Abstract
The adult human distal gut microbial community is typically dominated by 2 bacterial phyla (divisions), the Firmicutes and the Bacteroidetes. Little is known about the factors that govern the interactions between their members. Here, we examine the niches of representatives of both phyla in vivo. Finished genome sequences were generated from Eubacterium rectale and E. eligens, which belong to Clostridium Cluster XIVa, one of the most common gut Firmicute clades. Comparison of these and 25 other gut Firmicutes and Bacteroidetes indicated that the Firmicutes possess smaller genomes and a disproportionately smaller number of glycan-degrading enzymes. Germ-free mice were then colonized with E. rectale and/or a prominent human gut Bacteroidetes, Bacteroides thetaiotaomicron, followed by whole-genome transcriptional profiling, high-resolution proteomic analysis, and biochemical assays of microbial–microbial and microbial–host interactions. B. thetaiotaomicron adapts to E. rectale by up-regulating expression of a variety of polysaccharide utilization loci encoding numerous glycoside hydrolases, and by signaling the host to produce mucosal glycans that it, but not E. rectale, can access. E. rectale adapts to B. thetaiotaomicron by decreasing production of its glycan-degrading enzymes, increasing expression of selected amino acid and sugar transporters, and facilitating glycolysis by reducing levels of NADH, in part via generation of butyrate from acetate, which in turn is used by the gut epithelium. This simplified model of the human gut microbiota illustrates niche specialization and functional redundancy within members of its major bacterial phyla, and the importance of host glycans as a nutrient foundation that ensures ecosystem stability.
Keywords: human gut Firmicutes and Bacteroidetes, carbohydrate metabolism, gnotobiotic mice, gut microbiome, nutrient sharing
The adult human gut houses a bacterial community containing trillions of members comprising thousands of species-level phylogenetic types (phylotypes). Culture-independent surveys of this community have revealed remarkable interpersonal variations in these strain- and species-level phylotypes. Two bacterial phyla, the Firmicutes and the Bacteroidetes, commonly dominate this ecosystem (1), as they do in the guts of at least 60 mammalian species (2).
Comparative analysis of 5 previously sequenced human gut Bacteroidetes revealed that each genome contains a large repertoire of genes involved in acquisition and metabolism of polysaccharides. This repertoire includes (i) up to hundreds of glycoside hydrolases (GHs) and polysaccharide lyases (PLs); (ii) myriad paralogs of SusC and SusD, outer membrane proteins involved in recognition and import of specific carbohydrate structures (3); and (iii) a large array of environmental sensors and regulators (4). These genes are assembled in similarly organized, selectively regulated polysaccharide utilization loci (PULs) that encode functions necessary to detect, bind, degrade and import carbohydrate species encountered in the gut habitat–either from the diet or from host glycans associated with mucus and the surfaces of epithelial cells (5–7). Studies of gnotobiotic mice colonized only with human gut-derived Bacteroides thetaiotaomicron have demonstrated that this organism can vary its pattern of expression of PULs as a function of diet, e.g., during the transition from mother's milk to a polysaccharide-rich chow consumed when mice are weaned (5), or when adult mice are switched from a diet rich in plant polysaccharides to a diet devoid of these glycans and replete with simple sugars (under the latter conditions, the organism forages on host glycans) (6, 7).
Our previous functional genomic studies of the responses of B. thetaiotaomicron to cocolonization of the guts of gnotobiotic mice with Bifidobacterium longum, an Actinobacterium found in the intestines of adults and infants, or with Lactobacillus casei, a Firmicute present in a number of fermented diary products, have shown that B. thetaiotaomicron adapts to the presence of these other microbes by modifying expression of its PULs in ways that expand the breadth of its carbohydrate foraging activities (8).
These observations support the notion that gut microbes may live at the intersection of 2 forms of selective pressure: bottom-up selection, where fierce competition between members of a community that approaches a population density of 1011 to 1012 organisms per milliliter of colonic contents drives phylotypes to assume distinct functional roles (niches); and top-down selection, where the host selects for functional redundancy to ensure against the failure of bioreactor functions that could prove highly deleterious (9, 10).
The gene content, genomic arrangement and functional properties of PULs in sequenced gut Bacteroidetes illustrate the specialization and functional redundancy within members of this phylum. They also emphasize how the combined metabolic activities of members of the microbiota undoubtedly result in interactions that are both very dynamic and overwhelmingly complex (at least to the human observer), involving multiple potential pathways for the processing of substrates (including the order of substrate processing), varying patterns of physical partitioning of microbes relative to substrates within the ecosystem, plus various schemes for utilization of products of bacterial metabolism. Such a system likely provides multiple options for processing of a given metabolite, and for the types of bacteria that can be involved in these activities.
All of this means that the task of defining the interactions of members of the human gut microbiota is daunting, as is the task of identifying general principles that govern the operation of this system. In the present study, we have taken a reductionist approach to begin to define interactions between members of the Firmicutes and the Bacteroidetes that are commonly represented in the human gut microbiota. In the human colon, Clostridium cluster XIVa is 1 of 2 abundantly represented clusters of Firmicutes. Therefore, we have generated the initial 2 complete genome sequences for members of the genus Eubacterium in Clostridium cluster XIVa (the human gut-derived E. rectale strain ATCC 33656 and E. eligens strain ATCC 27750) and compared them with the draft sequences of 25 other sequenced human gut bacteria belonging to the Firmicutes and the Bacteroidetes. The interactions between E. rectale and B. thetaiotaomicron were then characterized by performing whole-genome transcriptional profiling of each species after colonization of gnotobiotic mice with each organism alone, or in combination under 3 dietary conditions. Transcriptional data were verified by mass spectrometry of cecal proteins, plus biochemical assays of carbohydrate metabolism. Last, we examined colonization and interactions between these microbes from a host perspective; to do so, we performed whole-genome transcriptional analysis of colonic RNA prepared from mice that were germ-free or colonized with one or both species. Our results illustrate how members of the dominant gut bacterial phyla are able to adapt their substrate utilization in response to one another and to host dietary changes, and how host physiology can be affected by changes in microbiota composition.
Results and Discussion
Comparative Genomic Studies of Human Gut-Associated Firmicutes and Bacteroidetes.
We produced finished genome sequences for Eubacterium rectale, which contains a single 3,449,685-bp chromosome encoding 3,627 predicted proteins, and Eubacterium eligens, which contains a 2,144,190-bp chromosome specifying 2,071 predicted proteins, plus 2 plasmids (Table S1). We also analyzed 25 recently sequenced gut genomes, including (i) 9 sequenced human gut-derived Bacteroidetes [includes the finished genomes of B. thetaiotaomicron, B. fragilis, B. vulgatus, and Parabacteroides distasonis, plus deep draft assemblies of the B. caccae, B. ovatus, B. uniformis, B. stercoris and P. merdae genomes generated as part of the human gut microbiome initiative (HGMI) (http://genome.wustl.edu/hgm/HGM_frontpage.cgi)], and (ii) 16 other human gut Firmicutes where deep draft assemblies were available through the HGMI (see Fig. S1 for a phylogenetic tree). We classified the predicted proteins in these 2 genomes using Gene Ontology (GO) terms generated via Interproscan, and according to the scheme incorporated into the Carbohydrate Active Enzymes (CAZy) database [www.cazy.org (11)], and then applied a binomial test to identify functional categories of genes that are either over- or under-represented between the Firmicutes and Bacteroidetes phyla. This analysis, described in SI Results, Figs. S2 and S3, and Table S2 and Table S3, emphasized among other things that the Firmicutes, including E. rectale and E. eligens, have significantly fewer polysaccharide-degrading enzymes and more ABC transporters and PTS systems than the Bacteroidetes (12). We subsequently chose E. rectale and B. thetaiotaomicron as representatives of these 2 phyla for further characterization of their niches in vivo, because of their prominence in culture-independent surveys of the distal human gut microbiota (13, 14), the pattern of representation of carbohydrate active enzymes in their glycobiomes and E. rectale's ability to generate butyrate as a major end product of fermentation (15, 16). These choices set the stage for an “arranged marriage” between a Firmicute and a Bacteroidetes, hosted by formerly germ-free mice.
Functional Genomic Analyses of the Minimal Human Gut Microbiome.
Creating a “minimal human gut microbiota” in gnotobiotic mice.
Young adult male germ-free mice belonging to the NMRI inbred strain were colonized with B. thetaiotaomicron or E. rectale alone (monoassociations) or cocolonized with both species (biassociation). Ten to fourteen days after inoculation by gavage, both species colonized the ceca of recipient mice, fed a standard chow diet rich in complex plant polysaccharides, to high levels (n = 4–5 mice per treatment group in each of 3 independent experiments; Fig. S4A). Moreover, cecal levels of colonization for both organisms were not significantly different between mono- and biassociated animals (Fig. S4A).
B. thetaiotaomicron's response to E. rectale.
A custom, multispecies, human gut microbiome Affymetrix GeneChip was designed (SI Methods), and used to compare the transcriptional profile of each bacterial species when it was the sole inhabitant of the cecum, and when it coexisted together with the other species. A significant number of B. thetaiotaomicron genes located in PULs exhibited differences in their expression upon E. rectale colonization [55 of 106; P < 10−15 (cumulative hypergeometric test); see SI Methods for the statistical criteria for defining significantly different levels of gene expression]. Of these 55 genes, 51 (93%) were up-regulated (Fig. S4B; see Table S4A for a complete list of differentially regulated B. thetaiotaomicron genes).
As noted in the Introduction, 2 previous studies from our lab examined changes in B. thetaiotaomicron's transcriptome in the ceca of monoassociated gnotobiotic mice when they were switched from a diet rich in plant polysaccharides to a glucose-sucrose chow (6), or in suckling mice consuming mother's milk as they transitioned to a standard chow diet (5). In both situations, in the absence of dietary plant polysaccharides, B. thetaiotaomicron adaptively forages on host glycans. The genes up-regulated in B. thetaiotaomicron upon cocolonization with E. rectale have a significant overlap with those noted in these 2 previous datasets (P < 10−14, cumulative hypergeometric test; Fig. S4C). In addition, they include several of the genes up-regulated during growth on minimal medium containing porcine mucosal glycans as the sole carbon source (7). For example, in cocolonized mice and in vitro, B. thetaiotaomicron up-regulates several genes (BT3787-BT3792; BT3774-BT3777) (Fig. S4D) used in degrading α-mannosidic linkages, a component of host N-glycans and the diet. (Note that E. rectale is unable to grow in defined medium containing α-mannan or mannose as the sole carbon sources; Table S3). B. thetaiotaomicron also up-regulates expression of its starch utilization system (Sus) PUL in the presence of E. rectale (BT3698–3704) (Fig. S4D). This well-characterized PUL is essential for degradation of starch molecules containing ≥6 glucose units (17).
Thus, it appears that B. thetaiotaomicron adapts to the presence of E. rectale by up-regulating expression of a variety of PULs so that it can broaden its niche and degrade an increased variety of glycan substrates, including those derived from the host that E. rectale is unable to access. There are a number of reasons why the capacity to access host glycans likely represents an important trait underpinning microbiota function and stability: (i) glycans in the mucus gel are abundant and are a consistently represented source of nutrients; (ii) mucus could serve as a microhabitat for Bacteroidetes spp. to embed in (and adhere to via SusD paralogs), thereby avoiding washout from the ecosystem; and (iii) the products of polysaccharide digestion/fermentation generated by Bacteroidetes spp. could be shared with other members of the microbiota that are also embedded in mucus (7).
E. rectale's response to B. thetaiotaomicron.
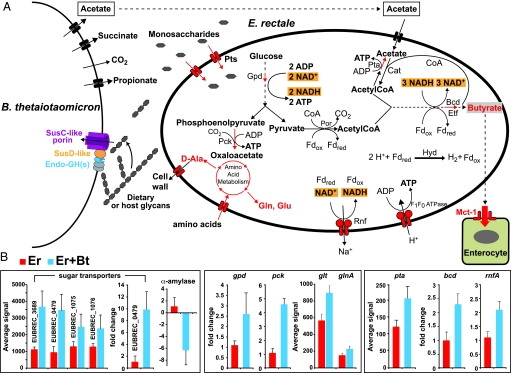
E. rectale's response to B. thetaiotaomicron in the mouse cecum stands in marked contrast to B. thetaiotaomicron's response to E. rectale. Carbohydrate metabolism genes, particularly GHs, are significantly overrepresented among the E. rectale genes that are downregulated in the presence of B. thetaiotaomicron compared with monoassociation; i.e., 12 of E. rectale's predicted 51 GHs have significantly reduced expression while only 2 are up-regulated (Fig. S4 E and F; see Table S4B for a complete list of E. rectale genes regulated by the presence of B. thetaiotaomicron). The 2 up-regulated GH genes (EUBREC_1072, a 6-P-β-glucosidase and EUBREC_3687, a cellobiose phosphorylase) are predicted to break down cellobiose. Three simple sugar transport systems with predicted specificity for cellobiose, galactoside, and arabinose/lactose (EUBREC_3689, EUBREC_0479, and EUBREC_1075–6, respectively) are among the most strongly up-regulated genes (Fig. S4G and Table S4B). Phosphoenolpyruvate carboxykinase (Pck EUBREC_2002) is also induced with cocolonization (Table S4B, GeneChip data were verified by qRT-PCR assays in 2 independent experiments involving 3–4 mice per treatment group; Fig. 1B). This enzyme catalyzes an energy conserving reaction that produces oxaloacetate from phosphoenolpyruvate. In a subsequent transaminase reaction, oxaloacetate can be converted to aspartate, linking this branching of the glycolytic pathway with amino acid biosynthesis (Fig. 1A).
Fig. 1.
Summary of metabolic responses of E. rectale to B. thetaiotaomicron. (A) Overview of metabolic pathways. (B) GeneChip probeset intensities and qRT-PCR validation assays are shown for a subset of genes. Mean values for triplicate qRT-PCR determinations (n = 4 mice per group) ± SD are plotted. Pts, phosphotransferase systems; Gpd, glycerol 3-phosphate dehydrogenase; Pck, phosphoenolpyruvate carboxykinase; Por, pyruvate:ferredoxin oxidoreductase; Hyd, hydrogenase; Rnf, NADH: ferredoxin oxidoreductase complex; Fdred, reduced ferredoxin; Fdox oxidized ferredoxin; Pta, phosphate acetyltransferase; Bcd, butyryl-CoA dehydrogenase; Etf electron transport flavoproteins; Cat, butyryl CoA: acetate CoA transferase; Glt, glutamate synthetase; GlnA, glutamine synthetase Gln, glutamine; Glu, glutamate; Mct1, monocarboxylate transporter 1.
Additional data support the notion that E. rectale is better able to access nutrients in the presence of B. thetaiotaomicron. For example, a number of peptide and amino acid transporters in E. rectale are up-regulated, as are the central carbon and nitrogen regulatory genes CodY (EUBREC_1812), glutamate synthase (EUBREC_1829) and glutamine synthetase (EUBREC_2543) (Fig. 1B and Fig. S4H; note that these genes are also up-regulated during growth in tryptone glucose medium; Table S4C).
Changes in E. rectale's fermentative pathways.
E. rectale possesses genes (EUBEC733–737; EUBEC1017) for the production of butyrate that show high similarity to genes from other Clostridia. This pathway involves condensation of 2 molecules of acetylCoA to form butyrate and is accompanied by oxidation of NADH to NAD+ (Fig. 1). Transcriptional and high-resolution proteomic analyses (see below) disclosed that the enzymes involved in production of butyrate are among the most highly expressed in cecal contents recovered from mono- and biassociated mice containing E. rectale (Table S4B and Table S6A).
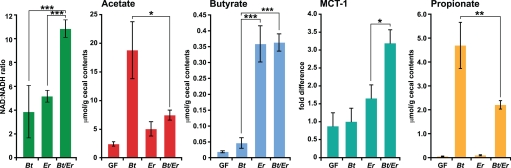
In vitro studies have shown that in the presence of carbohydrates, E. rectale consumes large amounts of acetate for butyrate production (18). Several observations indicate that E. rectale utilizes B. thetaiotaomicron-derived acetate to generate increased amounts of butyrate in the ceca of our gnotobiotic mice. First, E. rectale up-regulates a phosphate acetyltransferase (EUBREC_1443; EC 2.3.1.8)—1 of 2 enzymes involved in the interconversion of acetyl-CoA and acetate (Fig. 1B). Second, cecal acetate levels are significantly lower in cocolonized mice compared with B. thetaiotaomicron monoassociated animals (Fig. 2). Third, although cecal butyrate levels are similar in E. rectale mono- and biassociated animals (Fig. 2), expression of mouse Mct-1, encoding a monocarboxylate transporter whose inducer and preferred substrate is butyrate (19), is significantly higher in the distal gut of mice containing both E. rectale and B. thetaiotaomicron versus E. rectale alone (P < 0.05; Fig. 2). The cecal concentrations of butyrate we observed are similar to those known to up-regulate Mct-1 in colonic epithelial cell lines (19). Higher levels of acetate (i.e., those encountered in B. thetaiotaomicron monoassociated mice) were insufficient to induce any change in Mct-1 expression compared with germ-free controls (Fig. 2).
Fig. 2.
Cocolonization affects the efficiency of fermentation. Cecal contents from 4 mice in each treatment group were assayed for NAD+, NADH acetate, butyrate and proprionate levels. Expression of Mct-1 mRNA, a monocarboxylate transporter whose preferred substrate is butyrate was defined by qRT-PCR in the proximal colon. Cecal propionate concentrations. Mean values ± SEM are plotted; n = 4–5 mice per group; ∗, P < 0.05, ∗∗, P < 0.001 compared with cocolonization (Student's t test).
The last step in E. rectale's butyrate production pathway is catalyzed by the butyrylCoA dehydrogenase/electron transfer flavoprotein (Bcd/Etf) complex (EUBREC_0735–0737; EC 1.3.99.2), and offers a recently discovered additional pathway for energy conservation, via a bifurcation of electrons from NADH to crotonylCoA and ferredoxin (20). Reduced ferredoxin, in turn, can be reoxidized via hydrogenases, or via the membrane-bound oxidoreductase, Rnf, which generates sodium-motive force (Fig. 1A). The up-regulation and high level of expression of these key metabolic genes when E. rectale encounters B. thetaiotomicron (Fig. 1B; Table S4B and Table S6A) indicates that E. rectale not only employs this pathway to generate energy, but to also accommodate the increased demand for NAD+ in the glycolytic pathway. Consistent with these observations, we found that the NAD+/NADH ratio in cecal contents was significantly increased with cocolonization (Fig. 2).
The pathway for acetate metabolism observed in this simplified model human gut community composed of B. thetaiotaomicron and E. rectale differs markedly from what is seen in mice that harbor B. thetaiotaomicron and the principal human gut methanogenic archaeon, Methanobrevibacter smithii. When B. thetaiotaomicron encounters M. smithii in the ceca of gnotobiotic mice, there is increased production of acetate by B. thetaiotaomicron, no diversion to butyrate and no induction of Mct-1 (21), increased serum acetate levels, and increased adiposity compared with B. thetaiotaomicron mono-associated controls. In contrast, serum acetate levels and host adiposity (as measured by fat pad to body weight ratios) are not significantly different between B. thetaiotaomicron monoassociated and B. thetaiotaomicron-E. rectale cocolonized animals (n = 4–5 animals/group; n = 3 independent experiments; data not shown).
Colonic transcriptional changes evoked by E. rectale-B. thetaiotaomicron cocolonization.
We subsequently used Affymetrix Mouse 430 2 GeneChips to compare patterns of gene expression in the proximal colons of mice that were either germ-free, monoassociated with E.rectale or B. thetaiotaomicron, or cocolonized with both organisms (n = 4 mice per group; total of 16 GeneChip datasets). In contrast to the small number of genes whose expression was significantly changed (≥1.5-fold, FDR <1%) after colonization with either bacterium alone relative to germ-free controls (Table S7 A and B), cocolonization produced significant alterations in the expression of 508 host genes (Table S7C). Expression of many of these genes also changed with monoassociation with either organism, and in the same direction as seen after cocolonization, but in most cases the changes evoked by B. thetaiotaomicron or E. rectale alone did not achieve statistical significance. Unsupervised hierarchical clustering of average expression intensity values derived from each of the 4 sets of GeneChips/group, revealed that the E.rectale monoassociation and E.rectale-B.thetaiotaomicron biassociation profiles clustered separate from the germ-free and B. thetaiotaomicron monoassociation datasets (Fig. S5).
Ingenuity Pathway Analysis (www.ingenuity.com) disclosed that the list of 508 host genes affected by cocolonization was significantly enriched in functions related to cellular growth and proliferation (112 genes; Table S8A), and cell death (130 genes) (Table S8B). A number of components of the canonical wnt/β catenin pathway, which is known to be critically involved in controlling self-renewal of the colonic epithelium, were present in this list (Akt3, Axin2, Csnk1D, Dkk3, FrzB, Fzd2, Gja1, Mdm2, Ppp2r5e, Sfrp2, Tgfb3, Tgfbr1, and Tgfbr2). Many of the changes observed in biassociated mice are likely to be related to the increased influx of butyrate, generated by E. rectale, into colonic cells (Fig. 1A). Butyrate, a histone deacetylase inhibitor that evokes pronounced transcriptional changes in different types of cultured epithelial cell lines (22–25), is the preferred energy substrate for colonic enterocytes (26). While transcriptional changes caused by butyrate differ depending upon the cell lineage, state of cellular differentiation, and cellular energy status (23, 24, 27, 28), in vitro and in vivo studies have shown that it affects expression of genes involved in proliferation, differentiation and apoptosis (25, 28).
As mentioned above, as part of its adaptation to the presence of E. rectale, B. thetaiotaomicron up-regulates a number of genes involved in the harvest of host glycans. Included among these B. thetaiotaomicron genes are components of a fucose utilization operon linked to production of a bacterial signal that induces synthesis of intestinal mucosal fucosylated glycans, and microbial catabolism of fucose from O-glycans (29). GeneChip profiling of colonic gene expression disclosed that cocolonization results in increased expression of Fut2 (α-1,2 fucosyltransferase), Fut4 (α-1,3-fucosyltransferase), plus 10 other genes involved in the synthesis of mucosal glycans (glycosphingolipids and O-glycans) (Table S8C). Thus, by increasing host production of glycans, B. thetaiotaomicron can benefit itself, and through its metabolic products, E. rectale.
E. rectale's Colonization Levels and Production of Butyrate Are Affected by Host Diet.
In a final series of experiments, we assessed how E. rectale and B. thetaiotaomicron were affected by changes in host diet. Groups of age- and gender-matched cocolonized mice were fed 1 of 3 diets that varied primarily in their carbohydrate and fat content: (i) the standard low-fat, plant polysaccharide-rich diet used for the experiments described above (abbreviated “LF/PP” for low-fat/plant polysaccharide), (ii) a high-fat, “high-sugar” Western-type diet (abbreviated HF/HS) that contained sucrose, maltodextrin, and corn starch, plus complex polysaccharides (primarily cellulose) that were not digestible by B. thetaiotaomicron or E. rectale, and (iii) a control diet that was similar to (ii) except that the fat content was 4-fold lower (“LF/HS” for low-fat, high-sugar; n = 5 mice per group). Whereas B. thetaiotaomicron's colonization levels were similar in all 3 diets, colonization of E. rectale was significantly reduced (5-fold) in mice fed either the LF/HS or HF/HS diets (P < 0.01, heteroscedastic t test).
Whole-genome transcriptional profiling of both bacterial species showed that relative to the standard polysaccharide-rich chow diet (LF/PP), both the Western style HF/HS diet and its LF/HS control produced a significant up-regulation of B. thetaiotaomicron PULs involved in harvesting and degrading host polysaccharides, and a downregulation of several PULs involved in the degradation of dietary plant polysaccharides (Fig. S6A). E. rectale's response to the HF/HS and LF/HS diets was to down-regulate several of its GHs and a number of its sugar transporters (Fig. S6B). Moreover, levels of butyrate were 5-fold lower in cocolonized mice fed these compared with the standard chow (LF/PS) diet [0.496 ± 0.0051 μmol per gram of wet weight cecal contents; (LF/PP) vs. 0.095 ± 0.002 (HF/HS) vs. 0.080 ± 0.008 (LF/HS) (P < 0.05 ANOVA)].
These dietary manipulations lend further support to the view that B. thetaiotaomicron with its large repertoire of PUL-associated GHs functions in this model 2-member human microbiota to process complex dietary plant polysaccharides and to distribute to the products of digestion to E. rectale, which, in turn, synthesizes butyrate. The reduced colonization response of E. rectale to the HF/HS and LF/HS diets can be explained by a number of factors: (i) this Firmicute does not have predicted GHs and PLs that can process host glycans (Fig. S3); (ii) it cannot use most of the sugars we tested that are derived from mucosal polysaccharides (Table S3); and (iii) the host possesses enzymes in its glycobiome that can directly process the simple sugars present in these 2 diets. Indeed, human subjects that are fed diets deficient in complex polysaccharides harbor lower levels of butyrate-producing gut bacteria, including members of the E. rectale-containing clade (30). Our simplified gnotobiotic model of the microbiota underscores the functional implications of diet-associated changes in the representation of this clade, not only as they relate to the operations of the microbiota itself but also potentially as they relate to butyrate-mediated changes in gut epithelial homeostasis.
Proteomic Studies of This Simplified 2-Component Model of the Human Gut Microbiome.
Model communities such as the one described above, constructed in gnotobiotic mice, where microbiome gene content is precisely known and transcriptional data are obtained under conditions where potentially confounding host variables such as diet and host genotype can be constrained, provide a way to test the efficacy of high-resolution mass spectrometric methods for characterizing gut microbial community proteomes. Therefore, we assayed the proteins present in luminal contents, collected from the ceca of 8 gnotobiotic mice fed the standard polysaccharide-rich LF/PP diet (germ-free, monoassociated, and cocolonized; n = 2 mice per treatment group representing 2 independent biological experiments; see SI Methods for additional details).
The measured proteomes had high reproducibility in terms of total number of proteins observed and spectra matching to each species. Table S5 and SI Results provide a summary of our analyses, including the percentage of mRNAs called “Present” in the GeneChip datasets for which there was an identified protein product. The most abundant identified products from both microbes included ribosomal proteins, elongation factors, chaperones, and proteins involved in energy metabolism (for a full list of identified proteins, see Table S6; note that Table S4 A and B, which list differentially expressed genes in monoassociation versus biassociation experiments, also indicate whether protein products from their transcripts were identified in these mass spectrometry datasets). Many conserved hypothetical and pure hypothetical proteins were identified, as were proteins encoded by 10 genes in B. thetaiotaomicron whose presence had not been predicted in our initial annotation of the finished genome (Table S6A). Together, the results provide validation of experimental and computational procedures used for proteomic assays of a model gut microbiota, and illustrate some of the benefits in obtaining this type of information.
Prospectus.
These studies of a model 2-member human gut microbiota created in gnotobiotic mice support a view of the Bacteroidetes, whose genomes contain a disproportionately large number of glycan-degrading enzymes compared with sequenced Firmicutes, as responding to increasing microbial diversity in the distal intestine by modulating expression of their vast array of polysaccharide utilization loci. B. thetaiotaomicron adapts to the presence of E. rectale by up-regulating a variety of loci specific for host-derived mucin glycans that E. rectale is unable to use. E. rectale, which like other Firmicutes has a more specialized capacity for glycan degradation, broadly downregulates its available GHs in the presence of B. thetaiotaomicron, even though it does not grow efficiently in the absence of carbohydrates. It also becomes more selective in its harvest of sugars and its transcriptional profile suggests improved access to other nutrients (e.g., there is a generalized up-regulation of amino acid biosynthetic genes and a set of nutrient transporters that can harvest peptides). Thus, this simplified, model microbial community illustrates some of the basic ecologic principles that likely shape the operations of the human gut microbiota: nutrient interchange and the observed reciprocal effects on metabolism of these 2 organisms provide examples of classic syntrophy while “character displacement”, where cooccurrence drives (niche) divergence, also operates.
We have previously used gnotobiotic mice to show that the efficiency of fermentation of dietary polysaccharides to short chain fatty acids by B. thetaiotaomicron increases in the presence of M. smithii (21). Cocolonization increases the density of colonization of the distal gut by both organisms, increases production of formate and acetate by B. thetaiotaomicron and allows M. smithii to use H2 and formate to produce methane, thereby preventing the build-up of these fermentation end-products (and NADH) in the gut bioreactor, and improving the efficiency of carbohydrate metabolism (21). Removal of H2 by this methanogenic archaeon allows B. thetaiotaomicron to regenerate NAD+, which can then be used for glycolysis. This situation constitutes a mutualism, in which both members show a clear benefit. The present study, characterizing the cocolonization with B. thetaiotaomicron and E. rectale, describes a more nuanced interaction where both species colonize to similar levels if carbohydrate substrates are readily available. Moreover, certain aspects of bacterial-host mutualism become more apparent with cocolonization, including increased microbial production and host transport of butyrate, and increased host production and microbial consumption of mucosal glycans.
It seems likely that as the complexity of the gut community increases, interactions between B. thetaiotaomicron and E. rectale will either by subsumed or magnified by other “similar” phylogenetic types (as defined by their 16S rRNA sequence and/or by their glycobiomes). Synthesizing model human gut microbiotas of increasing complexity in gnotobiotic mice using sequenced members should be very useful for further testing this idea, as well as a variety of ecologic concepts and principles that may operate to influence the assembly and dynamic operations of our gut microbial communities.
Materials and Methods
Genome Comparisons.
All nucleotide sequences from all contigs of completed genome assemblies containing both capillary sequencing and pyrosequencer data, produced as part of the HGMI, were downloaded from the Washington University Genome Sequencing Center's website (http://genome.wustl.edu/pub/organism/Microbes/Human_Gut_Microbiome) on September 27, 2007. The finished genome sequences of B. thetaiotaomicron VPI-5482, Bacteroides vulgatus ATCC 8482, and B. fragilis NCTC9343 were obtained from GenBank.
For comparison purposes, protein-coding genes were identified in all genomes using YACOP (32). Each proteome was assigned InterPro numbers and GO terms using InterProScan release 16.1. Statistical comparisons between genomes were carried out as described in ref. 4, using perl scripts that are available upon request from the authors.
GeneChip Analysis.
Previously described methods were used to isolate RNA from a 100- to 300-mg aliquot of frozen cecal contents, synthesize cDNA, and to biotinylate and hybridize the cDNAs to a custom bacterial GeneChip (21). The only modification was that in RNA isolation protocol, 0.1 mm zirconia/silica beads (Biospec Products) were used for lysis of bacterial cells in a bead beater (Biospec; 4-min run at highest speed). Genes in a given bacterial species that were differentially expressed in mono- versus biassociation experiments were identified using CyberT (default parameters) after probe masking and scaling with the MAS5 algorithm (Affymetrix; for details about the methods used to create the mask, see the Methods section of SI Text).
RNA was purified from proximal colon using Mini RNeasy kit (Qiagen) with on-column DNase digestion. Biotinylated cRNA targets were prepared from each sample (n = 4 per treatment group). cRNA was hybridized to Affymetrix Mouse Genome Mo430 2 GeneChips, and the resulting datasets analyzed using Probe Logarithmic Error Intensity Estimate method (PLIER + 16). Fold-changes and p-values were calculated using Cyber-t. Significance was defined by maintaining a FDR <1% using Benjamini–Hochberg correction (33).
Other Methods.
Details about bacterial culture, genome sequencing and finishing, animal husbandry, quantitative PCR assays of the level of colonization of the ceca of gnotobiotic mice, GeneChip design and masking, plus proteomic and metabolite assays of cecal contents are provided in SI Methods.
Supplementary Material
Acknowledgments.
We thank Maria Karlsson and David O'Donnell for help with gnotobiotic husbandry; Jan Crowley, Janaki Guruge, Jill Manchester, and Sabrina Wagoner for technical assistance; Ruth Ley for valuable comments during the course of this work; and Manesh Shaw for proteomics data-mining and computational aspects related to mass spectrometry. This work was supported by National Science Foundation Grant 0333284 and National Institutes of Health Grants DK30292, DK70977, DK52574, GM07200, and T32-AI07172 and by the Laboratory Directed Research Program of Oak Ridge National Laboratory.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE14686, 14709, 14737). The sequence reported in this paper has been deposited in the GenBank database [accession nos. CP001107 (ATCC 33656, Eubacterium rectale) and CP001104–CP001106 (ATCC 27750, E. eligens)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0901529106/DCSupplemental.
References
- 1.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shipman JA, Berleman JE, Salyers AA. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J Bacteriol. 2000;182:5365–5372. doi: 10.1128/jb.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjursell MK, Martens EC, Gordon JI. Functional genomic and metabolic studies of the adaptations of a prominent adult human gut symbiont, Bacteroides thetaiotaomicron, to the suckling period. J Biol Chem. 2006;281:36269–36279. doi: 10.1074/jbc.M606509200. [DOI] [PubMed] [Google Scholar]
- 6.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 7.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances the fitness and transmission of a saccharolytic human gut symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozupone CA, et al. The convergence of carbohydrate active gene repertoires in human gut microbes. Proc Natl Acad Sci USA. 2008;105:15076–15081. doi: 10.1073/pnas.0807339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 11.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brigham CJ, Malamy MH. Characterization of the RokA and HexA broad-substrate-specificity hexokinases from Bacteroides fragilis and their role in hexose and N-acetylglucosamine utilization. J Bacteriol. 2005;187:890–901. doi: 10.1128/JB.187.3.890-901.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 15.Barcenilla A, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan SH, et al. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (London) 2008;32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- 17.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duncan SH, Flint HJ. Proposal of a neotype strain (A1–86) for Eubacterium rectale. Int J Syst Evol Microbiol. 2008;58:1735–1736. doi: 10.1099/ijs.0.2008/004580-0. [DOI] [PubMed] [Google Scholar]
- 19.Cuff MA, Lambert DW, Shirazi-Beechey SP. Substrate-induced regulation of the human colonic monocarboxylate transporter, MCT1. J Physiol. 2002;539:361–371. doi: 10.1113/jphysiol.2001.014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F, et al. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol. 2008;190:843–850. doi: 10.1128/JB.01417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Candido EP, Reeves R, Davie JR. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978;14:105–113. doi: 10.1016/0092-8674(78)90305-7. [DOI] [PubMed] [Google Scholar]
- 23.Tabuchi Y, et al. Genetic networks responsive to sodium butyrate in colonic epithelial cells. FEBS Lett. 2006;580:3035–3041. doi: 10.1016/j.febslet.2006.04.048. [DOI] [PubMed] [Google Scholar]
- 24.Joseph J, et al. Expression profiling of sodium butyrate (NaB)-treated cells: Identification of regulation of genes related to cytokine signaling and cancer metastasis by NaB. Oncogene. 2004;23:6304–6315. doi: 10.1038/sj.onc.1207852. [DOI] [PubMed] [Google Scholar]
- 25.Lecona E, et al. Upregulation of annexin A1 expression by butyrate in human colon adenocarcinoma cells: Role of p53, NF-Y, and p38 mitogen-activated protein kinase. Mol Cell Biol. 2008;28:4665–4674. doi: 10.1128/MCB.00650-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roediger WE. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterol. 1982;83:424–429. [PubMed] [Google Scholar]
- 27.Comalada M, et al. The effects of short-chain fatty acids on colon epithelial proliferation and survival depend on the cellular phenotype. J Cancer Res Clin Oncol. 2006;132:487–497. doi: 10.1007/s00432-006-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daly K, Shirazi-Beechey SP. Microarray analysis of butyrate regulated genes in colonic epithelial cells. DNA Cell Biol. 2006;25:49–62. doi: 10.1089/dna.2006.25.49. [DOI] [PubMed] [Google Scholar]
- 29.Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci USA. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duncan SH, GE, et al. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbell SP. Neutral theory and the evolution of ecological equivalence. Ecology. 2006;87:1387–1398. doi: 10.1890/0012-9658(2006)87[1387:ntateo]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 32.McHardy AC, Goesmann A, Puhler A, Meyer F. Development of joint application strategies for two microbial gene finders. Bioinformatics. 2004;20:1622–1631. doi: 10.1093/bioinformatics/bth137. [DOI] [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Methodol. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.