenching of HdeA(W35) was greatly reduced in the presence of bound MDH (Fig. S5), indicating that MDH binding effectively shields W35.
Additional evidence for the direct involvement of the HdeA dimer interface in substrate binding comes from HdeA(F35K), in which we introduced a charged amino acid into the normally hydrophobic dimer interface. This mutant exhibited a 40-fold decrease in affinity for MDH and severely impaired chaperone-like activity toward both MDH and rhodanese (Fig. S6).
Many small heat shock proteins form large chaperone oligomers (16). To test the possibility that substrate binding induces oligomerization of HdeA, which could similarly affect HdeA tryptophan fluorescence, we used our FRET system. We were unable to detect any evidence of energy transfer between mixtures of bimane- and AF532-labeled HdeA(C27) in the presence of excess MDH, suggesting that HdeA remains monomeric upon binding substrate (Fig. S7). These results support our conclusion that the hydrophobic residues that form the dimer interface in inactive HdeApH7.0 serve to bind substrate in active HdeApH2.2, thus giving them a dual role.
HdeA Adopts Different Conformations to Accommodate Different Substrates.
Activated HdeA exhibits many of the characteristics of an intrinsically disordered protein (10) (see also Fig. S1). This observation raises intriguing questions about the nature of interactions between a partially unfolded chaperone and its partially or largely unfolded substrate proteins. To determine how substrate binding affects HdeA conformation, we conducted intramolecular FRET experiments in which we determined distances between a specific donor and acceptor position within the HdeA monomer in the absence and presence of bound substrate proteins. We used W82 as donor because its fluorescence is unaffected by substrate binding (Fig. S4 D–F). We used C27, which lies ≈20 Å from the W82 donor (12) to attach the nonfluorescent thionitrobenzoic acid (TNB) acceptor via a reversible disulfide. Simply adding reductants such as DTT cleaves the TNB acceptor and thereby enables the facile measurement of donor fluorescence in the presence or absence of acceptor under otherwise identical conditions. This, along with their extensive spectral overlap, makes the tryptophan–TNB pair an excellent tool for measuring intramolecular distances by FRET (17).
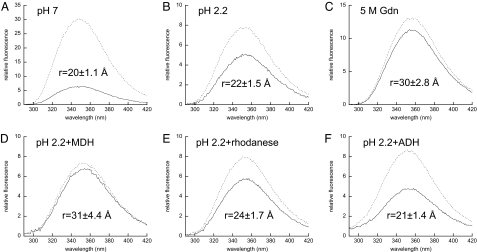
We first measured the emission spectrum of TNB-labeled HdeA(W82,C27) at pH 7 and observed a marked increase in fluorescence intensity upon removal of the TNB acceptor (Fig. 5A). We then used the observed change in fluorescence intensity to calculate the apparent distance between the tryptophan donor and TNB acceptor and obtained a value of 20 ± 1.1 Å. This distance is consistent with the 20-Å distance between the α-carbons of S27 and W82 observed in the HdeA crystal structure (12), making us confident that our measurements indeed reflected the intramolecular distance between these residues. When we performed the same experiments at pH 2.2, we calculated a distance of 22 ± 1.5 Å (Fig. 5B), in which the partial unfolding of HdeA results in a 2-Å increase in the apparent donor–acceptor distance. Complete unfolding of HdeA in 5 M Gdn yielded an apparent distance of 30 ± 2.8 Å (Fig. 5C). The difference between HdeApH2.2 and completely unfolded HdeA probably reflects the residual secondary structure of HdeA at low pH (Fig. S1A). It is likely that the calculated distances in 5 M Gdn, and to a lesser extent at pH 2.2, reflect an ensemble of unfolded and partially folded conformations and therefore represent an average of many actual distances. It is clear, however, that the progressive unfolding of HdeA leads to increased donor–acceptor distances, as expected.
Fig. 5.
Substrate binding induces substantial conformational changes in HdeA. HdeA conformation was assessed by determining the FRET efficiency and apparent distances between the W82 donor and TNB acceptor attached via C27. Emission spectra (λex = 280 nm) of 1 μM HdeA(W82,C27) were recorded in the absence (FD, dashed lines) or presence of the TNB acceptor (FDA, solid lines) attached via C27 at pH 7.0 (A), pH 2.2 (B), or in 5 M Gdn (C). The effects of substrate binding on the conformation of active 1 μM HdeApH2.2 were assessed in the presence of 4 μM MDH (D), 4 μM rhodanese (E), or 4 μM ADH (F). Integrated emission intensities were used to calculate FRET efficiency and apparent distances (r) as outlined in SI Materials and Methods. Triplicate samples were prepared for each condition, and the resulting standard deviations were used in conjunction with errors in R0 to determine the error for each distance reported (see SI Materials and Methods).
We then tested the effects of substrate binding on the conformation of chaperone-active HdeApH2.2. When we incubated HdeApH2.2 with MDH, we found that the apparent distance between the tryptophan donor and TNB acceptor increased by 9 Å to 31 ± 4.4 Å (Fig. 5D), indicating that MDH binding has a substantial effect on HdeA conformation. As an additional means of assessing donor–acceptor distances, we performed time-resolved fluorescence measurements and used the fluorescence lifetimes (τ) in the presence and absence of acceptor, rather than steady-state intensity, to calculate apparent distances. We observed a similar, large increase in donor–acceptor distance upon substrate binding when we used fluorescence lifetimes to calculate FRET efficiency and corresponding distances of 17 Å at pH 7, 21 Å at pH 2.2, and 29 Å at pH 2.2 in the presence of MDH (Fig. S8 and Table S2).
To exclude the possibility that this significant apparent change in distance is an orientation-related artifact, for example, in which MDH binding changes FRET efficiency by altering κ2, we repeated these experiments using AF350-C5 maleimide as the acceptor. This label contains a 5-carbon flexible linker between the attachment point and the fluorophore, which should enhance the rotational freedom of the acceptor and thereby minimize orientation-specific effects on energy transfer efficiency. The distances obtained by using the AF350 acceptor were similar to the distances calculated for the TNB-labeled protein (Fig. S9). The observations that W82 is completely solvent-exposed as judged by λmax (Fig. S4) and that λmax is unaffected by MDH binding suggest that W82 is mobile and therefore unlikely to exhibit orientational bias. Furthermore, fast anisotropy decays were observed for both the donor and acceptor, indicative of highly mobile probes even in the presence of bound MDH (Fig. S9). Taken together, these data indicate that we are indeed monitoring the effects of substrate binding on donor–acceptor distance, with negligible contributions caused by orientation of the probes.
To test whether HdeA adopts substrate-specific conformations, we tested the effects of two other model substrate proteins, rhodanese and alcohol dehydrogenase (ADH). Both proteins have been shown to interact with HdeA (10, 12), and we found that they do so with high affinity (Kd ≈200 nM). In contrast to MDH, we found that rhodanese binding induces only a small increase of 2 Å (to 24 ± 1.7 Å) in the apparent donor–acceptor distance (Fig. 5E), whereas ADH binding may actually lead to a slight compaction of the HdeA structure (21 ± 1.4 Å; Fig. 5F), although the apparent changes induced by rhodanese and ADH binding are within the error of the measurements. Even when we take these errors into account, we can clearly conclude, however, that the conformation of HdeA in the MDH-bound form at 31 ± 4.4 Å is significantly different from either the ADH-bound form at 21 ± 1.4 Å or the rhodanese-bound form at 24 ± 1.7 Å. These results suggest that substrate binding can, at least in some cases, shift the conformational equilibrium of HdeA, such that an ensemble of disordered conformations may be presented to the substrate, and the substrate can bind to particular conformation(s) on the basis of its structure. Thus, the final conformation of HdeA in the chaperone–substrate complex depends on the structure of the substrate protein.
Discussion
HdeA is a new member of a growing class of stress-sensing proteins that use stress-induced activation rather than transcriptional regulation to respond rapidly to stress conditions (18). When enteric bacteria are ingested, their periplasmic space is rapidly acidified, which can cause the acid-induced unfolding and aggregation of numerous periplasmic proteins (10, 11). Thus, survival in extreme acid conditions requires a very rapid response to protect acid-unfolded proteins from irreversible aggregation. As a molecular chaperone, HdeA protects periplasmic proteins from acid-induced aggregation (10, 11, 19). HdeA is also partially unfolded in response to acidic pH, but in a clever twist, it becomes activated by the very same low-pH conditions that lead to the inactivation of other proteins. In this work we find that HdeA adopts its active monomeric conformation within a fraction of a second after a pH downshift. This rapidity of HdeA activation enables presynthesized HdeA to remain inactive under nonstress conditions, but ready to bind to other unfolding proteins the instant that bacteria encounter extremely low pH. This type of regulation is orders of magnitude more rapid than transcriptional control. It ensures that HdeA is poised to deal with the potentially lethal problem of protein aggregation immediately after ingestion by the host organism.
The observation that HdeA becomes partially unfolded to achieve its chaperone active conformation seems to defy the traditional protein structure–function paradigm: most proteins require structure to be functional. Although structural disorder has been observed in numerous other chaperone proteins (20), the mechanistic relevance of disorder to chaperone function has not been entirely clear. In general, protein disorder is thought to allow proteins to bind more adaptively to diverse ligands (20, 21). Another proposed model suggests that disordered proteins are capable of binding the same targets with a substantially smaller protein scaffold than their well-folded counterparts (22). Both of these characteristics seem desirable for a general chaperone protein, and indeed, both characteristics seem to apply to HdeA. Our intramolecular FRET experiments suggest that upon substrate binding, HdeA populates different conformational ensembles that depend on the structure of the substrate protein. This mode of substrate recognition is likely to confer broad substrate specificity on HdeA. Some degree of structural plasticity has been proposed as important for chaperone–substrate interactions (23–25), but here we discuss a chaperone that adaptively binds to substrates such that different substrates influence the chaperone's conformational equilibrium in different ways. This mechanism provides an excellent explanation of how a protein as small as HdeA can adaptively bind a broad range of structurally diverse substrates (10–12).
HdeA was known to undergo a dimer-to-monomer transition upon activation, raising the possibility that the hydrophobic dimer interface of HdeA plays a role in substrate binding (12). Our results provide experimental support for this hypothesis. Apparently to regulate HdeA chaperone activity, residues that participate in substrate binding are masked by an adjacent HdeA monomer in the inactive dimeric form. Additional residues that are buried in the hydrophobic core in the inactive conformation of HdeA are also likely to contribute to substrate binding (14), in which case HdeA may use most if not all of its hydrophobic residues for substrate binding. The substrate-binding site may involve two largely hydrophobic stretches in the primary sequence of HdeA, which are joined together by the HdeA C18–C66 disulfide bond (14). This may help to explain why we found this disulfide to be crucial for HdeA activity.
Chaperones in general require not only substrate-binding activity but also the ability to control the binding and release of substrate proteins. This often involves large structures and complex mechanisms. The GroEL–GroES complex, for instance, is an ≈870-kDa multimeric machine with a well-characterized but complex allosteric catalytic cycle that involves ATP binding and hydrolysis (26, 27). Hsp70-class chaperones typically use J-domain cochaperones and nucleotide exchange factors that work in concert to modulate the affinities for substrates and nucleotides, resulting again in a complex cycle of ATP binding, hydrolysis, and nucleotide exchange that is essential to chaperone function (26, 28). Even the so-called “small” heat shock proteins typically form high-order oligomers containing from 12 to up to 50 or more subunits, and substrate release from the small Hsps requires the assistance of ATP-dependent chaperone systems (16). In stark contrast, HdeA is only 89 amino acids long with a mass of 9.7 kDa, making it the smallest proteinaceous chaperone of which we are aware. Unlike most chaperones, HdeA functions as a monomer and is not known to require any energy factors or cochaperones. How does this small chaperone both bind proteins and regulate their release? We found that acid-induced monomerization allows HdeA to use its hydrophobic dimer interface for substrate binding. This provides an elegant way to expose a high-affinity substrate-binding site rapidly and reversibly. By using this mechanism, HdeA exploits the unfolding effects of acid to become activated under the same stress conditions that it has evolved to protect cells against. In addition, it neatly sidesteps the requirement for energy cofactors, which are absent in the periplasm where HdeA functions and which many other chaperones use to regulate substrate binding affinity. The combination of the partially disordered structure of HdeA, the use of residues at the dimer interface, and adaptive binding of substrates provides a substantial hydrophobic surface area for substrate binding while maintaining a minimal chaperone size. These remarkably efficient features result in a chaperone that is rapidly activated by acid stress and capable of suppressing the aggregation of substrate proteins several times its own size. The fate of these substrate proteins after pH neutralization, which inactivates HdeA, is currently under investigation.
Materials and Methods
Protein Expression and Purification.
E. coli HdeA was expressed and purified essentially as described in ref. 12 with modifications detailed in SI Materials and Methods. Each mutant exhibited far-UV CD spectra and chaperone-like activity similar to that of wild-type HdeA (Fig. S10).
HdeA Activity Assays.
Rhodanese aggregation assays were performed as described in ref. 12 with minor modifications as outlined in SI Materials and Methods.
Stopped-Flow Fluorescence Measurements.
Kinetics of pH-dependent conformational changes of HdeA were monitored by measuring changes in tryptophan fluorescence. Measurements were performed by using a Kintek2004 stopped-flow instrument with a λex of 295 nm and an emission band pass filter of 340 ± 10 nm. Temperature was held constant at 37 °C with a circulating water bath. HdeA was diluted into 10 mM phosphate buffer, 150 mM NaCl (pH 7). This was rapidly mixed with an equal volume of 200 mM phosphate, 150 mM NaCl (pH 2) to achieve acidification (final pH 2.2). For all stopped-flow experiments, 8–10 kinetic traces were collected and fit independently.
Fluorescence Labeling and FRET Measurements.
Labeling of HdeA(C27) with monobromobimane or AF532 (C-5 maleimide) was performed essentially as recommended by Molecular Probes (see SI Materials and Methods for detailed protocol). HdeA(W82,C27) was labeled with 5,5′-dithiobis(2-nitrobenzoic acid) as described in ref. 17, which yielded a labeling stoichiometry of 1 ± 0.05. Fluorescence spectra were recorded and used to calculate apparent distances as described in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Stefan Walter for helpful discussions. This work was supported by National Institutes of Health Grants GM057039 (to J.C.A.B.) and GM065318 (to U.J.). J.C.A.B. is an investigator for the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811811106/DCSupplemental.
References
- 1.Chuang SE, Blattner FR. Characterization of twenty-six new heat shock genes of Escherichia coli. J Bacteriol. 1993;175:5242–5252. doi: 10.1128/jb.175.16.5242-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jakob U, Muse W, Eser M, Bardwell JC. Chaperone activity with a redox switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 3.Haslbeck M, et al. Hsp26: A temperature-regulated chaperone. EMBO J. 1999;18:6744–6751. doi: 10.1093/emboj/18.23.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boron WF, Boulpaep EL. Medical Physiology. Philadelphia: Saunders; 2003. [Google Scholar]
- 5.Smith JL. The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J Food Protect. 2003;66:1292–1303. doi: 10.4315/0362-028x-66.7.1292. [DOI] [PubMed] [Google Scholar]
- 6.Castanie-Cornet MP, et al. Control of acid resistance in Escherichia coli. J Bacteriol. 1999;181:3525–3535. doi: 10.1128/jb.181.11.3525-3535.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Biase D, Tramonti A, Bossa F, Visca P. The response to stationary-phase stress conditions in Escherichia coli: Role and regulation of the glutamic acid decarboxylase system. Mol Microbiol. 1999;32:1198–1211. doi: 10.1046/j.1365-2958.1999.01430.x. [DOI] [PubMed] [Google Scholar]
- 8.Koebnik R, Locher KP, Van Gelder P. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol Microbiol. 2000;37:239–253. doi: 10.1046/j.1365-2958.2000.01983.x. [DOI] [PubMed] [Google Scholar]
- 9.Fink AL, et al. Classification of acid denaturation of proteins: Intermediates and unfolded states. Biochemistry. 1994;33:12504–12511. doi: 10.1021/bi00207a018. [DOI] [PubMed] [Google Scholar]
- 10.Hong W, et al. Periplasmic protein HdeA exhibits chaperone-like activity exclusively within stomach pH range by transforming into disordered conformation. J Biol Chem. 2005;280:27029–27034. doi: 10.1074/jbc.M503934200. [DOI] [PubMed] [Google Scholar]
- 11.Kern R, et al. Escherichia coli HdeB is an acid stress chaperone. J Bacteriol. 2007;189:603–610. doi: 10.1128/JB.01522-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gajiwala KS, Burley SK. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J Mol Biol. 2000;295:605–612. doi: 10.1006/jmbi.1999.3347. [DOI] [PubMed] [Google Scholar]
- 13.Waterman SR, Small PL. Identification of σS-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol Microbiol. 1996;21:925–940. doi: 10.1046/j.1365-2958.1996.00058.x. [DOI] [PubMed] [Google Scholar]
- 14.Wu YE, et al. Conserved amphiphilic feature is essential for periplasmic chaperone HdeA to support acid resistance in enteric bacteria. Biochem J. 2008;412:389–397. doi: 10.1042/BJ20071682. [DOI] [PubMed] [Google Scholar]
- 15.Lakowicz JR. Principles of Fluorescence Spectroscopy. New York: Springer; 2006. [Google Scholar]
- 16.Haslbeck M, Franzmann T, Weinfurtner D, Buchner J. Some like it hot: The structure and function of small heat-shock proteins. Nat Struct Mol Biol. 2005;12:842–846. doi: 10.1038/nsmb993. [DOI] [PubMed] [Google Scholar]
- 17.Wu P, Brand L. Conformational flexibility in a staphylococcal nuclease mutant K45C from time-resolved resonance energy transfer measurements. Biochemistry. 1994;33:10457–10462. doi: 10.1021/bi00200a029. [DOI] [PubMed] [Google Scholar]
- 18.Winter J, Jakob U. Beyond transcription: New mechanisms for the regulation of molecular chaperones. Crit Rev Biochem Mol Biol. 2004;39:297–317. doi: 10.1080/10409230490900658. [DOI] [PubMed] [Google Scholar]
- 19.Malki A, et al. Solubilization of protein aggregates by the acid stress chaperones HdeA and HdeB. J Biol Chem. 2008;283:13679–13687. doi: 10.1074/jbc.M800869200. [DOI] [PubMed] [Google Scholar]
- 20.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 21.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Gunasekaran K, et al. Extended disordered proteins: Targeting function with less scaffold. Trends Biochem Sci. 2003;28:81–85. doi: 10.1016/S0968-0004(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 23.Ashcroft AE, et al. Structural plasticity and noncovalent substrate binding in the GroEL apical domain: A study using electrospray ionization mass spectrometry and fluorescence binding studies. J Biol Chem. 2002;277:33115–33126. doi: 10.1074/jbc.M203398200. [DOI] [PubMed] [Google Scholar]
- 24.Hu K, Galius V, Pervushin K. Structural plasticity of peptidyl-prolyl isomerase sFkpA is a key to its chaperone function as revealed by solution NMR. Biochemistry. 2006;45:11983–11991. doi: 10.1021/bi0607913. [DOI] [PubMed] [Google Scholar]
- 25.White HE, et al. Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26. Structure. 2006;14:1197–1204. doi: 10.1016/j.str.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 26.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 27.Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: Physiology and mechanism. Annu Rev Cell Mol Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- 28.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.