Abstract
Neural developmental programs require a high level of coordination between the decision to exit cell cycle and acquisition of cell fate. The Maf-family transcription factor NRL is essential for rod photoreceptor specification in the mammalian retina as its loss of function converts rod precursors to functional cones. Ectopic expression of NRL or a photoreceptor-specific orphan nuclear receptor NR2E3 completely suppresses cone development while concurrently directing the post-mitotic photoreceptor precursors towards rod cell fate. Given that NRL and NR2E3 have overlapping functions and NR2E3 expression is abolished in the Nrl−/− retina, we wanted to clarify the distinct roles of NRL and NR2E3 during retinal differentiation. Here, we demonstrate that NRL binds to a sequence element in the Nr2e3 promoter and enhances its activity synergistically with the homeodomain protein CRX. Using transgenic mice, we show that NRL can only partially suppress cone development in the absence of NR2E3. Gene profiling of retinas from transgenic mice that ectopically express NR2E3 or NRL in cone precursors reveals overlapping and unique targets of these two transcription factors. Together with previous reports, our findings establish the hierarchy of transcriptional regulators in determining rod versus cone cell fate in photoreceptor precursors during the development of mammalian retina.
Keywords: Retina, Development, Transcription Factor, Maf, Gene Regulation, Cell Fate Determination
INTRODUCTION
The central nervous system is assembled from thousands of distinct cell types that must be produced in correct numbers and within appropriate spatial and temporal constraints [19,20,48]. Unraveling the molecular mechanisms that control specification of diverse cell types during neurogenesis has been the subject of intense scrutiny. The vertebrate retina offers a convenient paradigm to examine cell fate decisions, as it is relatively less complex and more accessible to experimental manipulations. Birth-dating studies reveal that six neuronal cell types and one type of glia are produced from common pools of neuroepithelial progenitors in a conserved order of birth [1,7,34,35,53,54,57]. Retinal ganglion cells are generated first, followed by cone, amacrine and horizontal cells, whereas rods, bipolar cells and Muller glia are generated later [7,45,60]. A competence model has been proposed to explain the sequential birth order [10,34]; according to this, multipotent pools of progenitor cells go through discrete competence states during which they can only give rise to specific subsets of neurons. Restriction(s) in developmental potential can occur prior to or after exit from the final cell cycle through combinatorial regulatory mechanism(s) involving intrinsic factors and/or extrinsic signaling molecules [8,21,31]. As considerable overlap exists in the timings of cell birth, specific instructions and stringent controls are required to produce particular subtypes of neurons from a set of progenitor cells.
Rod and cone photoreceptors are specialized light-sensing neurons with highly structured membrane disks (called outer segments), which contain visual pigments and other phototransduction components. In most mammals (including humans and mice), rod photoreceptors greatly outnumber cones and generally account for over 70% of all cells in the retina [7]. Although the morphology and physiology of photoreceptors are well documented, the developmental pathways from a multipotent retinal progenitor to a committed precursor and a terminally-differentiated photoreceptor are only beginning to be elucidated.
NRL belongs to the basic motif-leucine zipper family of transcription factors and is an essential regulator of early events leading to the birth and development of rod photoreceptors [4,37,40,47,51]. It is preferentially expressed in rods (and not other retinal cells) and pineal gland [4,50]. In mice lacking NRL, rods are transformed to cones that share morphological, molecular, and electrophysiological characteristics similar to wild type cones [18,37,39]. More recently, gain of function studies reveal that NRL can influence all photoreceptor precursors to initiate a rod differentiation program at the expense of cones [40]. Despite the absence of cones, cone bipolar and horizontal interneurons are present in the adult retina but do not attain appropriate neuronal morphology during synaptogenesis [40,46]. CRX is a photoreceptor-specific homeodomain protein that plays a critical role in the maturation of photoreceptors [14,23], but it does not appear to be essential for initial specification events [24]. Mis-expression studies in adult iris cells [2] suggest that CRX acts as a photoreceptor competence factor before NRL defines rod identity. Control of photoreceptor cell fate also involves the participation of NR2E3, a photoreceptor-specific orphan nuclear receptor [3,12,16,26,42]. The rd7 mice carrying an antisense L1 insertion into exon 5 of the Nr2e3 gene exhibit a progressive photoreceptor degeneration accompanied by 1.5–2 fold increase in the number of S-cones [3,13,27,55]. Ectopic expression of NR2E3 or NRL [15,40] in the photoreceptor precursors of Nrl−/− mice results in the complete inhibition of cone developmental program [15]; however, in contrast to NRL [40], functional rods are not generated by NR2E3 expression alone [15].
Given that NRL and NR2E3 functions are overlapping and NR2E3 expression is undetectable in the Nrl−/− mice [15,36,37,40] it has been suggested that NR2E3 is downstream of NRL in transcriptional hierarchy controlling retinal development [37]. In this report, we have examined whether NR2E3 is a direct target of NRL and evaluated the precise role NRL in cone specification in the absence of NR2E3. We also present expression profiles of retinas from transgenic mice that ectopically express either NRL and NR2E3 or NR2E3 alone in cone precursors, with a goal to identify cone-enriched genes in mature photoreceptors.
RESULTS
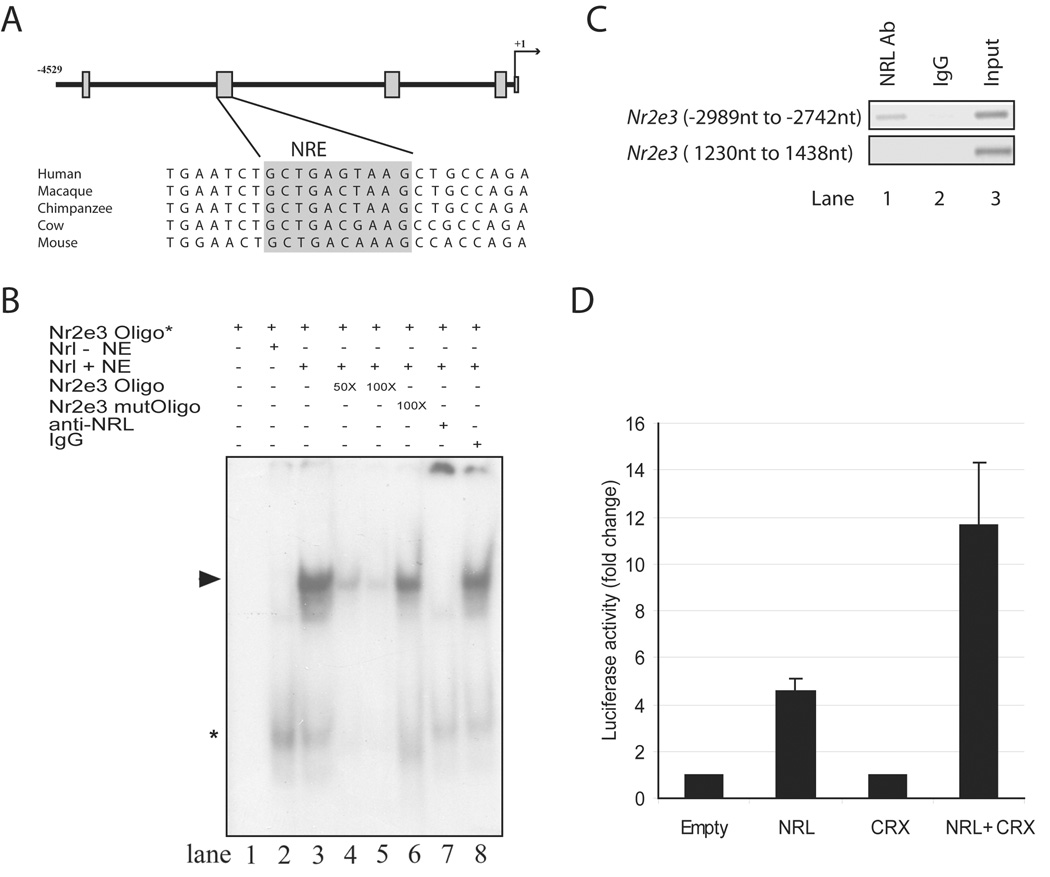
NRL directly binds to the Nr2e3 promoter
To determine whether NRL can modulate NR2E3 expression, we first analyzed the promoter of the Nr2e3 gene and identified four sequence regions that are conserved in mammals (Figure 1 A). In silico analysis revealed a putative NRL response element (NRE) in one of the conserved regions (see Figure 1 A, grey box). Addition of nuclear extracts from COS-1 cells expressing the NRL protein, but not from mock-transfected cells, to 32P-labeled NRE oligonucleotide resulted in band-shift in electrophoretic mobility shift assays (EMSA) (Figure 1 B; lanes 1–3), suggesting the binding of NRL to NRE sequence in the Nr2e3 promoter region. The specificity of binding was substantiated by competition with an excess of unlabeled oligonucleotide spanning the NRE but not with a mutant sequence (lanes 4–6). The major shifted band (shown by the arrowhead) was clearly detectable upon the addition of rabbit IgG but not anti-NRL antibody (lanes 7, 8), providing further evidence in support of NRL’s binding to Nr2e3-NRE. To determine whether NRL could bind the Nr2e3 promoter in vivo, we performed chromatin immunoprecipitation (ChIP) experiments. Cross-linked protein-DNA complexes from wild-type adult retinas were immunoprecipitated with anti-NRL antibody, and purified ChIP DNA was used for PCR with primers flanking the Nr2e3-NRE site. A strong enrichment of the Nr2e3-NRE promoter fragment was observed with anti-NRL antibody compared to rabbit IgG (Figure 1 C). Additionally, no significant enrichment was detected for another randomly-selected sequence in the Nr2e3 gene (negative control) (Figure 1 C).
Figure 1. Binding to and activation of the Nr2e3 promoter by NRL.
(A) Schematic of approximately 4.5 kb genomic DNA upstream of the Nr2e3 transcription start site (denoted as +1). The four boxes indicate sequence regions conserved in mammals. A comparison of sequences in the second conserved region including a putative NRE (highlighted in grey) is shown below the schema. (B) EMSA showing the binding of NRL to NRE site in the Nr2e3 promoter. Lanes are as indicated above the autoradiograph. Nr2e3 oligo* indicates 32P-labeled NRE oligonucleotide (−2820 nt to −2786 nt) (in all lanes). Nrl – NE shows 10 µg nuclear extract from untransfected COS-1 cells (lane 2), whereas Nrl + NE means 10 µg nuclear extract from COS-1 cells transfected with Nrl cDNA expression plasmid (lanes 3–8). Lane 4 and 5 included 50- or 100 fold molar excess of unlabeled NRE oligonucleotide. Lane 6 included 100-fold molar excess of unlabeled mutant NRE oligonucleotide. Lane 7 contains 2 µg anti-NRL antibody, whereas lane 8 has 2 µg normal rabbit IgG. Arrowhead represents the specific shifted band, which is undetectable when anti-NRL antibody is included. Asterisk indicates a shifted band (of low molecular mass) that does not seem to be altered by the addition of anti-NRL antibody. These experiments were performed three times, and similar results were obtained. (C) PCR assays using immunoprecipitated chromatin from adult C57BL/6J retinas. Lane1, NRL antibody used for IP; lane 2, normal rabbit IgG used for IP (negative control); lane 3, input DNA used as template. Top panel: primers amplifying the NRE containing region (−2989 nt to −2742 nt) in the Nr2e3 promoter region were used for PCR. Bottom panel: primers amplifying an irrelevant region (1230 nt to 1438 nt) in the Nr2e3 gene were used for PCR. (D) Luciferase reporter assays showing the activation of Nr2e3 promoter by NRL and CRX.
NRL induces the Nr2e3 promoter activity in transfected cells
We then examined the activity of a 4.5 kb Nr2e3 promoter fragment (encompassing the conserved NRE sequence; see Figure 1 A) in the presence of NRL. Transfection of HEK-293 cells with NRL, but not CRX, expression plasmid induced the luciferase reporter activity that was driven by the Nr2e3 promoter (Figure 1 D). Co-transfection of HEK-293 cells with both NRL and CRX plasmids resulted in further increase of the Nr2e3 promoter activity (Figure 1 D). This is consistent with previously-reported synergistic activation of several rod-specific genes by NRL and CRX [14,16,38,44].
Overlapping yet distinct gene profiles are generated by NRL and NR2E3
Recent investigations into the role of NRL and NR2E3 [12,15,29,40,42] and our findings reported here (Figure 1) suggest that NRL suppresses cone differentiation by directly signaling through NR2E3. This level of regulation also implies that many molecular defects observed in mice lacking functional NR2E3 (e.g., the rd7 mouse) are also present in the Nrl−/− mice [17,37]. To dissect the transcriptional activity of NRL versus NR2E3 in mature photoreceptors, we took advantage of two recently-generated transgenic mouse models – Crxp-Nrl/WT [40] and Crxp-Nr2e3/WT [15]. In these mice, a 2 kb Crx proximal promoter [22] leads to the expression of NRL or NR2E3 in photoreceptor precursors and transformation of cones to rod photoreceptors, without any obvious perturbation in retinal lamination or development of other cell types [15,40].
In the Crxp-Nrl/WT retinas, NRL and consequently NR2E3 ([40], see Fig. 1) are ectopically expressed in cone precursors; while only NR2E3 (and not NRL) is ectopically expressed in cone precursors of the Crxp-Nr2e3/WT retina. NRL and NR2E3 are also expressed in the developing rod precursors of both transgenic lines. Therefore, gene profiling of retinas from Crxp-Nrl/WT and Crxp-Nr2e3/WT mice can reveal expression changes induced by NRL+NR2E3 or NR2E3 alone in cone precursors, respectively. Retinal RNA from P28 adult mice was hybridized to Affymetrix MOE430.2.0 GeneChips, which contain 45,101 probesets for mouse transcripts. A comparative analysis of gene clusters from Crxp-Nrl/WT and Crxp-Nr2e3/WT retinas to WT samples revealed a number of genes involved in diverse signaling pathways and transcriptional regulation; Table 1 shows the genes with FDRCI P-value of <0.1 and a fold change >4. In addition to established cone-specific genes, we also discovered several new genes down-regulated in the Crxp-Nrl/WT and Crxp-Nr2e3/WT coneless groups which are potential cone-enriched target genes. We then compared Crxp-Nrl/WT and Crxp-Nr2e3/WT gene profiles to Nrl−/− (cone-only) and rd7 (1.5–2 fold more S-cones) profiles. Many cone phototransduction genes that are up-regulated in the Nrl−/− (cone-only, Table 2) and rd7 (1.5–2 fold more S-cones, Table 3) retinas are also significantly repressed in the Crxp-Nrl/WT and Crxp-Nr2e3/WT coneless samples. Gene expression changes showing FDRCI P-value < 0.1 and a fold change > 10 are listed in Table 2 and Table 3.
Table 1.
Non-redundant differentially expressed genes in Crxp-Nrl/WT or Crxp-Nr2e3/WT samples compared to WT retinas. Gene profiles of P28 retinal samples from Crxp-Nrl/WT or Crxp-Nr2e3/WT mice were compared to those from the WT retina. Common genes in Crxp-Nrl/WT and Crxp-Nr2e3/WT, or unique genes from Crxp-Nrl/WT or Crxp-Nr2e3/WT with a minimum fold change of 4 and FDRCI P-value of < 0.1 are shown. AFC, average fold change.
| Gene Symbol | Gene Title | AFC Crxp-Nrl/WT versus WT | AFC Crxp-Nr2e3/WT versus WT | GO biological process description |
|---|---|---|---|---|
| Overlapping genes in Crxp-Nrl/WT and Crxp-Nr2e3/WT versus WT group | ||||
| Gdpd3 | Glycerophosphodiester phosphodiesterase domain containing 3 | 21.4 | 4.6 | Alcohol metabolic process |
| Sgcg | Sarcoglycan, gamma (dystrophin-associated glycoprotein) | 14.8 | 12.1 | Cytoskeleton organization and biogenesis |
| Cap1 | CAP, adenylate cyclase-associated protein 1 (yeast) | 10.2 | 2.1 | Actin cytoskeleton organization and biogenesis |
| Plekhk1 | Pleckstrin homology domain containing, family K member 1 | 6.3 | 4.7 | Regulation of cell proliferation |
| Tsc22d1 | TSC22 domain family, member 1 | 6.0 | 6.5 | Regulation of transcription |
| Paip1 | Polyadenylate binding protein-interacting protein 1 | 4.4 | 2.6 | Regulation of translation |
| Huwe1 | HECT, UBA and WWE domain containing 1 | 4.1 | 3.0 | DNA packaging |
| Abca13 | ATP-binding cassette, sub-family A (ABC1), member 13 | −4.0 | −6.5 | Intracellular protein transport |
| Clca3 | Chloride channel calcium activated 3 | −4.5 | −4.9 | Chloride transport |
| Camk2b | Calcium/calmodulin-dependent protein kinase II, beta | −5.1 | −20.0 | Regulation of phosphorylation |
| Gnat2 | Guanine nucleotide binding protein, alpha transducing 2 | −8.3 | −10.1 | Phototransduction |
| Pttg1 | Pituitary tumor-transforming 1 | −10.3 | −4.2 | Meiotic chromosome segregation |
| Pde6c | Phosphodiesterase 6C, cGMP specific, cone, alpha prime | −15.9 | −13.8 | Activation of MAPK activity/visual perception |
| Opn1mw | Opsin 1 (cone pigments), medium-wave- sensitive (color blindness, deutan) | −38.7 | −37.6 | Phototransduction |
| Arr3 | Arrestin 3, retinal | −56.3 | −31.1 | Regulation of phosphorylation |
| Pde6h | Phosphodiesterase 6H, cGMP-specific, cone, gamma | −75.6 | −70.9 | Activation of MAPK activity/visual perception |
| Opn1sw | Opsin 1 (cone pigments), short-wave- sensitive (color blindness, tritan) | −124.0 | −98.4 | Phototransduction |
| Unique genes in Crxp-Nrl/WT versus WT group | ||||
| Rds | Retinal degeneration, slow (retinitis pigmentosa 7) | 56.5 | 1.5 | Visual perception |
| Nudt21 | Nudix (nucleoside diphosphate linked moiety X)-type motif 21 | 10.3 | 1.1 | |
| Deadc1 | Deaminase domain containing 1 | 6.6 | 1.9 | tRNA processing |
| Atp2b2 | ATPase, Ca++ transporting, plasma membrane 2 | 5.6 | −1.1 | Inner ear development |
| Scd2 | Stearoyl-Coenzyme A desaturase 2 | 5.5 | −1.2 | Fatty acid biosynthetic process |
| Atp6v0a1 | ATPase, H+ transporting, lysosomal V0 subunit A1 | 5.2 | −1.1 | Transcription initiation |
| Stk35 | Serine/threonine kinase 35 | 5.2 | 1.2 | Protein amino acid phosphorylation |
| Uhmk1 | U2AF homology motif (UHM) kinase 1 | 4.3 | −1.3 | Protein amino acid phosphorylation |
| Gucy2e | Guanylate cyclase 2e | 4.3 | 1.4 | Visual perception |
| Thbs1 | Thrombospondin 1 | 4.2 | 1.1 | Blood vessel morphogenesis |
| Fabp4 | Fatty acid binding protein 4, adipocyte | −4.0 | 1.2 | Regulation of protein kinase activity |
| 5730410E15Rik | RIKEN cDNA 5730410E15 gene | −4.1 | −1.2 | |
| 5330426P16Rik | RIKEN cDNA 5330426P16 gene | −4.1 | −1.4 | |
| Ssbp2 | Single-stranded DNA binding protein 2 | −4.3 | −1.0 | Regulation of transcription |
| Ing3 | Inhibitor of growth family, member 3 | −4.4 | 1.0 | Regulation of transcription |
| Dmd | Dystrophin, muscular dystrophy | −4.4 | 1.1 | Peptide biosynthetic process |
| A930033H14Rik | RIKEN cDNA A930033H14 gene | −4.7 | 1.4 | |
| Aff1 | AF4/FMR2 family, member 1 | −5.0 | −1.3 | Regulation of transcription |
| Vapb | Vesicle-associated membrane protein, associated protein B and C | −5.1 | −1.2 | |
| Scn2b | Sodium channel, voltage-gated, type II, beta | −8.2 | −1.1 | Sodium ion transport |
| Unique genes in Crxp-Nr2e3/WT versus WT group | ||||
| Sox30 | SRY-box containing gene 30 | 1.8 | 6.8 | Regulation of transcription |
| 2900056M20Rik | RIKEN cDNA 2900056M20 gene | −1.9 | −6.6 | |
| LOC552908 | Hypothetical LOC552908 | −1.7 | −10.1 | |
Table 2.
Non-redundant differentially expressed genes in Crxp-Nrl/WT or Crxp-Nr2e3/WT samples compared to Nrl−/− retinas. Gene profiles of P28 retinal samples from Crxp-Nrl/WT or Crxp-Nr2e3/WT were compared to the profiles from the Nrl−/− retina. We show common differentially expressed genes in Crxp-Nrl/WT and Crxp-Nr2e3/WT retina, or unique genes from Crxp-Nrl/WT or Crxp-Nr2e3/WT, with a minimum fold change of 10 and FDRCI P-value of < 0.1. AFC, average fold change.
| Gene Symbol | Gene Title | AFC Crxp-Nrl/WT versus Nrl−/− | AFC Crxp- Nr2e3/WT versus Nrl−/− | GO biological process description |
|---|---|---|---|---|
| Overlapping genes in Crxp-Nrl/WT and Crxp-Nr2e3/WT versus Nrl−/− group | ||||
| Nrl | Neural retina leucine zipper gene | 386.9 | 381.8 | Photoreceptor cell development |
| Rho | Rhodopsin | 347.1 | 332.5 | Phototransduction |
| Nr2e3 | Nuclear receptor subfamily 2, group E, member 3 | 115.8 | 83.9 | Photoreceptor cell development |
| Gnb1 | Guanine nucleotide binding protein, beta 1 | 69.7 | 46.1 | Phototransduction |
| Slc24a1 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 | 50.9 | 43.3 | Calcium ion transport/visual perception |
| A930036K24Rik | RIKEN cDNA A930036K24 gene | 50.1 | 88.7 | |
| BC016201 | cDNA sequence BC016201 | 40.8 | 33.8 | |
| Esrrb | Estrogen related receptor, beta | 32.4 | 26.9 | Intracellular protein transport |
| Susd3 | Sushi domain containing 3 | 23.4 | 21.5 | |
| Aqp1 | Aquaporin 1 | 21.3 | 31.4 | Water transport |
| BC038479 | cDNA sequence BC038479 | 20.4 | 17.5 | |
| Reep6 | Receptor accessory protein 6 | 17.9 | 20.9 | Protein binding |
| Mef2c | Myocyte enhancer factor 2C | 16.7 | 10.8 | Regulation of transcription |
| Pde6b | Phosphodiesterase 6B, cGMP, rod receptor, beta polypeptide | 16.7 | 19.4 | Activation of MAPK activity/Visual perception |
| Wisp1 | WNT1 inducible signaling pathway protein 1 | 16.1 | 20.3 | Regulation of cell growth |
| Sh2d1a | SH2 domain protein 1A | 15.8 | 18.7 | Intracellular signaling cascade |
| Sgcg | Sarcoglycan, gamma (dystrophin-associated glycoprotein) | 14.6 | 10.6 | Cytoskeleton organization and biogenesis |
| Samd11 | Sterile alpha motif domain containing 11 | 13.9 | 16.5 | Negative regulation of transcription |
| Plekha2 | Pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 | 11.4 | 14.7 | Phosphatidylinositol binding |
| Vax2os1 | Vax2 opposite strand transcript 1 | 10.3 | 15.6 | |
| Guca1b | Guanylate cyclase activator 1B | 10.2 | 10.6 | Phototransduction |
| Gulo | Gulonolactone (L-) oxidase | −10.0 | −31.3 | L-ascorbic acid biosynthetic process |
| Pik3ap1 | Phosphoinositide-3-kinase adaptor protein 1 | −11.1 | −11.3 | Transmembrane receptor protein tyrosine kinase signaling protein activity |
| Gngt2 | Guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 | −11.7 | −11.9 | G-protein coupled receptor protein signaling pathway |
| En2 | Engrailed 2 | −12.0 | −10.4 | Regulation of transcription |
| Myocd | Myocardin | −12.5 | −12.7 | Regulation of cell growth |
| Kcne2 | Potassium voltage-gated channel, Isk-related subfamily, gene 2 | −12.7 | −12.4 | Potassium ion transport |
| Arhgdib | Rho, GDP dissociation inhibitor (GDI) beta | −13.8 | −12.8 | Rho GDP-dissociation inhibitor activity |
| Parvb | Parvin, beta | −13.9 | −14.0 | Actin binding |
| Cckbr | Cholecystokinin B receptor | −17.4 | −33.3 | G-protein coupled receptor protein signaling pathway |
| Klhl4 | Kelch-like 4 (Drosophila) | −19.8 | −17.4 | |
| A930009A15Rik | RIKEN cDNA A930009A15 gene | −24.4 | −28.9 | |
| Otop3 | Otopetrin 3 | −39.2 | −39.3 | |
| Cngb3 | Cyclic nucleotide gated channel beta 3 | −40.8 | −49.4 | Potassium ion transport |
| Gnat2 | Guanine nucleotide binding protein, alpha transducing 2 | −46.0 | −56.2 | Phototransduction |
| Fabp7 | Fatty acid binding protein 7, brain | −60.1 | −48.8 | Lipid binding |
| Opn1mw | Opsin 1 (cone pigments), medium-wave- sensitive (color blindness, deutan) | −77.9 | −76.2 | Phototransduction |
| Clca3 | Chloride channel calcium activated 3 | −91.6 | −93.5 | Chloride transport |
| Pde6c | Phosphodiesterase 6C, cGMP specific, cone, alpha prime | −158.3 | −136.8 | Activation of MAPK activity/visual perception |
| Arr3 | Arrestin 3, retinal | −271.2 | −166.0 | Regulation of phosphorylation |
| Pde6h | Phosphodiesterase 6H, cGMP-specific, cone, gamma | −429.1 | −409.3 | Activation of MAPK activity/visual perception |
| Opn1sw | Opsin 1 (cone pigments), short-wave- sensitive (color blindness, tritan) | −559.2 | −546.8 | Phototransduction |
| Unique genes in Crxp-Nrl/WT versus Nrl−/− group | ||||
| Rds | Retinal degeneration, slow (retinitis pigmentosa 7) | 149.7 | 3.3 | Sensory perception of light stimulus |
| Stk35 | Serine/threonine kinase 35 | 11.7 | 2.3 | Protein amino acid phosphorylation |
| Mtmr7 | Myotubularin related protein 7 | −11.8 | 1.8 | Phospholipid dephosphorylation |
| Pcdh15 | Protocadherin 15 | −12.6 | −3.3 | Sensory perception of light stimulus |
| Pip5k2b | Phosphatidylinositol-4-phosphate 5-kinase, type II, beta | −12.6 | 1.3 | Glycerophospholipid metabolic process |
| Unique genes in Crxp-Nr2e3/WT versus Nrl−/− group | ||||
| A930003C13Rik | RIKEN cDNA A930003C13 gene | 3.5 | 11.5 | |
| Skiv2l2 | Superkiller viralicidic activity 2-like 2 (S. cerevisiae) | −4.4 | −17.0 | RNA splicing |
Table 3.
Non-redundant differentially expressed genes in Crxp-Nrl/WT or Crxp-Nr2e3/WT samples compared to rd7 retinas. Gene profiles of P28 retinal samples from Crxp-Nrl/WT or Crxp-Nr2e3/WT were compared to those of rd7 retina. Common genes in Crxp-Nrl/WT and Crxp-Nr2e3/WT, or unique genes from Crxp-Nrl/WT or Crxp-Nr2e3/WT with a minimum fold change of 10 and FDRCI P-value of < 0.1 are shown. AFC, average fold change.
| Gene Symbol | Gene Title | AFC Crxp-Nrl/WT versus rd7 | AFC Crxp-Nr2e3/WT versus rd7 | GO biological process description |
|---|---|---|---|---|
| Overlapping genes in Crxp-Nrl/WT and Crxp-Nr2e3/WT versus rd7 group | ||||
| Eif2s3y | Eukaryotic translation initiation factor 2, subunit 3, structural gene Y-linked | 65.3 | 79.1 | Macromolecule biosynthetic process |
| Ddx3y | DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked | 62.1 | 74.0 | |
| Sgcg | Sarcoglycan, gamma (dystrophin-associated glycoprotein) | 14.8 | 10.7 | Cytoskeleton organization and biogenesis |
| Jarid1d | Jumonji, AT rich interactive domain 1D (Rbp2 like) | 11.8 | 11.2 | Regulation of transcription |
| LOC640072 /// | Hypothetical protein LOC640072 /// | −11.3 | −12.6 | |
| LOC677194 | Hypothetical protein LOC677194 | |||
| A230097K15Rik | RIKEN cDNA A230097K15 gene | −12.7 | −11.9 | |
| Arhgdib | Rho, GDP dissociation inhibitor (GDI) beta | −12.8 | −11.9 | Rho GDP-dissociation inhibitor activity |
| Gulo | Gulonolactone (L-) oxidase | −13.6 | −42.6 | L-ascorbic acid biosynthetic process |
| Socs3 | Suppressor of cytokine signaling 3 | −15.8c | −11.6 | Regulation of phosphorylation |
| Bub1b | Budding uninhibited by benzimidazoles 1 homolog, beta (S. cerevisiae) | −20.1 | −14.7 | Regulation of phosphorylation |
| Edn2 | Endothelin 2 | −20.1 | −11.1 | Regulation of vasoconstriction |
| Otop3 | Otopetrin 3 | −28.1 | −28.1 | |
| Fabp7 | Fatty acid binding protein 7, brain | −37.7 | −30.6 | Lipid binding |
| A930009A15Rik | RIKEN cDNA A930009A15 gene | −38.0 | −45.1 | |
| Gnat2 | Guanine nucleotide binding protein, alpha transducing 2 | −39.7 | −48.5 | Phototransduction |
| Opn1mw | Opsin 1 (cone pigments), medium-wave- sensitive (color blindness, deutan) | −40.5 | −39.6 | Phototransduction |
| Arr3 | Arrestin 3, retinal | −50.1 | −25.3 | Regulation of phosphorylation |
| Clca3 | Chloride channel calcium activated 3 | −78.5 | −80.1 | Chloride transport |
| Pde6c | Phosphodiesterase 6C, cGMP specific, cone, alpha prime | −127.0 | −109.7 | Activation of MAPK activity/visual perception |
| Opn1sw | Opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) | −223.2 | −218.3 | Phototransduction |
| Pde6h | Phosphodiesterase 6H, cGMP-specific, cone, gamma | −365.1 | −348.2 | Activation of MAPK activity/visual perception |
| Unique genes in Crxp-Nrl/WT versus rd7 group | ||||
| Rds | Retinal degeneration, slow (retinitis pigmentosa 7) | 74.7 | 1.6 | Sensory perception of light stimulus |
| Cap1 | CAP, adenylate cyclase-associated protein 1 (yeast) | 11.3 | 2.3 | Actin cytoskeleton organization and biogenesis |
| Scn2b | Sodium channel, voltage-gated, type II, beta | −10.2 | −1.3 | Sodium ion transport |
| Fabp4 | Fatty acid binding protein 4, adipocyte | −11.3 | −2.4 | Regulation of protein kinase activity |
| Mtmr7 | Myotubularin related protein 7 | −27.7 | −1.3 | Phospholipid dephosphorylation |
| Unique genes in Crxp-Nr2e3/WT versus rd7 group | ||||
| Camk2b | Calcium/calmodulin-dependent protein kinase II, beta | −3.1 | −12.2 | Regulation of phosphorylation |
| LOC552908 | Hypothetical LOC552908 | −2.4 | −14.0 | |
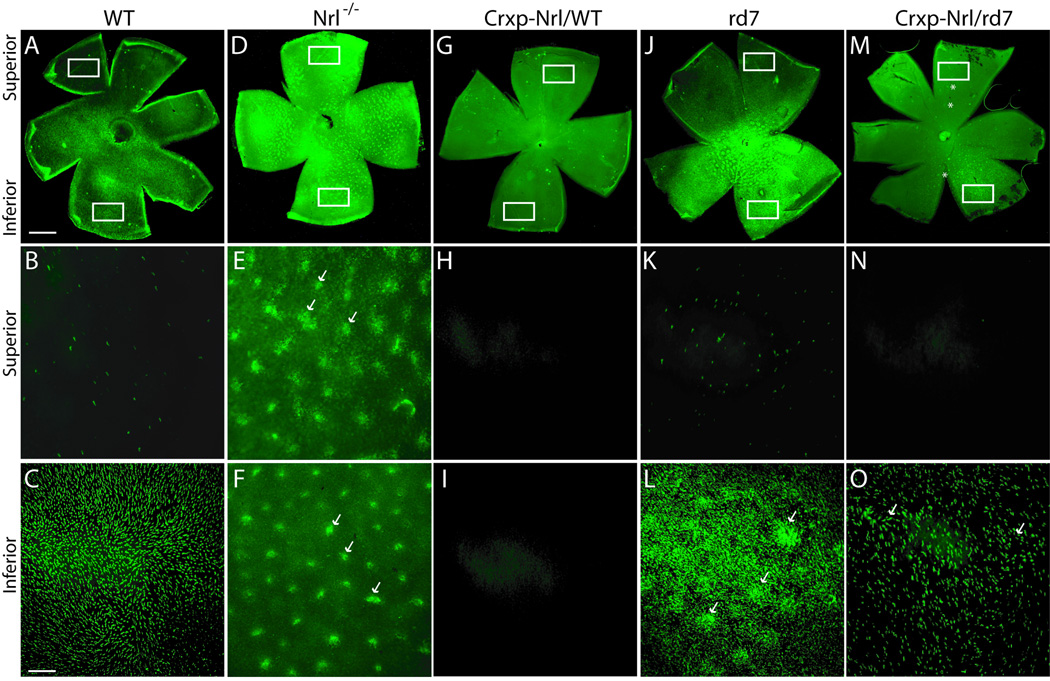
Expression of NRL can only suppress a subset of S-cones in the absence of NR2E3
Similarities in gene profiles of Crxp-Nrl/WT and Crxp-Nr2e3/WT retinas raise the question whether NRL can suppress cone gene expression and differentiation even in the absence of NR2E3. To evaluate this, we mated Crxp-Nrl/WT mice to rd7 mice to generate a transgenic mouse line (Crxp-Nrl/rd7) that expresses NRL, but not NR2E3, in both cone and rod precursors. We first analyzed cone markers, such as S- and M-opsin, in retinal whole mounts. As previously demonstrated [5], we observed an inferior to superior gradient of S-opsin expression (Figure 2 A–C) and a superior to inferior gradient of M-opsin in the WT mice (data not shown). As predicted, S-opsin was detected throughout in the Nrl−/− retinal whole mounts (Figure 2 D–F) and increased S-opsin staining was observed in the rd7 retinas (Figure 2 J–L); however, both S-opsin and M-opsin could not be detected in Crxp-Nrl/WT retinas (Figure 2 G–I, and data not shown). In both Nrl−/− and rd7 mice, whorls are detected in the whole mount preparations (Figure 2 D–F and J–K). In Crxp-Nrl/rd7 retinal whole mounts, we observed a large absence of S-opsin staining in the superior domain (Figure 2 M, O) yet detected a small population of S-opsin positive cells in the inferior retina (Figure 2 M, N). The expression of M-opsin was unaltered (data not shown), and whorls could be detected throughout the retinas (Figure 2 M–O).
Figure 2. Incomplete suppression of S-opsin expression by NRL in the absence of NR2E3.
(A–C) WT retina, showing superior to inferior gradient of S-opsin expression. (D–F) Nrl−/− retina. In the absence of NRL and NR2E3, whorls (arrows) and S-opsin can be detected throughout the retina. (G–I) Crxp-Nrl/WT retina. Ectopic expression of NRL in early cone precursors results in the complete absence of S-opsin. (J–L) rd7 retina. In the absence of functional NR2E3, enhanced S-opsin expression and whorls (arrows) are observed in both superior and inferior domain. (M–O) Crxp-Nrl/rd7 retina. In the presence of NRL but absence of NR2E3, expression of S-opsin is reduced but detectable in the inferior domain. Asterisks are positioned at 3’o clock relative to the whorls (M). Arrows indicate the irregular S-opsin staining of whorls (O).
Scale bar: 200 µm (A, D, G, J, M), and 50 µm (B, C, E, F, H, I, K, L, N, O).
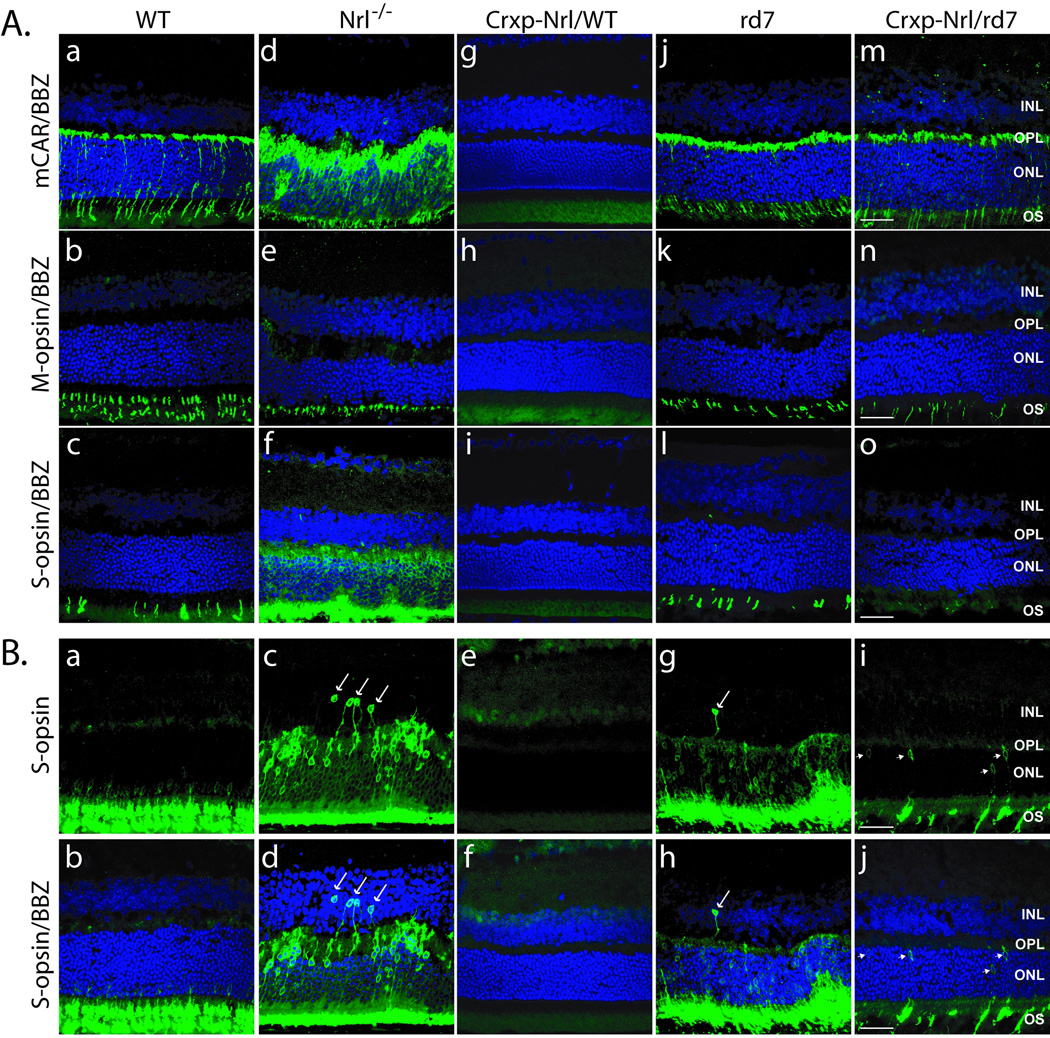
As shown previously [27,37,40], the number of cone arrestin (mCAR) and S-opsin positive cells in retinal cross-sections from Nrl−/− and rd7 retinas were increased compared to WT, and there is an absence of cone-specific markers in Crxp-Nrl/WT mice (Figure 3 A: a–o). In Crxp-Nrl/rd7 sections, we observed normal cone arrestin and M-opsin staining but an absence of S-opsin in the superior domain (Figure 3 A: m–o). In the inferior domain, we identified a few S-opsin positive cones and many S-opsin positive cell bodies at the inner portion of the ONL (Figure 3 B: i, j). These findings were in contrast to S-opsin positive cell bodies distributed throughout the ONL and INL in Nrl−/− and rd7 retinas (Figure 3 B: c–d and g–h). The expression of cone arrestin and M-opsin in the Crxp-Nrl/rd7 mice (harboring the Crxp-Nrl transgene in rd7 background with no NR2E3 function) but not in the Crxp-Nrl/WT mice (harboring the Crxp-Nrl transgene in wild-type background) demonstrates that NR2E3 is the primary suppressor of cone gene expression and cone differentiation.
Figure 3. Expression of cone-specific markers and targeting of photoreceptors to the ONL.
(A) Superior, and (B) inferior regions of the retina showing staining for cone arrestin (mCAR), S-opsin, and M-opsin antibodies (as shown on the left). Mouse strains are indicated. Compared to WT (B: a–b) and Crxp-Nrl/WT (B: e–f), targeting of S-cones (arrows) to the ONL is perturbed in Nrl−/− (B: c–d) and rd7 retinas (B: g–h), and S-opsin positive nuclei are present in the INL. S-cone staining (arrowheads) in the Crxp-Nrl/rd7 retinas (B: i–j) is observed in cells closest to the outer plexiform layer. OS, outer segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; BBZ, bisbenzamide. Scale bar: 25 µm.
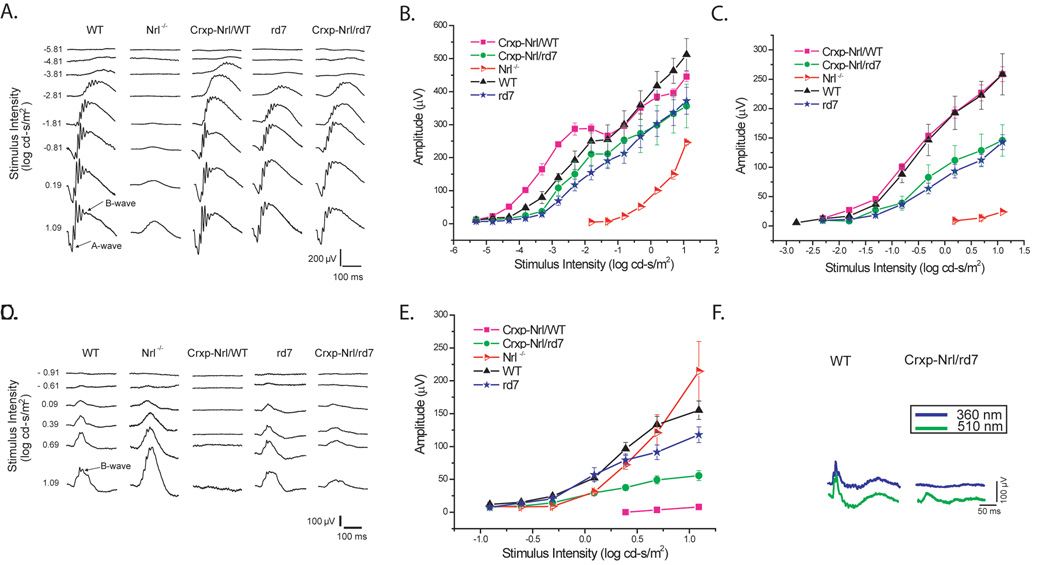
Cone function is detected but reduced in the Crxp-Nrl/rd7 mice
We performed electroretinography (ERG) recordings to measure the massed-field potential across the retina in the different transgenic lines. As reported previously [40], the ectopic expression of NRL in cone precursors (Crxp-Nrl/WT) resulted in an absence of cone-driven responses, whereas rod-driven components were preserved (Figure 4 A–E). To characterize the functionality of cone-driven neurons in the absence of NR2E3, we analyzed the photopic response from Crxp-Nrl/rd7 mice (Figure 4 C, D). In response to brief flashes of white light, we first detected a cone-driven b-wave at 0.09 log cd-s/m2. At the higher flash intensity of 1.09 log cd-s/m2 the maximum b-wave amplitude was about 40% of the WT and rd7 response amplitude (Figure 4 C, D).
Figure 4. Absence of normal cone function in cone photoreceptors expressing NRL but not NR2E3.
(A) Representative dark-adapted ERGs for increasing stimulus intensities are shown for WT, Nrl−/−, Crxp-Nrl/WT, rd7 and Crxp-Nrl/rd7 mice at two months age. Intensity-response functions for the (B) a-wave and (C) b-wave amplitude were plotted on log-linear coordinates. (D) Representative light-adapted ERGs waveforms with increasing stimulus intensity for WT, Nrl−/−, Crxp-Nrl/WT, rd7 and Crxp-Nrl/rd7 mice, as indicated. (E) Plots of the b-wave amplitude as a function of stimulus intensity for light-adapted conditions. At 2 months of age, there was no significant difference in the photopic response between WT and rd7 mice. (D–E) B-wave amplitude at the maximum intensity for the Crxp-Nrl/rd7 mice. A reduction of about 40% is observed from the WT and rd7 mice. (F) Representative light-adapted S- (360 nm) and M- (510 nm) cone ERGs showing a smaller M-cone response and undetectable S-cone response in Crxp-Nrl/rd7 mice compared to WT mice. Bars indicate ± standard error.
We further examined the photopic ERG in the Crxp-Nrl/rd7 transgenic mice by recording light-adapted cone-mediated responses at 360 nm and 510 nm to isolate S-cone and M-cone function, respectively (Figure 4 F). As predicted, the Nrl−/− mice showed an enhanced S-cone response when compared with WT mice [18,37]. There was no significant difference in the M-cone response amplitude between Nrl−/− and WT mice. We then recorded from Crxp-Nrl/rd7 mice and found that while the M-cone response was reduced by 40% from WT mice, S-cone responses were undetectable (Figure 4 F).
DISCUSSION
Regulatory networks defining rod versus cone identity are under the direct control of bZIP transcription factor NRL [37,40]. In this report, we demonstrate that NR2E3 is a direct transcriptional target of NRL and that specification of rod cell fate over cone differentiation is dictated by the activation of NR2E3 in response to NRL. Restricted expression of these two key transcriptional regulators in photoreceptor precursors is essential for proper development of rods. Ectopic expression of either protein in cone precursors can reprogram the cone development pathway to generate rod photoreceptors [15,40]. We had shown previously that ectopic NR2E3 expression can inhibit the development of functional S and M-cones in the Nrl−/− retina [15]. The current data suggest that NR2E3 is necessary to completely repress the development of M and some S-cones, and NRL alone can only repress a subset of S-cones. These genetic models therefore raise the possibility of heterogeneity within S-cones.
Several studies have indicated the association of NRL and NR2E3 with promoter elements of cone-specific genes [40,42,43]. In this report, we analyzed the relationship of NRL and NR2E3 in modulating the cone developmental program. Data from immunohistochemical and physiological studies presented herein suggest that NRL modulates the development of S-cones, and its gain or loss of function primarily results in alterations of the S-cone pathway. One possibility is that S-cones represent the “default fate” for photoreceptors in mice [9,52] and that the expression of NRL controls an important node for this process.
The presence of ectopic S-opsin cells in the INL of rd7 and Nrl−/− retinas is reminiscent of previous findings showing opsin-like immunoreactive cells in the developing retina [25]. Our study reveals the presence of ectopic S-opsin positive cells that persist and survive in the adult retinas from Nrl−/− and rd7 mice. What can account for the existence and survival of these neurons outside of their normal retinal photoreceptor layer? It is possible that NRL and NR2E3 dictate the expression of specific guidance cues that facilitate photoreceptor pathfinding to the vicinity of their appropriate target regions in a highly stereotyped and directed manner. Several candidate proteins that show an altered expression profile in the Nrl−/− retina appear to match the role of an axonal guidance cue [59,61]. These include members of families of secreted signaling molecules, such as Wingless/Wnt and Decapentaplegic/Bone Morphogenic Protein/Transforming Growth Factor B (Dpp/BMP/TGFb) [11], which appear to have important functions during retinal development [6,32,33,41,56,58,61]. We hypothesize that in the absence of NRL, and consequently NR2E3, changes in Wnt and BMP pathway may create noise in a homing signal that is required to (i) bring all photoreceptors to the ONL, and/or (ii) promote the appropriate wiring of rods and cones to bipolar and horizontal neurons. Although our current microarray experiments of P28 retinas did not reveal significant changes in classical pathfinding genes, future efforts will focus on profiling early postnatal stages of retinal development.
The absence of cones in the Crxp-Nrl/WT and Crxp-Nr2e3/WT retinas resulted in normal architecture and lamination features, similar to the WT. A lack of structural abnormalities has allowed us to profile expression changes that may be specifically due to the absence of one class of neurons (i.e., cones). While the retinal profiles of Crxp-Nrl/WT and Crxp-Nr2e3/WT mice had many common genes, the Crxp-Nrl/WT profile contained more unique changes in gene expression, consistent with NRL being upstream of NR2E3 in transcriptional hierarchy. One interesting novel gene revealed from the gene profiling experiments is PTTG1, which is down-regulated in the coneless retinas. The inhibitory chaperone PTTG1 has been implicated as a mitotic checkpoint gene involved at the metaphase-anaphase interface [63]. Elucidation of specific roles of PTTG1 and other cone-enriched genes will require further investigation.
In conclusion, our work refines the roles of NRL and NR2E3 during photoreceptor differentiation. We show, for the first time, that NR2E3 is a direct downstream target of NRL and that the correct sequential expression of these transcriptional regulators may be required for appropriate expression of rod-specific opsin and suppression of cone phototransduction genes during normal retinal development. Additional studies are needed to precisely define how specific down-stream targets of NRL and NR2E3 fine-tune the differentiation of functional photoreceptors from post-mitotic committed precursors.
EXPERIMENTAL PROCEDURES
Transgenic mice
The Crxp-Nrl/WT and Crxp-Nr2e3/WT mice were generated previously [15,40]. We mated Crxp-Nrl/WT mice with the rd7 mice (procured from Jackson Laboratory) to generate Crxp-Nrl/rd7 mice. The mice, used for analysis reported here, were in a mixed background of 129X1/SvJ and C57BL/6J. PCR primers for genotyping the Crxp-Nrl/WT allele are: F: 5’-AGCCAATGTCACCTCCTGTT-3’ and R: 5’-GGGCTCCCTGAATAGTAGCC-3’. PCR primers for genotyping the rd7 allele are as reported [27]. All studies involving mice were performed in accordance with institutional and federal guidelines and approved by the University Committee on Use and Care of Animals at the University of Michigan.
Gene profiling and analysis
The details of microarray analysis have been described earlier [59,61,62]. Briefly, total retinal RNA was isolated (Trizol, Invitrogen) from one P28 mice (two eyes) and used to generate double-stranded cDNA for hybridization to mouse GeneChips MOE430.2.0, per guidelines (Affymetrix). Retinal RNA from four independent WT, Nrl−/−, rd7, Crxp-Nrl/WT, and Crxp-Nr2e3/WT mice was used for each evaluation. RMA (Robust Multichip Average) was used to normalize and obtain gene expression scores [30]. Normalized data were subjected to two-stage analysis based on false discovery rate with confidence interval (FDRCI) which controls for both statistical and biological significance when identifying differentially expressed genes [62]. The differentially expressed genes were further classified into distinct functional categories using NCBI and FatiGO (http://fatigo.bioinfo.cipf.es).
Immunohistochemistry
Retinal whole mounts and 10 µm sections were probed with the following antibodies [16,49]: rabbit S-opsin, rabbit M-opsin, and rabbit cone-arrestin (mCAR; generous gift from C. Craft, University of Southern California, Los Angeles, CA, and Chemicon), mouse anti-rhodopsin (1D4 and 4D2; generous gift from R. Molday, University of British Columbia, Vancouver, Canada). The secondary antibodies for fluorescent detection were AlexaFluor 488 and 546 (Molecular probes, Invitrogen). Sections were visualized using an Olympus FluoView 500 laser scanning confocal microscope. Images were subsequently digitized using FluoView software version 5.0.
EMSA
The electrophoretic mobility shift assays were performed using established methods [28], with minor modifications. Nuclear protein extracts from transfected COS-1 cells were prepared using a commercial kit (Active motif, Carlsbad, CA), and expression of NRL protein was confirmed by SDS-PAGE followed by immunoblotting. Nuclear extracts were incubated with 1 µg poly (dI-dC) at 4°C for 15 min in the binding buffer (12 mM HEPES [N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid], pH 7.9; 60 mM KCl; 4 mM MgCl2; 1 mM EDTA [ethylenediaminetetra acetic acid]; 12% glycerol; 1 mM dithiothreitol; and 0.5 mM phenylmethylsulfonyl fluoride [PMSF]). Then, 32P-labeled double-stranded oligonucleotide (40,000 cpm) was added and the reaction was incubated at 4°C for 20 min. The DNA oligonucleotide (−2820 nt to −2786 nt: NRE F5’-TGGCCTCTGGTGGCTTTGTCAGCAGTTCCAAGGCT-3’, NRE R 5’-AGCCTTGGAACTGCTGACAAAGCCACCAGAGGCCA-3’) contains a putative NRL-response element (NRE) (underlined) that is predicted by Genomatix. In competition studies, nuclear extracts were pre-incubated with 50 or 100X unlabeled oligonucleotide for 30 min at room temperature and incubated with labeled oligonucleotide at room temperature for 20 min. A mutant oligonucleotide (F: 5’-TGGCCTCTGGTGGCTTTATTTGCAGTTCCAAGGCT-3’, R: 5’-AGCCTTGGAACTGCAAATAAAGCCACCAGAGGCCA-3’) with three nucleotides changed in the NRE site was also used to compete for the protein binding to the oligonucleotide. In order to immunologically identify the components in protein-DNA complexes, nuclear extracts were incubated with 2 µg of the anti-NRL antibody or normal rabbit IgG for 30 min at room temperature, followed by the addition of labeled oligonucleotide and a further incubation for 20 min at room temperature. The reaction mixtures were electrophoresed on 6% polyacrylamide gels at 175 volts for 2.5 hr and subjected to autoradiography.
ChIP
The chromatin immunoprecipitation assays were performed using a commercial kit (Active motif, Carlsbad, CA). Briefly, four snap-frozen retinas from wild type C57BL/6J mice were cross-linked for 15 min at room temperature with 1% formaldehyde in PBS containing protease inhibitors [40]. The reaction was stopped by adding glycine (125 mM), followed by 5 min incubation at room temperature. The remaining steps were performed according to the manufacturer’s instructions, using anti-NRL polyclonal antibody or normal rabbit IgG. ChIP DNAs were used for PCR amplification of a 248-bp fragment (−2989nt to −2742nt), containing a putative NRE (as determined by Genomatix), with primers 5’-GCATGCACTGTTCAAACACC-3’ and 5’-GATAGGCTGTGCAGGGGTTA-3’. PCR with another pair of primers (5’-TGTCCTGAGTCTCC CTGCTT -3’ and 5’- TAAGGCTGGCCAT AAAGTGG -3’) that amplifies a 209-bp fragment (1230 nt to 1438 nt) located about 4 kb downstream from the NRE site, served as a negative control.
ERG
Electroretinograms (ERGs) were recorded from 2–3 month old adult mice. Animals were dark-adapted for at least 12 hours before intraperitonial administration of ketamine (93 mg/kg) and xylazine (8 mg/kg). After pupil dilation with topical 1% atropine and 0.5% tropicamide, corneal ERGs were recorded from both eyes using gold wire loops with 0.5% tetracaine topical anesthesia and a drop of 2% methylcellulose for corneal hydration. A gold wire loop placed in the mouth was used as reference, and the ground electrode was attached to the tail. Body temperature was maintained at 37°C with a heating pad. ERGs were recorded to single xenon white flashes (PS22 Photic Stimulator, Grass Telefactor, West Warwick, RI) presented in a Ganzfeld bowl. Responses were amplified at 10,000 gain at 1 to 1000 Hz (CP511 AC amplifier, Grass Telefactor), and digitized at a rate of 32 KHz. A notch filter was used to remove 60 Hz line noise. Stimulus intensity was attenuated with neutral density filters and ERGs were recorded to increasing intensity (−6.0 to 1.09 log cd-s/m2). Scotopic ERGs were recorded at 3 to 60 second interstimulus intervals depending on the stimulus intensity and responses were computer averaged with at least 20 averages at the lower intensities. Animals were then light adapted for 10 min by exposure to a white 32 cd/m2 rod saturating background, and photopic ERGs were recorded for single flash white stimuli over a 2 log unit range. The a-wave was measured from the pre-stimulus baseline to the initial trough. B-waves were measured from the trough of the a-wave when present or from the baseline to the b-wave maximum. A second recording system was used to record S- and M-cone ERGs (Espion e2, Diagnosys LLC, Lowell, MA). Light-adapted ERGs were recorded on a 40 cd/m2 background to a xenon flash and a UV filter (360 nm peak; Hoya U-360 filter, Edmund Optics, Barrington, NJ). M-cone ERGs were isolated using a green light-emitting diode (510 nm peak) on the Espion e2. The flash energy was adjusted to elicit responses of approximately equal amplitude for the two wavelengths in WT mice. These stimuli were then used to record S- and M-cone ERGs in Nrl−/− and in Crxp-Nrl/rd7 mice.
ACKNOWLEDGMENTS
We thank Matthew Brooks and Ritu Khanna for assistance with microarray analysis, Thom Saunders for advice with transgenic mice, and Hemant Khanna for discussions and comments on the manuscript. We acknowledge S. Lentz, M. Gillett, and M. Van Keuren for technical assistance and S. Ferrara for administrative support. This research was supported by grants from the National Institutes of Health (EY011115, EY007003; NEI intramural support), The Foundation Fighting Blindness, Research to Prevent Blindness, Organogenesis Training Program (T-32-HD007505), and a Rackham Predoctoral Fellowship. The core facilities of the Michigan Diabetes Research and Training Center (NIH5P60 DK20572) were also used for this work.
ABBREVIATIONS
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
- ERG
electroretinography
- NRE
NRL-response element
- rd
retinal degeneration
- nt
nucleotide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
URLs
Genomatix: www.genomatix.de/index.html
LITERATURE REFERENCES
- 1.Adler R, Raymond PA. Have we achieved a unified model of photoreceptor cell fate specification in vertebrates? Brain Res. 2007 doi: 10.1016/j.brainres.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akagi T, Akita J, Haruta M, Suzuki T, Honda Y, Inoue T, Yoshiura S, Kageyama R, Yatsu T, Yamada M, Takahashi M. Iris-derived cells from adult rodents and primates adopt photoreceptor-specific phenotypes. Invest Ophthalmol Vis Sci. 2005;46:3411–3419. doi: 10.1167/iovs.04-1112. [DOI] [PubMed] [Google Scholar]
- 3.Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, Nusinowitz S, Heckenlively JR, Roderick TH, Kozak CA, Danciger M, Davisson MT, Farber DB. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci U S A. 2000;97:5551–5556. doi: 10.1073/pnas.97.10.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akimoto M, Cheng H, Zhu D, Brzezinski JA, Khanna R, Filippova E, Oh EC, Jing Y, Linares JL, Brooks M, Zareparsi S, Mears AJ, Hero A, Glaser T, Swaroop A. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc Natl Acad Sci U S A. 2006;103:3890–3895. doi: 10.1073/pnas.0508214103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Applebury ML, Antoch MP, Baxter LC, Chun LL, Falk JD, Farhangfar F, Kage K, Krzystolik MG, Lyass LA, Robbins JT. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000;27:513–523. doi: 10.1016/s0896-6273(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 6.Belecky-Adams T, Adler R. Developmental expression patterns of bone morphogenetic proteins, receptors, and binding proteins in the chick retina. J Comp Neurol. 2001;430:562–572. [PubMed] [Google Scholar]
- 7.Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J Comp Neurol. 1979;188:263–272. doi: 10.1002/cne.901880205. [DOI] [PubMed] [Google Scholar]
- 8.Cayouette M, Barres BA, Raff M. Importance of intrinsic mechanisms in cell fate decisions in the developing rat retina. Neuron. 2003;40:897–904. doi: 10.1016/s0896-6273(03)00756-6. [DOI] [PubMed] [Google Scholar]
- 9.Cepko C. Giving in to the blues. Nat Genet. 2000;24:99–100. doi: 10.1038/72887. [DOI] [PubMed] [Google Scholar]
- 10.Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charron F, Tessier-Lavigne M. Novel brain wiring functions for classical morphogens: a role as graded positional cues in axon guidance. Development. 2005;132:2251–2262. doi: 10.1242/dev.01830. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, Rattner A, Nathans J. The rod photoreceptor-specific nuclear receptor Nr2e3 represses transcription of multiple cone-specific genes. J Neurosci. 2005;25:118–129. doi: 10.1523/JNEUROSCI.3571-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Rattner A, Nathans J. Effects of L1 retrotransposon insertion on transcript processing, localization and accumulation: lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum Mol Genet. 2006;15:2146–2156. doi: 10.1093/hmg/ddl138. [DOI] [PubMed] [Google Scholar]
- 14.Chen S, Wang QL, Nie Z, Sun H, Lennon G, Copeland NG, Gilbert DJ, Jenkins NA, Zack DJ. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron. 1997;19:1017–1030. doi: 10.1016/s0896-6273(00)80394-3. [DOI] [PubMed] [Google Scholar]
- 15.Cheng H, Aleman TS, Cideciyan AV, Khanna R, Jacobson SG, Swaroop A. In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum Mol Genet. 2006;15:2588–2602. doi: 10.1093/hmg/ddl185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng H, Khanna H, Oh EC, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563–1575. doi: 10.1093/hmg/ddh173. [DOI] [PubMed] [Google Scholar]
- 17.Corbo JC, Cepko CL. A hybrid photoreceptor expressing both rod and cone genes in a mouse model of enhanced S-cone syndrome. PLoS Genet. 2005;1:e11. doi: 10.1371/journal.pgen.0010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniele LL, Lillo C, Lyubarsky AL, Nikonov SS, Philp N, Mears AJ, Swaroop A, Williams DS, Pugh EN., Jr Cone-like morphological, molecular, and electrophysiological features of the photoreceptors of the Nrl knockout mouse. Invest Ophthalmol Vis Sci. 2005;46:2156–2167. doi: 10.1167/iovs.04-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donovan SL, Dyer MA. Regulation of proliferation during central nervous system development. Semin Cell Dev Biol. 2005;16:407–421. doi: 10.1016/j.semcdb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 20.Dyer MA, Cepko CL. Regulating proliferation during retinal development. Nat Rev Neurosci. 2001;2:333–342. doi: 10.1038/35072555. [DOI] [PubMed] [Google Scholar]
- 21.Edlund T, Jessell TM. Progression from extrinsic to intrinsic signaling in cell fate specification: a view from the nervous system. Cell. 1999;96:211–224. doi: 10.1016/s0092-8674(00)80561-9. [DOI] [PubMed] [Google Scholar]
- 22.Furukawa A, Koike C, Lippincott P, Cepko CL, Furukawa T. The mouse Crx 5′-upstream transgene sequence directs cell-specific and developmentally regulated expression in retinal photoreceptor cells. J Neurosci. 2002;22:1640–1647. doi: 10.1523/JNEUROSCI.22-05-01640.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furukawa T, Morrow EM, Cepko CL. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell. 1997;91:531–541. doi: 10.1016/s0092-8674(00)80439-0. [DOI] [PubMed] [Google Scholar]
- 24.Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999;23:466–470. doi: 10.1038/70591. [DOI] [PubMed] [Google Scholar]
- 25.Gunhan E, van der List D, Chalupa LM. Ectopic photoreceptors and cone bipolar cells in the developing and mature retina. J Neurosci. 2003;23:1383–1389. doi: 10.1523/JNEUROSCI.23-04-01383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S, Duhl D, Carmi R, Bennett J, Weleber RG, Fishman GA, Wright AF, Stone EM, Sheffield VC. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24:127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- 27.Haider NB, Naggert JK, Nishina PM. Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. Hum Mol Genet. 2001;10:1619–1626. doi: 10.1093/hmg/10.16.1619. [DOI] [PubMed] [Google Scholar]
- 28.Hao H, Qi H, Ratnam M. Modulation of the folate receptor type beta gene by coordinate actions of retinoic acid receptors at activator Sp1/ets and repressor AP-1 sites. Blood. 2003;101:4551–4560. doi: 10.1182/blood-2002-10-3174. [DOI] [PubMed] [Google Scholar]
- 29.Hsiau TH, Diaconu C, Myers CA, Lee J, Cepko CL, Corbo JC. The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS ONE. 2007;2:e643. doi: 10.1371/journal.pone.0000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 31.Levine EM, Fuhrmann S, Reh TA. Soluble factors and the development of rod photoreceptors. Cell Mol Life Sci. 2000;57:224–234. doi: 10.1007/PL00000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H, Thurig S, Mohamed O, Dufort D, Wallace VA. Mapping canonical Wnt signaling in the developing and adult retina. Invest Ophthalmol Vis Sci. 2006;47:5088–5097. doi: 10.1167/iovs.06-0403. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, Wilson S, Reh T. BMP receptor 1b is required for axon guidance and cell survival in the developing retina. Dev Biol. 2003;256:34–48. doi: 10.1016/s0012-1606(02)00115-x. [DOI] [PubMed] [Google Scholar]
- 34.Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- 35.Marquardt T, Gruss P. Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci. 2002;25:32–38. doi: 10.1016/s0166-2236(00)02028-2. [DOI] [PubMed] [Google Scholar]
- 36.McIlvain VA, Knox BE. Nr2e3 and Nrl can reprogram retinal precursors to the rod fate in Xenopus retina. Dev Dyn. 2007;236:1970–1979. doi: 10.1002/dvdy.21128. [DOI] [PubMed] [Google Scholar]
- 37.Mears AJ, Kondo M, Swain PK, Takada Y, Bush RA, Saunders TL, Sieving PA, Swaroop A. Nrl is required for rod photoreceptor development. Nat Genet. 2001;29:447–452. doi: 10.1038/ng774. [DOI] [PubMed] [Google Scholar]
- 38.Mitton KP, Swain PK, Chen S, Xu S, Zack DJ, Swaroop A. The leucine zipper of NRL interacts with the CRX homeodomain. A possible mechanism of transcriptional synergy in rhodopsin regulation. J Biol Chem. 2000;275:29794–29799. doi: 10.1074/jbc.M003658200. [DOI] [PubMed] [Google Scholar]
- 39.Nikonov SS, Daniele LL, Zhu X, Craft CM, Swaroop A, Pugh EN., Jr Photoreceptors of Nrl −/− mice coexpress functional S- and M-cone opsins having distinct inactivation mechanisms. J Gen Physiol. 2005;125:287–304. doi: 10.1085/jgp.200409208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oh EC, Khan N, Novelli E, Khanna H, Strettoi E, Swaroop A. Transformation of cone precursors to functional rod photoreceptors by bZIP transcription factor NRL. Proc Natl Acad Sci U S A. 2007;104:1679–1684. doi: 10.1073/pnas.0605934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osakada F, Ooto S, Akagi T, Mandai M, Akaike A, Takahashi M. Wnt signaling promotes regeneration in the retina of adult mammals. J Neurosci. 2007;27:4210–4219. doi: 10.1523/JNEUROSCI.4193-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng GH, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Hum Mol Genet. 2005;14:747–764. doi: 10.1093/hmg/ddi070. [DOI] [PubMed] [Google Scholar]
- 43.Peng GH, Chen S. Chromatin immunoprecipitation identifies photoreceptor transcription factor targets in mouse models of retinal degeneration: new findings and challenges. Vis Neurosci. 2005;22:575–586. doi: 10.1017/S0952523805225063. [DOI] [PubMed] [Google Scholar]
- 44.Pittler SJ, Zhang Y, Chen S, Mears AJ, Zack DJ, Ren Z, Swain PK, Yao S, Swaroop A, White JB. Functional analysis of the rod photoreceptor cGMP phosphodiesterase alpha-subunit gene promoter: Nrl and Crx are required for full transcriptional activity. J Biol Chem. 2004;279:19800–19807. doi: 10.1074/jbc.M401864200. [DOI] [PubMed] [Google Scholar]
- 45.Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM. Timing and topography of cell genesis in the rat retina. J Comp Neurol. 2004;474:304–324. doi: 10.1002/cne.20134. [DOI] [PubMed] [Google Scholar]
- 46.Raven MA, Oh EC, Swaroop A, Reese BE. Afferent control of horizontal cell morphology revealed by genetic respecification of rods and cones. J Neurosci. 2007;27:3540–3547. doi: 10.1523/JNEUROSCI.0372-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rehemtulla A, Warwar R, Kumar R, Ji X, Zack DJ, Swaroop A. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc Natl Acad Sci U S A. 1996;93:191–195. doi: 10.1073/pnas.93.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevens CF. Neuronal diversity: too many cell types for comfort? Curr Biol. 1998;8:R708–R710. doi: 10.1016/s0960-9822(98)70454-3. [DOI] [PubMed] [Google Scholar]
- 49.Strettoi E, Porciatti V, Falsini B, Pignatelli V, Rossi C. Morphological and functional abnormalities in the inner retina of the rd/rd mouse. J Neurosci. 2002;22:5492–5504. doi: 10.1523/JNEUROSCI.22-13-05492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swain PK, Hicks D, Mears AJ, Apel IJ, Smith JE, John SK, Hendrickson A, Milam AH, Swaroop A. Multiple phosphorylated isoforms of NRL are expressed in rod photoreceptors. J Biol Chem. 2001;276:36824–36830. doi: 10.1074/jbc.M105855200. [DOI] [PubMed] [Google Scholar]
- 51.Swaroop A, Xu JZ, Pawar H, Jackson A, Skolnick C, Agarwal N. A conserved retina-specific gene encodes a basic motif/leucine zipper domain. Proc Natl Acad Sci U S A. 1992;89:266–270. doi: 10.1073/pnas.89.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szel A, Lukats A, Fekete T, Szepessy Z, Rohlich P. Photoreceptor distribution in the retinas of subprimate mammals. J Opt Soc Am A Opt Image Sci Vis. 2000;17:568–579. doi: 10.1364/josaa.17.000568. [DOI] [PubMed] [Google Scholar]
- 53.Turner DL, Cepko CL. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- 54.Turner DL, Snyder EY, Cepko CL. Lineage-independent determination of cell type in the embryonic mouse retina. Neuron. 1990;4:833–845. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- 55.Ueno S, Kondo M, Miyata K, Hirai T, Miyata T, Usukura J, Nishizawa Y, Miyake Y. Physiological function of S-cone system is not enhanced in rd7 mice. Exp Eye Res. 2005;81:751–758. doi: 10.1016/j.exer.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 56.Van Raay TJ, Vetter ML. Wnt/frizzled signaling during vertebrate retinal development. Dev Neurosci. 2004;26:352–358. doi: 10.1159/000082277. [DOI] [PubMed] [Google Scholar]
- 57.Wetts R, Fraser SE. Multipotent precursors can give rise to all major cell types of the frog retina. Science. 1988;239:1142–1145. doi: 10.1126/science.2449732. [DOI] [PubMed] [Google Scholar]
- 58.Xu Q, Wang Y, Dabdoub A, Smallwood PM, Williams J, Woods C, Kelley MW, Jiang L, Tasman W, Zhang K, Nathans J. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004;116:883–895. doi: 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- 59.Yoshida S, Mears AJ, Friedman JS, Carter T, He S, Oh E, Jing Y, Farjo R, Fleury G, Barlow C, Hero AO, Swaroop A. Expression profiling of the developing and mature Nrl−/− mouse retina: identification of retinal disease candidates and transcriptional regulatory targets of Nrl. Hum Mol Genet. 2004;13:1487–1503. doi: 10.1093/hmg/ddh160. [DOI] [PubMed] [Google Scholar]
- 60.Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- 61.Yu J, He S, Friedman JS, Akimoto M, Ghosh D, Mears AJ, Hicks D, Swaroop A. Altered expression of genes of the Bmp/Smad and Wnt/calcium signaling pathways in the cone-only Nrl−/− mouse retina, revealed by gene profiling using custom cDNA microarrays. J Biol Chem. 2004;279:42211–42220. doi: 10.1074/jbc.M408223200. [DOI] [PubMed] [Google Scholar]
- 62.Zhu D, Hero AO, Qin ZS, Swaroop A. High throughput screening of co-expressed gene pairs with controlled false discovery rate (FDR) and minimum acceptable strength (MAS) J Comput Biol. 2005;12:1029–1045. doi: 10.1089/cmb.2005.12.1029. [DOI] [PubMed] [Google Scholar]
- 63.Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–422. doi: 10.1126/science.285.5426.418. [DOI] [PubMed] [Google Scholar]