Abstract
Knocking out myostatin activity during development increases the rate of muscle protein synthesis. The present study was done to determine whether postdevelopmental loss of myostatin activity stimulates myofibrillar protein synthesis and the phosphorylation of some of the proteins involved in regulation of protein synthesis rate. Myostatin activity was inhibited for 4 days, in 4- to 5-mo-old male mice, with injections of an anti-myostatin antibody (JA16). The mean myofibrillar synthesis rate increased 19% (P < 0.01) relative to the mean rate in saline-treated mice, as determined by incorporation of deuterium-labeled phenylalanine. JA16 increased phosphorylation of p70 S6 kinase (S6K) and ribosomal protein S6 (rpS6) 1.9-fold (P < 0.05). It did not affect phosphorylation of eukaryotic initiation factor 4E-binding protein-1 or Akt. Microarrays and real-time PCR analyses indicated that JA16 administration did not selectively enrich levels of mRNAs encoding myofibrillar proteins, ribosomal proteins, or translation initiation and elongation factors. Rapamycin treatment did not affect the rate of myofibrillar protein synthesis whether or not the mice received JA16 injections, although it eliminated the phosphorylation of S6K and rpS6. We conclude that the normal level of myostatin activity in mature muscle is sufficient to inhibit myofibrillar synthesis rate and phosphorylation of S6K and rpS6. Reversal of the inhibition of myofibrillar synthesis with an anti-myostatin antibody is not dependent on mTOR activation.
Keywords: rapamycin, mammalian target of rapamycin, Akt, eukaryotic initiation factor 4E-binding protein-1, translation, JA16 anti-myostatin antibody
genetic mutations causing loss of myostatin activity lead to marked hypermuscularity in several mammalian species, including humans (3, 12, 13, 19, 20). When myostatin activity is inhibited in mice several weeks or months after birth, there is a more modest muscle growth (2, 9, 23, 26–28). The postdevelopmental effects of myostatin are of great interest with respect to potential clinical applications of myostatin inhibitors. The early responses to myostatin inhibition are important in understanding the mechanism of hypertrophy and could be useful biomarkers for preclinical or initial clinical trials of the efficacy of potential anti-myostatin agents. Thus the present study was done to search for early molecular changes in adult skeletal muscle after inhibition of myostatin activity with an anti-myostatin antibody.
For muscle fiber enlargement to be functionally useful, the mass of myofibrils must increase along with the overall muscle size. Increased myofibrillar mass can occur only if the rate of myofibrillar synthesis exceeds the rate of degradation. In cultured myoblasts and myotubes, myostatin has an inhibitory effect on overall protein synthesis but does not affect the rate of proteolysis (24). In neonatal rats, infusion of follistatin, an inhibitor of myostatin, increases muscle protein synthesis (22). Mice with constitutive myostatin knockout have increased myofibrillar protein synthesis but normal myofibrillar half-life (25). There have been no studies of myofibrillar or total muscle protein synthesis after myostatin activity in mature animals has been reduced following normal muscle development. Because effects of postdevelopmental myostatin inhibition should not be inferred from studies of mice with constitutive myostatin knockout or addition of myostatin to cultured myotubes, we examined the effect of the anti-myostatin antibody on myofibrillar protein synthesis.
In principle, reducing myostatin activity could affect the rate of protein synthesis by several different mechanisms. One way would be to add new nuclei to the myofibers, thereby increasing the rate of RNA production per fiber. However, postnatal inhibition of myostatin activity causes fiber enlargement primarily by increasing the fiber volume per myonucleus rather than by increasing the number of nuclei per fiber (2, 26, 28). Another way to increase protein synthesis would be to increase the rate of transcription of genes encoding translation initiation or elongation factors or other components of the protein synthetic machinery. A microarray study indicated that mice with constitutive myostatin knockout have elevated expression of several genes encoding proteins involved in translation (21). The same approach was used in the present study to examine expression of these genes. Yet another potential mechanism for increasing the rate of protein synthesis is activation of the mammalian target of rapamycin (mTOR), a key integrator of nutrient and growth factor signals that determine cell size and protein metabolism (5, 18). Activated mTOR promotes phosphorylation of p70 S6 kinase (S6K) and translational initiation factor 4E-binding protein-1 (4E-BP1). Therefore, effects of the anti-myostatin antibody on phosphorylation of S6K, its target ribosomal protein S6 (rpS6), and 4E-BP1 were examined in the present study. Rapamycin was used to determine whether the effect of the anti-myostatin antibody on myofibrillar synthesis is mTOR dependent. Akt (also known as protein kinase B), which is in a signaling pathway that can activate mTOR, was examined because there is evidence that myostatin inhibits Akt phosphorylation (1, 11, 15).
MATERIALS AND METHODS
Male mice with a predominantly C57BL/6 background were used for this study. They were siblings of mice being generated for studies of Cre-mediated postdevelopmental knockout of a floxed myostatin exon (26). The mice used in the present study did not have the Cre transgene. Mice homozygous for the floxed exon (Mstnf/f) have normal myostatin expression if they do not have a Cre transgene (26). The study included 20 mice with the wild-type myostatin gene (Mstnw/w), six Mstnf/w mice, and six Mstnf/f mice. Treatment groups were balanced with respect to the Mstn genotype to eliminate the possibility of bias due to this factor. The use of mice for this research was approved by the University of Rochester Animal Research Committee and was compliant with all applicable federal and state regulations for humane use of animals in research. The mice were housed in microisolator cages with food and water available at all times.
The mice received two intraperitoneal (ip) injections of JA16 antibody (n = 16) or vehicle (PBS; n = 16) at 4–5 mo of age (JA16 anti-myostatin antibody was generously provided by Wyeth Pharmaceuticals). To ensure effective inhibition of myostatin activity, they were injected with larger doses (0.12 mg/g body wt on day 1, 0.09 mg/g body wt on day 3) of the antibody than the dose reported to induce muscle hypertrophy (0.06 mg/g) when administered once/wk for several weeks (27). Two days after the second antibody injection (day 5, between 0900 and 1030), the mice received an ip injection of l-[ring-2H5]phenylalanine (D5Phe, purchased from Cambridge Isotope Laboratories; 0.02 ml/g body wt of 150 mM D5Phe in 75 mM NaCl). They were killed 30 min after the tracer injection by CO2 inhalation and then cervical dislocation after cessation of breathing (∼3 min after start of CO2 inhalation). The gastrocnemius muscles were removed as quickly as possible and immersed immediately in liquid nitrogen. Tissue was stored at −70°C.
Some of the mice (n = 6 JA16 treated, 6 PBS treated) also received ip injections of the mTOR inhibitor rapamycin (purchased from LC Laboratories), 0.06 mg/mouse daily from day 1 to day 4. The final injection was made ∼16 h before the D5Phe injection. The concentrated rapamycin solution in ethanol (40 mg/ml), stored at −15°C, was diluted to 0.1 mg/ml on the day of injection with 0.2% sodium carboxymethylcellulose/0.25% polysorbate 80 in water.
The rate of myofibrillar protein synthesis was determined by tracer incorporation, as described in detail previously (25). This involved determining the ratio of D5Phe to unlabeled phenylalanine for both the free amino acid pool of the muscle tissue and a myofibrillar protein preparation that was washed of all free amino acids before acid hydrolysis. t-Butyldimethylsilyl amino acid derivatives were separated by gas chromatography and analyzed with a quadrupole mass spectrometer to determine the ratio of D5Phe to unlabeled phenylalanine. The D5Phe enrichment of water-soluble proteins of several muscle samples also was determined as follows. Muscles were homogenized in water and centrifuged to pellet the myofibrils and other insoluble proteins. The supernatant was mixed with an equal volume of 10% perchloric acid to precipitate sarcoplasmic proteins. Pellets were washed four times in 5% perchloric acid to remove free amino acids, then in water and ethanol to remove perchloric acid. The pellets were dried in a vacuum centrifuge, and the proteins were hydrolyzed and analyzed with the same procedures used to determine D5Phe enrichment in myofibrillar proteins.
The rate of protein synthesis was calculated with the following formula: fractional synthesis rate (%/day) = 100 × (D5Phe/total Phe in protein) ÷ (D5Phe/total Phe in free amino acid pool) × (1,440 min/day) ÷ (30 min D5Phe incorporation period).
It is assumed that there is only trivial recycling of the tracer from newly synthesized proteins back to the free amino acid pool during the brief incorporation period, which is reasonable given that the mass of myofibrillar proteins already present is ∼1,000-fold greater than the amount synthesized in 30 min. Even if newly formed myofibrillar proteins are degraded preferentially, the net isotope incorporation would reflect the productive rate of synthesis. It also is assumed that the %D5Phe in the free amino acid pool of the tissue is the same as that of the phenylalanyl-tRNA pool used for peptide synthesis. This is a reasonable assumption because there is so much tracer flooding the tissue that endogenous (unlabeled) phenylalanine becomes a minor part of the free phenyalanine pool [the ratio of D5Phe to endogenous Phe in muscle rapidly increases to ∼7:1 within the first few minutes and declines very gradually after that (25)]. It is assumed that the %D5Phe in the free amino acid pool at the end of the 30-min tracer incorporation period is representative of the mean value over this period. The detailed time course data presented in a previous report validates this assumption (25). The %D5Phe in the free amino acid pool was nearly identical under all experimental conditions in the present study (84–86%), suggesting that none of the conditions affected tracer uptake or dilution of tracer by protein breakdown.
Proteins for Western blot analysis were extracted by homogenizing muscle tissue in 10 volumes of ice-cold buffer (20 mM HEPES, 100 mM KCl, 4.2 mM EGTA, 50 mM NaF, 0.2 mM EDTA, 50 mM disodium glycerol 2-phosphate, 1 mM DTT, 0.5 mM Na3VO4, 1 mM PMSF, 1% vol/vol Calbiochem protease arrest reagent, pH 7.4). Insoluble proteins were removed by centrifugation. The protein concentration of the soluble fraction was determined with the Bradford assay (Pierce). Proteins were denatured and reduced by heating at 95°C in Laemmli buffer with 5% 2-mercaptoethanol. Equal amounts of total protein from each sample were subjected to SDS-PAGE and transferred to nitrocellulose paper by semidry electrophoresis. Equal loading was confirmed by reversible staining of the blots with Memcode reagents (Pierce). The blots were blocked with 5% nonfat milk in Tris-buffered saline plus Tween-20 (TBST; pH 7.6 with 0.1% Tween-20), washed in TBST (5 min), incubated overnight at 4°C with primary antibody (1,000:1 dilution of stock antibody in TBST with 5% BSA; all antibodies purchased from Cell Signaling Technology), washed with TBST (4 × 5 min), incubated for 1 h at room temperature with secondary antibody [2,000:1 dilution of horseradish peroxidase (HRP)-tagged goat anti-rabbit IgG in TBST with 5% nonfat milk], and then washed in TBST (4 × 5 min) and water (10 min). Blots were covered with HRP-dependent chemiluminescence reagents (SuperSignal West Dura; Pierce) and photographed in a dark box with a cooled charge-coupled device camera (Quantum Scientific Imaging, Model 504s) with a Fujinon HF9HA-1B lens. Pixel intensities were adjusted for bias, dark current, and position relative to lens. Background light intensity was subtracted from the sum of pixel intensities for each band. Values were converted to a percentage of the mean value for the three to four control samples (i.e., muscles from PBS-treated mice) on each blot.
RNA was extracted by homogenizing muscles in Trizol (Invitrogen) followed by precipitation and washing as recommended by instructions provided with the Trizol reagent. The RNA pellet was dissolved in RNase-free water. The total RNA concentration was determined by absorbance of 260-nm light (Nanodrop 1000 spectrophotometer). RNA integrity was confirmed by detection of strong 18S and 28S rRNA signals with an Agilent Bioanalyzer.
For microarray analysis, RNA was converted to biotin-labeled, fragmented cDNA with kits from NuGEN. The cDNA was hybridized overnight with Affymetrix Mouse Genome 430 2.0 arrays. The arrays were washed, stained with phycoerythrin-streptavidin, and scanned with standard protocols recommended by Affymetrix. Data were normalized to total mRNA mass, and expression levels were computed with Gene Chip Operating Software (Affymetrix). All microarray data, including the Affymetrix CEL files, are available from the Gene Expression Omnibus web site under accession no. GSE13707 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13707).
For quantitative RT-PCR, 2 μg of RNA was reverse transcribed with the high-capacity cDNA reverse transcription kit (Applied Biosystems). Triplicate PCR reactions per cDNA were monitored with an ABI 7900HT thermal cycler/fluorescence monitor using the standard conditions recommended for Taqman PCR. Data were analyzed with the SDS 2.2 software (Applied Biosystems). Primer and probe sequences for Myh4 cDNA were published previously (26). Primers and probes for 18S rRNA cDNA (product no. 4308329) and Acta1 cDNA (Mm00808218_g1) were purchased from Applied Biosystems.
Levels of statistical significance for differences between treatment groups were determined by analysis of variance and t-tests.
RESULTS
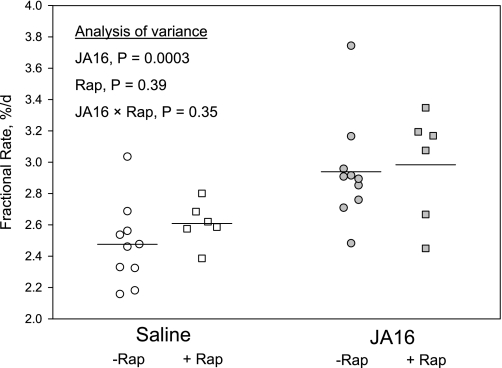
Administration of the anti-myostatin antibody increased the mean rate of myofibrillar synthesis by 18.7% in mice that did not receive rapamycin (P = 0.003; Fig. 1). Synthesis of soluble proteins (not shown in Fig. 1) was 72% more rapid than myofibrillar synthesis regardless of treatment, so it was increased by JA16 administration to the same extent as myofibrillar synthesis (18.8%, P = 0.003). Rapamycin did not cause any reduction in mean myofibrillar synthesis rate in either saline-treated or JA16-treated mice (Fig. 1). Thus the stimulatory effect on myofibrillar synthesis of the anti-myostatin treatment was not significantly different in rapamycin-treated mice (14.4%, P = 0.03) than it was in the mice that did not receive rapamycin. Analysis of variance indicated that JA16 treatment was a highly significant source of variance, whereas rapamycin treatment and the JA16 × rapamycin interaction did not contribute significantly to variance (P values shown in Fig. 1).
Fig. 1.
Myofibrillar synthesis rates based on D5Phe incorporation in gastrocnemius muscles of male mice given ip injections of saline (open symbols) or JA16 anti-myostatin antibody (gray symbols) 4 and 2 days before administration of the D5Phe tracer. Each point represents an individual mouse, and lines represent mean values. Some of the mice (gray squares) were given daily injections of rapamycin (Rap) for 4 days before tracer administration.
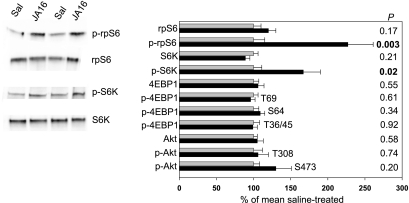
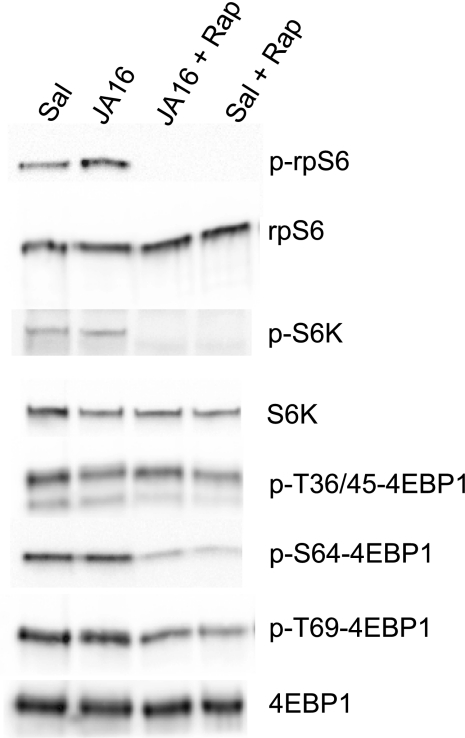
Blocking myostatin activity did not significantly affect muscle concentrations of S6K, rpS6, 4E-BP1, or Akt (P > 0.10; Fig. 2). The JA16 antibody increased the level of phosphorylation of phospho-Thr389-S6K (1.7-fold, P = 0.02) and phospho-Ser235/236-rpS6 (2.3-fold, P = 0.003) but did not affect phosphorylation of 4E-BP1 (phospho-Thr36/45, phospho-Ser64, and phospho-Thr69) or Akt (phospho-Thr308 and phospho-Ser473) (Fig. 2). The proportions of S6K and rpS6 in the phosphorylated form were 1.9-fold greater in JA16-treated mice (P < 0.05). In mice that received rapamycin, there was no detectable phospho-S6K or phospho-rpS6 in skeletal muscle, and there was less phosphorylation of 4E-BP1 at Thr64 (∼35% of normal) and Ser69 (∼60% of normal) (Fig. 3).
Fig. 2.
Mean ± SE relative amounts of total and phosphorylated (p) p70 S6 kinase (S6K), ribosomal protein S6 (rpS6), 4E-binding protein-1 (4E-BP1), and Akt in 10 saline (Sal)-treated (gray bars) and 10 JA16-treated mice (black bars), as determined by immunoblotting. Representative bands are shown for proteins (S6K, rpS6) with increased phosphorylation (P < 0.05) in JA16-treated mice (P values for all proteins shown on right side of figure). The phosphorylated amino acids detected by the various anti-phospho 4E-BP1 and anti-phospho-Akt antibodies are shown next to the bars. The anti-phospho-S6K antibody detected phosphorylation at Thr (T)389. The anti-phospho-rpS6 antibody detected phosphorylation at Ser (S)235/236.
Fig. 3.
Efficacy of rapamycin treatment was demonstrated by elimination of S6K and rpS6 phosphorylation (representative blots are shown; no p-S6K or p-rpS6 was detected in any of the 12 rapamycin-treated mice). Phosphorylation of 4E-BP1 was attenuated but not eliminated by rapamycin.
Although levels of total RNA, ribosomal RNA, Acta1 mRNA (which encodes skeletal muscle α-actin), and Myh4 mRNA (which encodes the most abundant myosin heavy chain in mouse gastrocnemius, type 2b) were not consistently affected by JA16 treatment (P > 0.2), the mean levels per milligram tissue increased to approximately the same extent as the mean increase in myofibrillar synthesis rate (Table 1). Microarrays did not reveal any significant differences between saline-treated and JA16-treated mice in the expression of genes encoding other myofibrillar proteins (various isoforms of myosin heavy and light chains, troponins, tropomyosins, titin, and titin cap) relative to expression of either Acta1 or Myh4.
Table 1.
RNA analyses
| Saline | JA16 | %Δ | P | |
|---|---|---|---|---|
| Total RNA, μg/mg tissue | 0.69±0.05 | 0.77±0.05 | +11 | 0.32 |
| 18S rRNA/μg total RNA, arbitrary units | 1.00±0.16 | 1.06±0.13 | +6 | 0.79 |
| 18S rRNA/mg tissue, arbitrary units | 1.00±0.14 | 1.21±0.17 | +21 | 0.36 |
| 28S rRNA/18S rRNA | 1.38±0.08 | 1.47±0.06 | +7 | 0.37 |
| Acta1 mRNA/μg total RNA, arbitrary units | 1.00±0.08 | 1.08±0.08 | +8 | 0.46 |
| Acta1 mRNA/mg tissue, arbitrary units | 1.00±0.11 | 1.20±0.13 | +20 | 0.25 |
| Myh4 mRNA/μg total RNA, arbitrary units | 1.00±0.13 | 1.07±0.18 | +7 | 0.76 |
| Myh4 mRNA/mg tissue, arbitrary units | 1.00±0.17 | 1.20±0.23 | +20 | 0.51 |
Values are means ± SE. Items expressed in arbitrary units were measured by real-time PCR with cDNA templates generated by reverse transcription of total RNA. Mean value in saline-treated mice defined as 1 unit. Total RNA was determined by UV absorbance. 28S/18S rRNA ratios were determined with Agilent Bioanalyzer.
There were five genes encoding ribosomal proteins that were expressed at higher levels (normalized to total mRNA mass) in JA16-treated mice at nominal P < 0.05, but only three were increased >7% (Rpsa, +46%; Rpl22l1, +19%; Rps27l, +16%). Rpsa is expressed at a very low level relative to other ribosomal proteins. The protein it encodes is also known as the 67-kDa laminin receptor, and it is unlikely to have a significant role in global protein synthesis. Because there were more than 200 probe sets for ribosomal protein genes, these effects might be “false positives” and must be considered tentative without confirmation in a prospective study. Except for a slight (7%) increase in expression of Eif5a, genes encoding translation initiation factors, translation elongation factors, and tRNA synthetases were not expressed at higher levels in JA16-treated mice at P < 0.05.
DISCUSSION
The present study indicates that myostatin exerts a tonic inhibitory influence on the rate of myofibrillar protein synthesis even after muscles are fully developed. Increased protein synthesis is probably the major mechanism for the muscle fiber hypertrophy, because previous studies suggested that myostatin does not influence muscle proteolysis (24, 25). The magnitude of the stimulatory effect of the anti-myostatin antibody on the fractional rate of myofibrillar synthesis was similar to the effect observed in young mice with constitutive myostatin knockout (25). However, the effect on the synthesis rate per muscle, which is what determines the myofibrillar mass when protein synthesis and breakdown are in equilibrium, was quantitatively much greater with constitutive knockout. The greater effect of constitutive knockout on protein synthesis per whole muscle reflects an increase in the number of muscle fibers during development in myostatin-deficient mice.
With the method used in the present study, myofibrillar synthesis rate depends not only on the rate of tracer incorporation into the individual proteins of the myofibrils but also on their assembly into structures that render them insoluble in the buffer used to wash away nonmyofibrillar proteins. Thus it is possible theoretically that the increase in D5Phe incorporation into myofibrils in JA16-treated mice reflects more rapid assembly of insoluble myofibrillar protein complexes rather than, or in addition to, more rapid production of the individual myofibrillar proteins. However, analysis of proteins soluble in water (including any myofibrillar components not yet incorporated into insoluble complexes) indicated that inhibition of myostatin activity stimulated tracer incorporation into these proteins to the same extent as tracer incorporation into insoluble myofibrils. Reports that myostatin inhibits protein synthesis in cultured myoblasts and myotubes (24), and that follistatin stimulates protein synthesis in neonatal muscle (22), also suggest that myostatin activity influences the rate of protein synthesis rather than myofibrillar assembly, because the global protein synthesis measurements in these studies did not depend on myofibrillar assembly.
The anti-myostatin antibody increased phosphorylation of S6K and one of its targets, rpS6. This is consistent with reports that S6K phosphorylation is reduced in rat muscle with elevated myostatin expression (1) and is increased in neonatal rats treated with follistatin, which binds to myostatin and inhibits its activity (22). The anti-myostatin antibody did not stimulate phosphorylation of 4E-BP1, which often increases in concert with S6K phosphorylation because both are phosphorylated by mTOR. However, an increase in S6K phosphorylation without a similar effect on 4E-BP1 is not unprecedented (4, 7). Moreover, when myostatin is overexpressed in rat muscle, the modest depression of 4E-BP1 phosphorylation is approximately threefold less than the depression of S6 phosphorylation (1). S6K phosphorylation could have increased in JA16-treated mice without an increase in mTOR activity. Another potential explanation is that myostatin inhibits the activity of a phosphatase that normally offsets some of the S6K phosphorylation caused by other growth factors and nutrients signaling through mTOR.
Alhough rapamycin eliminated phosphorylation of S6K and rpS6 and attenuated phosphorylation of 4E-BP1, it did not block the stimulation of myofibrillar synthesis induced by anti-myostatin treatment. Thus mTOR activation does not appear to be necessary to overcome the tonic inhibitory effect of myostatin on myofibrillar synthesis. We recognize that the null hypothesis cannot be proven and that sampling and technical variance could have caused us to underestimate the effect of rapamycin on protein synthesis. On the basis of statistical confidence intervals, there is <5% probability that a repeat experiment would demonstrate complete inhibition of the effect of the JA16 antibody on myofibrillar synthesis and ∼80% probability that a repeat experiment would demonstrate <50% inhibition of the JA16 effect.
The failure of rapamycin to influence baseline protein synthesis (i.e., synthesis in mice that did not receive the anti-myostatin antibody) is consistent with previous reports that rapamycin does not affect the baseline rate of total protein synthesis in muscle (8) or liver (16). Thus mTOR-mediated phosphorylation of S6K is not essential to sustain the normal maintenance level of either global or myofibrillar protein synthesis. This conclusion is reinforced by the fact that myotubes derived from S6K-null mice have normal fractional rates of protein synthesis and global translation initiation (14). The only S6K target examined in the present study was rpS6. This target was chosen because its phosphorylation status is a marker of S6K activity, not because of compelling evidence that it promotes myofibrillar or global protein synthesis in muscle. The role of rpS6 in regulating protein synthesis is uncertain. Although it was originally postulated that phosphorylation of rpS6 promoted translation of mRNAs with 5′ terminal oligopyrimidine tracts, including mRNAs encoding ribosomal proteins, this notion has been challenged by more recent studies (17). S6K has several other substrates in addition to rpS6, including upstream binding factor, which promotes rRNA transcription (6), and two proteins directly involved in translation, eEF2 kinase (which inhibits elongation by phosphorylating eEF2; eEF2 kinase is inactivated by p-S6K) and eIF4B (which enhances the processivity of eIF4A and could accelerate translation of mRNAs with long, structured 5′ ends) (17). Activation of upstream binding factor and inhibition of eEF2 kinase should have global effects on protein synthesis, including myofibrillar synthesis. If the increase in S6K phosphorylation did activate upstream binding factor, the effect was insufficient to consistently increase rRNA levels (although a trend in that direction was observed). We did not examine these other S6K targets because elimination of S6K phosphorylation with rapamycin did not affect protein synthesis.
Myostatin has influenced the phosphorylation of Akt, an activator of mTOR, in some experiments. In cultured myotubes and cardiomyocytes and in rat muscle in vivo, myostatin has an inhibitory effect on Akt phosphorylation (1, 15). In contrast, myostatin stimulates Akt phosphorylation in cultured skeletal muscle fibroblasts (10). In the present study, we observed no significant effect of inhibiting myostatin on total levels or phosphorylation of Akt. Thus it does not appear that Akt activation is required for the initial increase in myofibrillar protein synthesis.
Although there was not a statistically significant difference in RNA abundance per milligram muscle tissue in JA16-treated and saline-treated mice, there was a trend for higher total RNA, rRNA, and myofibrillar mRNA levels in JA16-treated mice. Thus the rate of protein synthesis normalized to RNA levels was not significantly affected by inhibiting myostatin, and we cannot conclude that translational efficiency was increased.
It has been reported that several genes encoding translation initiation and elongation factors are expressed at higher levels in muscle of young mice with constitutive myostatin deficiency (21). The early response to myostatin inhibition in mature mice did not include altered expression of these same genes, except for a slight increase in expression of Eif5a (7% increase in expression in JA16-treated mice, 26% increase in mice with constitutive myostatin knockout). Mice with constitutive myostatin knockout also have increased expression of several ribosomal protein genes. The only one that also was expressed at higher levels in the present study was Rps27l (16% increase with JA16 treatment, 20% increase with myostatin knockout). Overall, the microarray data indicate that the increase in muscle protein synthesis induced by the anti-myostatin antibody cannot be explained by a concerted upregulation of genes encoding the translational machinery of the muscle fibers.
GRANTS
This study was supported by grants from the National Institutes of Health (AR-054366 and RR-024160).
Acknowledgments
We thank Andrew Cardillo, Michelle Zanche, and Meghann McBennett for technical assistance.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Amirouche A, Durieux AC, Banzet S, Koulmann N, Bonnefoy R, Mouret C, Bigard X, Peinnequin A, Freyssenet D. Down-regulation of Akt/mammalian target of rapamycin signaling pathway in response to myostatin overexpression in skeletal muscle. Endocrinology 150: 286–294, 2009. [DOI] [PubMed] [Google Scholar]
- 2.Bogdanovich S, Krag TO, Barton ER, Morris LD, Whittemore LA, Ahima RS, Khurana TS. Functional improvement of dystrophic muscle by myostatin blockade. Nature 420: 418–421, 2002. [DOI] [PubMed] [Google Scholar]
- 3.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibe B, Bouix J, Caiment F, Elsen JM, Eychenne F, Larzul C, Laville E, Meish F, Milenkovic D, Tobin J, Charlier C, Georges M. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet 38: 813–818, 2006. [DOI] [PubMed] [Google Scholar]
- 4.Dreyer HC, Glynn EL, Lujan HL, Fry CS, DiCarlo SE, Rasmussen BB. Chronic paraplegia-induced muscle atrophy downregulates the mTOR/S6K1 signaling pathway. J Appl Physiol 104: 27–33, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glass DJ Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol 37: 1974–1984, 2005. [DOI] [PubMed] [Google Scholar]
- 6.Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 23: 8862–8877, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang YP, Ballou LM, Lin RZ. Rapamycin-insensitive regulation of 4e-BP1 in regenerating rat liver. J Biol Chem 276: 10943–10951, 2001. [DOI] [PubMed] [Google Scholar]
- 8.Kubica N, Bolster DR, Farrell PA, Kimball SR, Jefferson LS. Resistance exercise increases muscle protein synthesis and translation of eukaryotic initiation factor 2Bepsilon mRNA in a mammalian target of rapamycin-dependent manner. J Biol Chem 280: 7570–7580, 2005. [DOI] [PubMed] [Google Scholar]
- 9.Lee SJ, Reed LA, Davies MV, Girgenrath S, Goad ME, Tomkinson KN, Wright JF, Barker C, Ehrmantraut G, Holmstrom J, Trowell B, Gertz B, Jiang MS, Sebald SM, Matzuk M, Li E, Liang LF, Quattlebaum E, Stotish RL, Wolfman NM. Regulation of muscle growth by multiple ligands signaling through activin type II receptors. Proc Natl Acad Sci USA 102: 18117–18122, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li ZB, Kollias HD, Wagner KR. Myostatin directly regulates skeletal muscle fibrosis. J Biol Chem 283: 19371–19378, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McFarlane C, Plummer E, Thomas M, Hennebry A, Ashby M, Ling N, Smith H, Sharma M, Kambadur R. Myostatin induces cachexia by activating the ubiquitin proteolytic system through an NF-κB-independent, FoxO1-dependent mechanism. J Cell Physiol 209: 501–514, 2006. [DOI] [PubMed] [Google Scholar]
- 12.McPherron AC, Lawler AM, Lee SJ. Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature 387: 83–90, 1997. [DOI] [PubMed] [Google Scholar]
- 13.McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA 94: 12457–12461, 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mieulet V, Roceri M, Espeillac C, Sotiropoulos A, Ohanna M, Oorschot V, Klumperman J, Sandri M, Pende M. S6 kinase inactivation impairs growth and translational target phosphorylation in muscle cells maintaining proper regulation of protein turnover. Am J Physiol Cell Physiol 293: C712–C722, 2007. [DOI] [PubMed] [Google Scholar]
- 15.Morissette MR, Cook SA, Foo S, McKoy G, Ashida N, Novikov M, Scherrer-Crosbie M, Li L, Matsui T, Brooks G, Rosenzweig A. Myostatin regulates cardiomyocyte growth through modulation of Akt signaling. Circ Res 99: 15–24, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reiter AK, Anthony TG, Anthony JC, Jefferson LS, Kimball SR. The mTOR signaling pathway mediates control of ribosomal protein mRNA translation in rat liver. Int J Biochem Cell Biol 36: 2169–2179, 2004. [DOI] [PubMed] [Google Scholar]
- 17.Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 31: 342–348, 2006. [DOI] [PubMed] [Google Scholar]
- 18.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol 17: 596–603, 2005. [DOI] [PubMed] [Google Scholar]
- 19.Schuelke M, Wagner KR, Stolz LE, Hubner C, Riebel T, Komen W, Braun T, Tobin JF, Lee SJ. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med 350: 2682–2688, 2004. [DOI] [PubMed] [Google Scholar]
- 20.Shelton GD, Engvall E. Gross muscle hypertrophy in whippet dogs is caused by a mutation in the myostatin gene. Neuromuscul Disord 17: 721–722, 2007. [DOI] [PubMed] [Google Scholar]
- 21.Steelman CA, Recknor JC, Nettleton D, Reecy JM. Transcriptional profiling of myostatin-knockout mice implicates Wnt signaling in postnatal skeletal muscle growth and hypertrophy. FASEB J 20: 580–582, 2006. [DOI] [PubMed] [Google Scholar]
- 22.Suryawan A, Frank JW, Nguyen HV, Davis TA. Expression of the TGF-beta family of ligands is developmentally regulated in skeletal muscle of neonatal rats. Pediatr Res 59: 175–179, 2006. [DOI] [PubMed] [Google Scholar]
- 23.Tang L, Yan Z, Wan Y, Han W, Zhang Y. Myostatin DNA vaccine increases skeletal muscle mass and endurance in mice. Muscle Nerve 36: 342–348, 2007. [DOI] [PubMed] [Google Scholar]
- 24.Taylor WE, Bhasin S, Artaza J, Byhower F, Azam M, Willard DH Jr, Kull FC Jr, Gonzalez-Cadavid N. Myostatin inhibits cell proliferation and protein synthesis in C2C12 muscle cells. Am J Physiol Endocrinol Metab 280: E221–E228, 2001. [DOI] [PubMed] [Google Scholar]
- 25.Welle S, Bhatt K, Pinkert CA. Myofibrillar protein synthesis in myostatin-deficient mice. Am J Physiol Endocrinol Metab 290: E409–E415, 2006. [DOI] [PubMed] [Google Scholar]
- 26.Welle S, Bhatt K, Pinkert CA, Tawil R, Thornton CA. Muscle growth after postdevelopmental myostatin gene knockout. Am J Physiol Endocrinol Metab 292: E985–E991, 2007. [DOI] [PubMed] [Google Scholar]
- 27.Whittemore LA, Song K, Li X, Aghajanian J, Davies M, Girgenrath S, Hill JJ, Jalenak M, Kelley P, Knight A, Maylor R, O'Hara D, Pearson A, Quazi A, Ryerson S, Tan XY, Tomkinson KN, Veldman GM, Widom A, Wright JF, Wudyka S, Zhao L, Wolfman NM. Inhibition of myostatin in adult mice increases skeletal muscle mass and strength. Biochem Biophys Res Commun 300: 965–971, 2003. [DOI] [PubMed] [Google Scholar]
- 28.Zhu X, Hadhazy M, Wehling M, Tidball JG, McNally EM. Dominant negative myostatin produces hypertrophy without hyperplasia in muscle. FEBS Lett 474: 71–75, 2000. [DOI] [PubMed] [Google Scholar]