Abstract
Liver fibrosis is characterized by excessive deposition of extracellular matrix in the liver during chronic injury. During early stages of this disease, cells begin to synthesize and secrete profibrotic proteins that stimulate matrix production and inhibit matrix degradation. Although it is clear that these proteins are important for development of fibrosis, what remains unknown is the mechanism by which chronic liver injury stimulates their production. In the present study, the hypothesis was tested that hypoxia-inducible factor-1α (HIF-1α) is activated in the liver during chronic injury and regulates expression of profibrotic proteins. To investigate this hypothesis, mice were subjected to bile duct ligation (BDL), an animal model of liver fibrosis. HIF-1α protein was increased in the livers of mice subjected to BDL by 3 days after surgery. To test the hypothesis that HIF-1α is required for the development of fibrosis, control and HIF-1α-deficient mice were subjected to BDL. Levels of type I collagen and α-smooth muscle actin mRNA and protein were increased in control mice by 14 days after BDL. These levels were significantly reduced in HIF-1α-deficient mice. Next, the levels of several profibrotic mediators were measured to elucidate the mechanism by which HIF-1α promotes liver fibrosis. Platelet-derived growth factor (PDGF)-A, PDGF-B, and plasminogen activator inhibitor-1 mRNA levels were increased to a greater extent in control mice subjected to BDL compared with HIF-1α-deficient mice at 7 and 14 days after BDL. Results from these studies suggest that HIF-1α is a critical regulator of profibrotic mediator production during the development of liver fibrosis.
Keywords: cholestasis, bile duct ligation, platelet-derived growth factor, collagen, α-smooth muscle actin
liver fibrosis is characterized by excessive deposition of extracellular matrix, such as collagen, in the liver during chronic injury. This disease is initiated when liver injury stimulates cells to synthesize and secrete growth factors, such as platelet-derived growth factor (PDGF) and other soluble mediators that activate hepatic stellate cells and stimulate them and other cell types, such as peribiliary fibroblasts to produce collagen (10, 21). In addition, other mediators are produced, such as matrix metalloproteinases that modulate matrix turnover (14, 22). Although it is clear that numerous mediators are important for the initiation and propagation of fibrosis, what remains largely unknown is the mechanism by which liver injury stimulates their production. A greater understanding of the mechanism(s) that regulate profibrotic mediator production may lead to the development of novel treatments for this disease. One factor that may promote production of profibrotic mediators is hepatocellular hypoxia.
Several studies have demonstrated that the liver becomes hypoxic when injured (8, 9, 18, 28). Hypoxia likely results from disrupted hepatic blood flow and sinusoidal fibrin deposition (8). Furthermore, changes occur in patients with liver disease that may promote hepatocellular hypoxia, such as systemic hypoxemia, sinusoidal capillarization, and formation of portal-systemic collateral vessels and intrahepatic shunts. Hypoxia has been shown to modulate gene expression by activating a number of transcription factors, including hypoxia-inducible factors (HIFs). HIFs are composed of an alpha subunit, typically HIF-1α or HIF-2α, and a beta subunit, HIF-1β, also called the arylhydrocarbon receptor nuclear translocater (7, 11). In normoxic cells, HIF-α subunits are constitutively produced and immediately targeted for proteolytic degradation. When cells become hypoxic, however, the mechanisms that target HIF-α subunits for degradation are inhibited, allowing HIF-α protein levels to increase (5). HIF-α subunits then translocate to the nucleus where they heterodimerize with HIF-1β and regulate expression of genes involved in glycolysis, angiogenesis, iron metabolism, pH control, and other functions. Interestingly, many of the genes that HIFs regulate, including various growth factors, have been implicated in the pathogenesis of liver fibrosis. Whether HIFs are activated in liver during fibrosis and regulate expression of genes that promote this disease, however, has not been examined.
The overall goal of the studies presented in this manuscript was to determine the mechanism by which profibrotic mediators, such as PDGF, fibroblasts growth factor-2 (FGF-2), and plasminogen activator inhibitor-1 (PAI-1) are upregulated in the liver during chronic injury. The hypothesis was tested that during chronic liver injury, HIF-1α is activated and promotes liver fibrosis by regulating expression of genes such as PDGF, FGF-2, and PAI-1 that are critical for the genesis of fibrosis.
MATERIALS AND METHODS
Mice.
Deletion of HIF-1α in mice causes embryonic lethality (17). To selectively reduce HIF-1α levels in adult mice, HIF-1αfl/fl mice, described in detail previously (31) were crossed with mice expressing Cre recombinase under control of the Mx interferon-inducible promoter (23) (Mx-Cre+/− mice; Jackson Laboratories, Bar Harbor, ME). Offspring of this breeding were HIF-1αfl/fl-Mx-Cre+ (i.e., HIF-1α-deletable) or HIF-1αfl/fl-Mx-Cre− (i.e., HIF-1α-nondeletable littermate controls). PCR of genomic DNA was used to detect the floxed HIF-1α gene and the Cre transgene as described previously (31). To activate the MxCre promoter, HIF-1αfl/fl-Mx-Cre+ and HIF-1αfl/fl-Mx-Cre− mice were treated with 500 μg of polyinosinic-polycytidylic acid (pIpC, Sigma Chemical, St. Louis, MO) dissolved in sterile saline by intraperitoneal injection every 3 days for a total of three injections. In mice containing the MxCre transgene, this treatment causes a near complete deletion of loxP-containing genes in liver and immune organs and a partial deletion in other tissues (23, 30). In HIF-1αfl/fl-Mx-Cre+ mice, this treatment results in the deletion of exons 13–15 from HIF-1α that contain the COOH-terminal transactivation domain and a portion of the nuclear localization sequence, both of which are essential for hypoxia responsiveness by HIF-1α (31). Mice were used for experiments 1 wk after the final pIpC injection.
All mice were maintained on a 12-h light-dark cycle under controlled temperature (18–21°C) and humidity. Food (Rodent Chow; Harlan-Teklad, Madison, WI) and tap water were allowed ad libitum. All procedures on animals were carried out in accordance with the Guide for the Care and Use of Laboratory Animals promulgated by the National Institutes of Health. These studies were approved by the Institutional IACUC at the University of Kansas Medical Center.
Analysis of HIF-1α deletion in liver.
Genomic DNA was isolated from the livers of mice and PCR was used to determine the degree of HIF-1α deletion. PCR was performed by using the following primers: primer H1, 5′-CTG TCT TCC CTG CTT AGG TCT TTC TAA C-3′; primer H2, 5′-GAG ATG GAG AAG GAG GTT AGT GTA TCC-3′; and primer H3, 5′-ACG TTG GCT CAT GGT GTA CTT TG-3′ as described previously (31).
Bile duct ligation.
Male, C57BL/6 (Harlan, Indianapolis, IN), HIF-1αfl/fl-Mx-Cre+, and HIF-1αfl/fl-Mx-Cre− mice 8–12 wk of age were anesthetized with isoflurane. A midline laparotomy was performed and the bile duct was ligated with 3-0 surgical silk. The abdominal incision was closed with sutures, and the mice received 0.2 mg/kg Buprenex by subcutaneous injection.
Real-time PCR.
Total liver RNA was isolated by use of TRI Reagent (6) (Sigma Chemical, St. Louis, MO) and reverse transcribed into cDNA as described by us previously (20). Real-time PCR was used to quantify the mRNA levels of collagen type Iα1, α-smooth muscle actin (α-SMA), 18S, PDGF-A, PDGF-B, connective tissue growth factor (CTGF), FGF-2, and PAI-1 on an Applied Biosystems Prism 7300 real-time PCR instrument (Applied Biosystems, Foster City, CA) with the SYBRgreen DNA PCR kit (Applied Biosystems) as described (20). The sequences of the primers were as follows: 18S forward 5′-TTG ACG GAA GGG CAC CAC CAG-3′; 18S reverse, 5′-GCA CCA CCA CCC ACG GAA TCG-3′; α-SMA forward 5′-CCA CCG CAA ATG CTT CTA AGT-3′; α-SMA reverse, 5′-GGC AGG AAT GAT TTG GAA AGG-3′; collagen type Iα1 forward 5′-TGT GTT CCC TAC TCA GCC GTC T-3′; collagen type Iα1 reverse, 5′-CAT CGG TCA TGC TCT CTC CAA-3′; CTGF forward 5′-CTG CCA GTG GAG TTC AAA TGC-3′; CTGF reverse, 5′-TCA TTG TCC CCA GGA CAG TTG-3′; FGF-2 forward 5′-AGC GAC CCA CAC GTC AAA CTA C-3′; FGF-2 reverse, 5′-CAG CCG TCC ATC TTC CTT CAT A-3′; PAI-1 forward 5′-AGT CTT TCC GAC CAA GAG CA-3′; PAI-1 reverse, 5′-ATC ACT TGC CCC ATG AAG AG-3′; PDGF-A forward 5′-GAG ATA CCC CGG GAG TTG AT-3′; PDGF-A reverse, 5′-AAA TGA CCG TCC TGG TCT TG-3′; PDGF-B forward 5′-CCC ACA GTG GCT TTT CAT TT-3′; PDGF-B reverse, 5′-GTG GAG GAG CAG ACT GAA GG-3′.
Western blot analysis.
A portion of liver was homogenized in 10 mM HEPES, pH 7.9, containing 100 mM KCl, 1.5 mM MgCl2, 0.1 mM EGTA, 0.5 mM DTT, 0.5% NP-40, and Halt protease inhibitor cocktail (Pierce Biotechnology, Rockford, IL) and incubated on ice. After 15 min, the homogenate was vortexed and centrifuged for 10 s at 10,000 g. The pellet was resuspended in 10 mM HEPES, pH 7.9, containing 420 mM NaCl, 1.5 mM MgCl2, 0.1 mM EGTA, 0.5 mM DTT, 5% glycerol, and Halt protease inhibitor cocktail mixed for 1 h at 4°C. The samples were centrifuged at 10,000 g for 10 min at 4°C and the concentration of protein determined in the supernatant. Aliquots (15 μg) of nuclear extracts were subjected to 10% SDS-PAGE, and proteins were transferred to Immobilon polyvinylidene difluoride transfer membranes (Millipore, Bedford, MA). The membranes were then probed with rabbit polyclonal anti-HIF-1α antibody (NB100-449, Novus Biologicals, Littleton, CO) diluted 1:1,000, followed by incubation with goat anti-rabbit antibody conjugated to horseradish peroxidase (Santa Cruz Biotechnology). Immunoreactive bands were visualized by using the Immun-Star HRP Substrate Kit (Bio-Rad Laboratories, Hercules, CA).
HIF-1α immunohistochemistry.
For HIF-1α immunostaining, livers were frozen in isopentane (Sigma Chemical) immersed in liquid nitrogen for 8 min. Sections of frozen liver were fixed in 4% formalin in phosphate-buffered saline (PBS) for 10 min at room temperature. Sections were incubated with rabbit polyclonal anti-HIF-1α antibody (NB100-449, Novus Biologicals, Littleton, CO) diluted 1:50 in PBS containing 3% goat serum at room temperature for 3 h. The sections were washed with PBS and then incubated with secondary antibody conjugated to Alexa 488 (green staining; Invitrogen). The sections were counterstained with Alexa Fluor 594-conjugated phalloidin to stain actin. For double immunohistochemistry for HIF-1α and macrophages, sections of liver were incubated with anti-HIF-1α antibody as described above and with rat anti-mouse F4/80 and rat anti-mouse CD68. The sections were washed with PBS and then incubated with anti-rabbit secondary antibody conjugated to Alexa 594 (red staining; Invitrogen) and anti-rat secondary antibody conjugated to Alexa 488 (green staining; Invitrogen). For double immunohistochemistry for HIF-1α and hepatocytes, sections of liver were incubated with anti-HIF-1α antibody as described above and goat anti-mouse albumin. The sections were washed with PBS and then incubated with anti-rabbit secondary antibody conjugated to Alexa 594 (red staining; Invitrogen) and anti-goat secondary antibody conjugated to Alexa 488 (green staining; Invitrogen).
Pimonidazole immunohistochemistry.
To detect regions of hypoxia in the liver, mice were treated with pimonidazole (120 mg/kg, Chemicon, Temecula, CA) dissolved in sterile saline by intraperitoneal injection 1, 3, 7, or 14 days after bile duct ligation and were killed 1 h later. Immunostaining for pimonidazole was performed as described previously (8).
Hypoxia in the liver was quantified morphometrically by analyzing the area of immunohistochemical staining of pimonidazole in a section of liver using Scion Image software (Scion, Frederick, MD) as described (8). An increase in the area of pimonidazole staining in the liver indicates hypoxia. The staining is expressed as a fraction of the total area. The random fields analyzed for each liver section were averaged and counted as a replicate, i.e., each replicate represents a different mouse. For the time course studies, data from mice subjected to sham operation at various times were combined into one group for statistical analysis, since no differences occurred among sham-operated groups.
Cytokeratin 19 immunohistochemistry.
For cytokeratin 19 (CK19) immunostaining, livers were frozen in isopentane (Sigma Chemical) immersed in liquid nitrogen for 8 min. Sections of frozen liver were fixed in 4% formalin in PBS for 10 min at room temperature. Sections were incubated with rat anti-CK19 antibody (Developmental Studies Hybridoma Bank, Iowa City, IA) diluted 1:1,000 in PBS containing 3% goat serum at room temperature for 3 h. The sections were washed with PBS and then incubated with secondary antibody conjugated to Alexa 488 (green staining; Invitrogen). The area of CK19 immunostaining was quantified morphometrically as described previously (8).
Assessment of biomarkers of hepatic injury and cholestasis.
Hepatocyte injury and cholestasis were evaluated by measuring the activity of alanine aminotransferase (ALT) and alkaline phosphatase (ALP) in the serum (Pointe Scientific, Brussels, Belgium). Serum bile acid concentrations were determined by using a commercially available kit (Colorimetric Total Bile Acids Assay Kit; Bioquant, San Diego, CA).
Quantification of type I collagen in the liver.
type I collagen in the liver was detected by immunohistochemistry and quantified morphometrically by analyzing the area of immunohistochemical staining of type I collagen as described by us previously (20). An increase in the area of type I collagen staining in the liver is an indicator of fibrosis. The staining is expressed as a fraction of the total area. The random fields analyzed for each liver section were averaged and counted as a replicate, i.e., each replicate represents a different mouse.
Statistical analysis.
Results are presented as means ± SE. Data were analyzed by ANOVA. Data expressed as a fraction were transformed by arc sine square root prior to analysis. Comparisons among group means were made using the Student-Newman-Keuls test. The criterion for significance was P < 0.05 for all studies.
RESULTS
Hypoxia in the livers of mice after bile duct ligation.
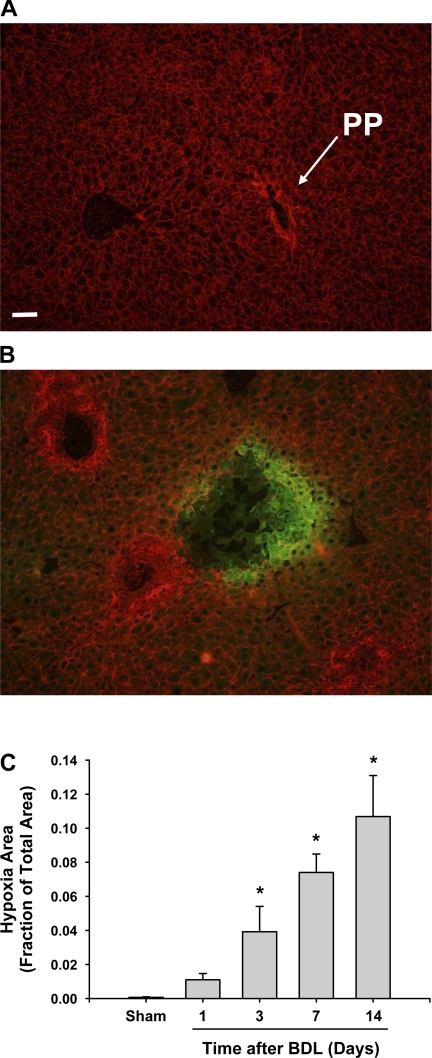
To determine whether cells in the liver become hypoxic during the development of liver fibrosis, mice were injected with pimonidazole. In cells that are hypoxic (i.e., <10 mmHg oxygen), pimonidazole is metabolized to a reactive intermediate that binds to sulfur-containing constituents within cells (1). Immunohistochemistry can then be used to detect the pimonidazole-sulfur adducts, which only occur in hypoxic cells. In liver sections from mice subjected to sham operation, no immunohistochemical staining for hypoxia (i.e., positive pimonidazole staining) was observed at 3 days after surgery (Fig. 1A). Staining for hypoxia was present in the livers of mice subjected to bile duct ligation 3 days earlier (Fig. 1B). At this time, hypoxia was primarily present at the periphery of bile infarcts (i.e., regions of hepatocellular necrosis extending from periportal areas). The extent of hypoxia increased at later times. At no time point, however, was hypoxia observed in periportal regions where bile duct proliferation occurred (i.e., areas of ductular reaction). Hypoxia was restricted to the hepatic parenchyma.
Fig. 1.
Hypoxia in the livers of bile duct-ligated mice. Wild-type mice were subjected to sham operation or bile duct ligation (BDL). One, 3, 7, and 14 days later, the mice received 120 mg/kg pimonidazole. One hour later, the mice were killed and immunohistochemistry was used to detect pimonidazole-sulfur adducts (i.e., hypoxia) in the liver, which appears green in the photomicrographs. The liver sections were counterstained with phalloidin (red staining) to more clearly define the cells in the liver. A: liver section from a mouse subjected to sham operation 3 days earlier. B: liver section from a mouse subjected to bile duct ligation 3 days earlier. PP: periportal region. C: total area of hypoxia staining was analyzed morphometrically. Data are expressed as means ± SE. *Significantly different (P < 0.05) from sham-operated mice; n = 6 where each n represents a liver section from a different mouse.
Morphometric analysis revealed that bile duct ligation caused a time-dependent increase in the area of hypoxia staining in the liver that was significant by 3 days after surgery (Fig. 1C).
Activation of HIF-1α in the liver after bile duct ligation.
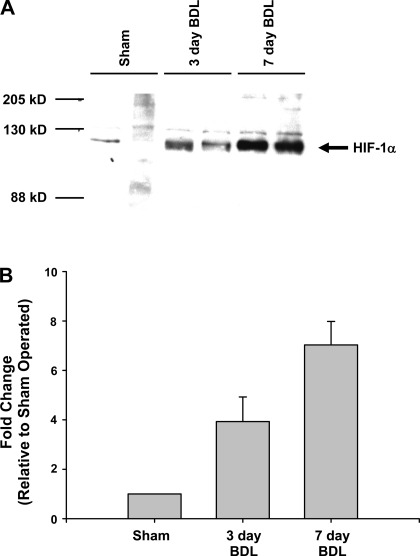
Nuclear extracts were isolated from the livers of mice subjected to sham operation or bile duct ligation, and Western blot analysis was performed to detect HIF-1α. Bile duct ligation increased HIF-1α protein by fourfold at 3 days after bile duct ligation and sevenfold by 7 days after bile duct ligation (Fig. 2).
Fig. 2.
Activation of hypoxia-inducible factor-1α (HIF-1α) in the livers of bile duct-ligated mice. Wild-type mice were subjected to sham operation or bile duct ligation. Three and 7 days later, HIF-1α was detected in nuclear extracts from liver by Western blot. A: representative Western blot; n = 4. B: intensity of the HIF-1α band was quantified, and the fold change relative to sham-operated mice was determined. Data are expressed as means ± SE; n = 4, where each n represents a different mouse.
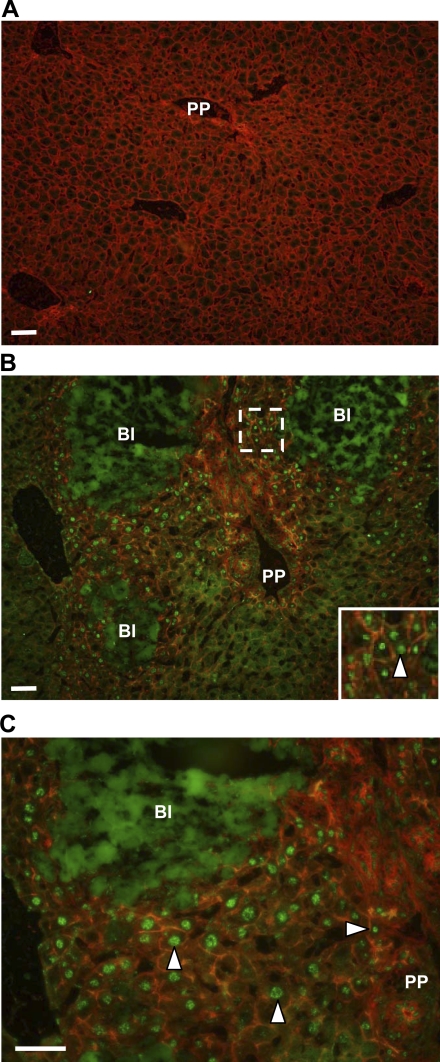
Immunohistochemistry revealed minimal HIF-1α in the livers of sham-operated mice (Fig. 3A). Occasionally, positively stained hepatocytes were observed adjacent to central veins. Numerous cells in the livers of mice subjected to bile duct ligation 7 days earlier showed positive nuclear staining for HIF-1α (Fig. 3, B and C). These cells were both adjacent to bile infarcts and present within periportal regions where bile duct proliferation occurred. Green immunofluorescence occurring within the bile infarcts was nonspecific and occurred in tissue sections incubated with only the fluorescent secondary antibody.
Fig. 3.
HIF-1α immunostaining in the livers of bile duct-ligated mice. C57Bl/6 mice were subjected to sham operation or bile duct ligation. Seven days later, immunohistochemistry was used to detect HIF-1α (green fluorescence). The liver sections were counterstained with phalloidin (red staining) to more clearly define the cells in the liver. Representative photomicrographs from mice subjected to sham operation (A) or bile duct ligation (B) 7 days earlier. The region from which the higher power image in the inset was obtained in B is outlined in a dotted line. C: high-power image of HIF-1α immunostaining of a liver section from a mouse subjected to bile duct ligation 7 days earlier showing positive nuclear HIF-1α staining in cells adjacent to bile infarcts and within periportal regions. BI, bile infarct. Arrowheads point to nuclei staining positive for HIF-1α. Representative images, n = 7.
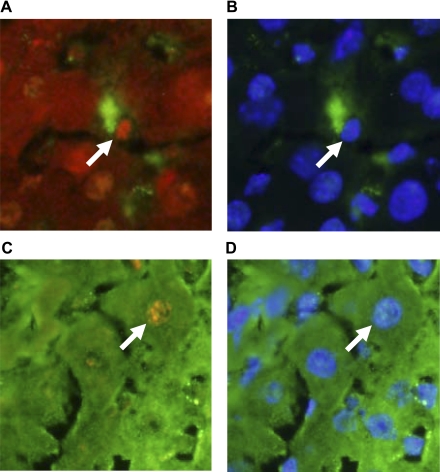
Next, double immunohistochemistry was used to identify cell types in which HIF-1α was activated. HIF-1α was activated in macrophages (Fig. 4, A and B) and hepatocytes (Fig. 4, C and D) in sections of liver from bile duct-ligated mice.
Fig. 4.
HIF-1α immunostaining in macrophages and hepatocytes in the livers of bile duct-ligated mice. C57Bl/6 mice were subjected to sham operation or bile duct ligation. Seven days later, immunohistochemistry was used to detect HIF-1α, macrophages, and hepatocytes. A: immunohistochemistry for HIF-1α (red staining) and macrophages (anti-F4/80 and anti-CD68; green staining). Arrow indicates positive HIF-1α immunostaining in the nucleus of a macrophage. B: the same field of liver shown in A showing macrophage staining (green staining) and 4′,6-diamidino-2-phenylindole (DAPI) staining that stains DNA (blue staining). Arrows in A and B point to the same region of liver (i.e., macrophage nucleus, which is positive for HIF-1α). C: immunohistochemistry for HIF-1α (red staining) and hepatocytes (anti-albumin; green staining). Arrow indicates positive HIF-1α immunostaining in the nucleus of a hepatocyte. D: the same field of liver shown in C showing hepatocyte staining (green staining) and DAPI staining (blue staining). Arrows in C and D point to the same region of liver (i.e., hepatocyte nucleus, which is positive for HIF-1α).
Effect of HIF-1α deletion on markers of cholestasis and liver injury.
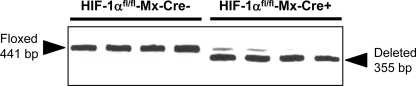
To evaluate the role of HIF-1α in the development of liver fibrosis, we first determined the deletion efficiency of HIF-1α in HIF-1αfl/fl-Mx-Cre+ mice and HIF-1αfl/fl-Mx-Cre− mice treated with pIpC. One week after the final pIpC injection, DNA was isolated from liver and the extent of HIF-1α deletion analyzed by PCR. As shown in Fig. 5, HIF-1α was deleted from 90–100% in HIF-1αfl/fl-Mx-Cre+ mice treated with pIpC. As expected, no HIF-1α deletion was detected in HIF-1αfl/fl-Mx-Cre− mice treated with pIpC. For the remainder of this article, HIF-1αfl/fl-Mx-Cre+ mice treated with pIpC will be referred to as HIF-1α-deficient mice and HIF-1αfl/fl-Mx-Cre− mice treated with pIpC will be referred to as control mice.
Fig. 5.
Livers were obtained from HIF-1αfl/fl-Mx-Cre+ mice and HIF-1αfl/fl-Mx-Cre− mice treated with polyinosinic-polycytidylic acid. PCR was used to estimate the extent of HIF-1α deletion in the livers. The floxed, undeleted HIF-1α gene produces a 441-bp PCR fragment, whereas the floxed, deleted HIF-1α gene produces a 355-bp PCR fragment.
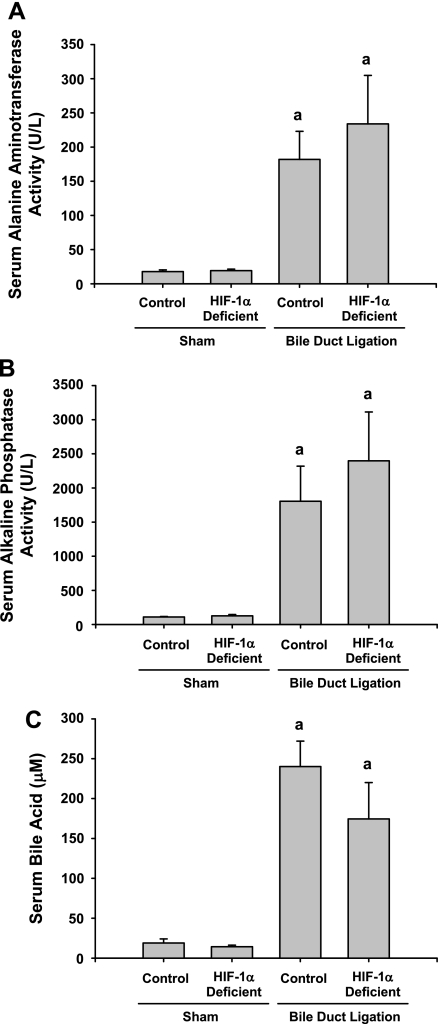
These mice were used to evaluate the role of HIF-1α in the development of liver fibrosis. For this study, control and HIF-1α-deficient mice were subjected to bile duct ligation. Fourteen days later, markers of cholestasis and liver injury were analyzed. Bile duct ligation in control and HIF-1α-deficient mice increased serum levels of ALT, ALP, and bile acids to a similar extent (Fig. 6).
Fig. 6.
Control and HIF-1α-deficient mice were subjected to bile duct ligation or sham operation. Fourteen days after surgery, serum levels of alanine aminotransferase, alkaline phosphatase, and bile acids were measured. Data are expressed as means ± SE; n = 8 mice/group. aSignificantly different (P < 0.05) from sham-operated mice.
Effect of HIF-1α deletion on histological changes in the liver after bile duct ligation.
Analysis of liver histology indicated no difference between sham-operated control and HIF-1α-deficient mice (data not shown). By 14 days after bile duct ligation, livers of control mice showed extensive bile duct proliferation and bile infarcts infiltrated by inflammatory cells (Fig. 7A). Similar changes were observed in bile duct-ligated HIF-1α-deficient mice (Fig. 7B).
Fig. 7.
Control mice and HIF-1α-deficient mice were subjected to bile duct ligation. Fourteen days later, the livers were removed and sections stained with hematoxylin and eosin. Representative photomicrograph of a section of liver from a control mouse (A) and HIF-1α-deficient mouse (B). Representative images of an n = 6 in each group.
Effect of HIF-1α deletion on bile duct proliferation in the liver after bile duct ligation.
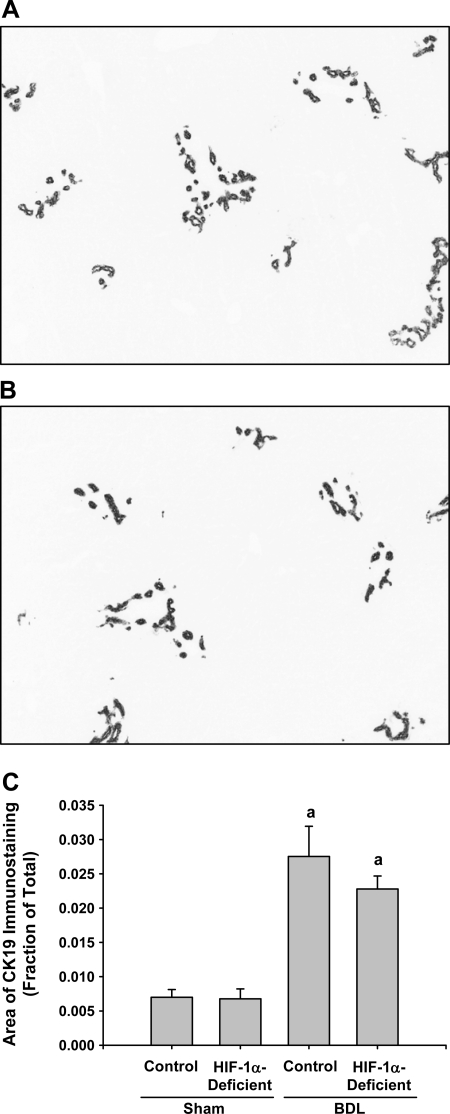
Bile duct epithelial cells were detected in sections of liver by immunohistochemistry for CK19. CK19 immunostaining was not different between sham-operated control and HIF-1α-deficient mice (data not shown). By 14 days after bile duct ligation, extensive CK19 immunostaining was observed in the livers of control mice (Fig. 8A) and HIF-1α-deficient mice (Fig. 8B) after bile duct ligation. Similar changes were observed in bile duct-ligated HIF-1α-deficient mice (Fig. 8B).
Fig. 8.
Control mice and HIF-1α-deficient mice were subjected to bile duct ligation or sham operation. Fourteen days later, CK19 was detected in the liver by immunohistochemistry. Representative photomicrograph of a section of liver from a control mouse (A) and HIF-1α-deficient mouse (B) stained for CK19. Positive staining for CK19 appears dark gray in the photomicrographs. C: total area of CK19 immunostaining was analyzed morphometrically. aSignificantly different from sham-operated mice (P < 0.05). Data are expressed as means ± SE; n = 6.
Morphometric analysis revealed that bile duct ligation caused a time-dependent increase in the area of CK19 staining in the livers of control mice and HIF-1α-deficient mice (Fig. 8C). There was no difference between the area of CK19 staining in control mice and HIF-1α-deficient mice after bile duct ligation.
Effect of HIF-1α deletion on levels of type I collagen and α-SMA in the liver after bile duct ligation.
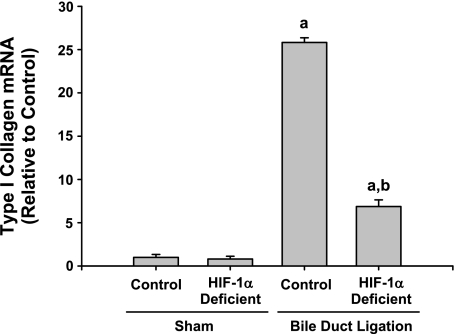
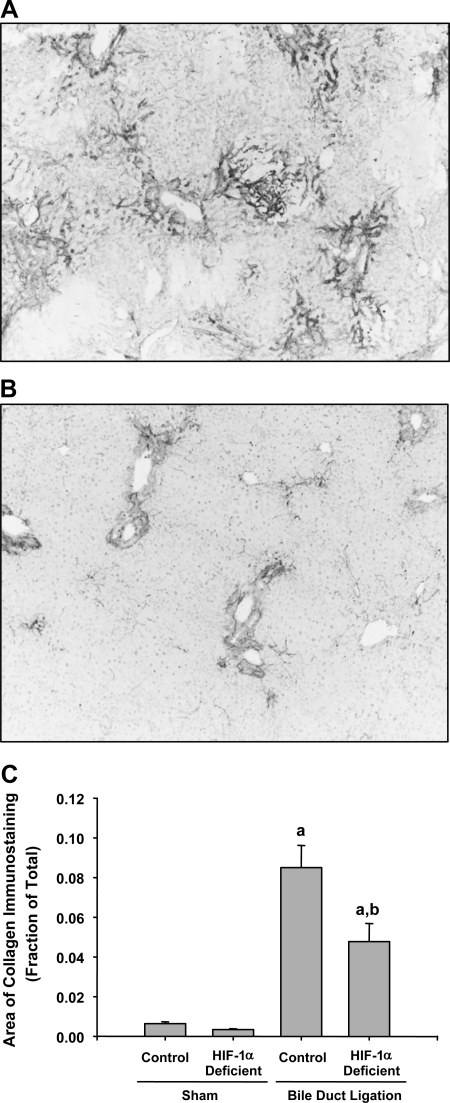
After bile duct ligation, activated hepatic stellate cells and peribiliary fibroblasts synthesize and secrete collagen (21). Accordingly, we determined whether levels of type I collagen were affected in the livers of HIF-1α-deficient mice after bile duct ligation. Type Iα1 collagen mRNA (Fig. 9) and type I collagen protein (Fig. 10) were significantly increased in the livers of control and HIF-1α-deficient mice by 14 days after bile duct ligation. Levels of type I collagen mRNA and protein were significantly reduced by ∼70 and 50%, respectively, in HIF-1α-deficient mice after bile duct ligation compared with control mice subjected to bile duct ligation (Figs. 9 and 10).
Fig. 9.
Control mice and HIF-1α-deficient mice were subjected to bile duct ligation or sham operation. Fourteen days later, levels of type I collagen were quantified in the liver by real-time PCR. aSignificantly different from sham-operated mice (P < 0.05). bSignificantly different from control mice subjected to bile duct ligation (P < 0.05). Data are expressed as means ± SE; n = 6.
Fig. 10.
Control mice and HIF-1α-deficient mice were subjected to bile duct ligation or sham operation. Fourteen days later, type I collagen was detected in the liver by immunohistochemistry. Representative photomicrograph of a section of liver from a control mouse (A) and HIF-1α-deficient mouse (B) stained for type I collagen 14 days after BDL. Positive staining for collagen appears dark gray in the photomicrographs. C: total area of type I collagen immunostaining was analyzed morphometrically. aSignificantly different from sham-operated mice (P < 0.05). bSignificantly different from control mice subjected to bile duct ligation (P < 0.05). Data are expressed as means ± SE; n = 6.
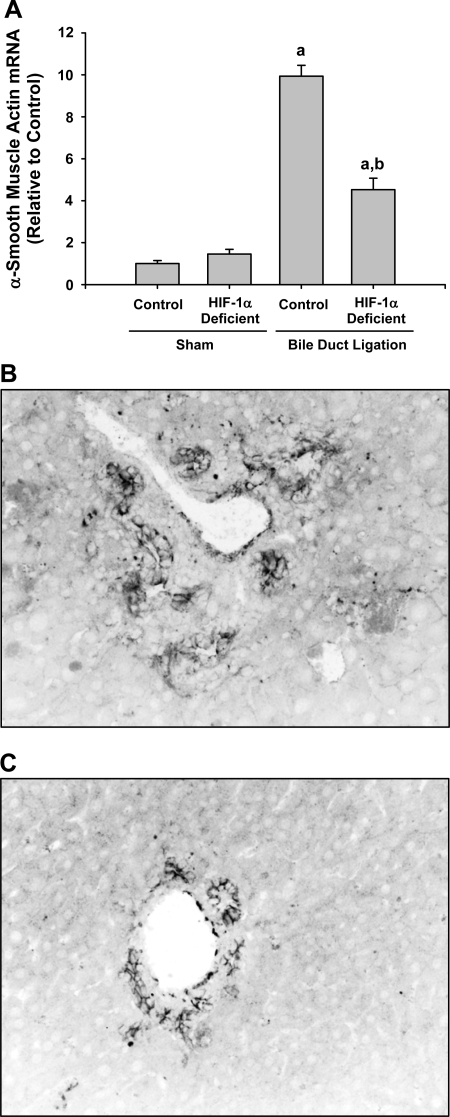
During the pathogenesis of fibrosis induced by bile duct ligation, hepatic stellate cells, and peribiliary cells differentiate into myofibroblasts and proliferate (21). When this occurs, levels of α-SMA, expressed by these cells, increase. Therefore, α-SMA levels were measured as an indirect indicator of myofibroblast activation and numbers in liver. α-SMA mRNA was significantly increased in control and HIF-1α-deficient mice 14 days after bile duct ligation (Fig. 11A). Similar to type I collagen mRNA levels, levels of α-SMA were significantly lower in HIF-1α-deficient mice compared with control mice after bile duct ligation (Fig. 11A). Immunohistochemical staining for α-SMA in livers of control mice 14 days after bile duct ligation showed extensive positive α-SMA staining around proliferating bile ducts and within the hepatic parenchyma in periportal regions (Fig. 11B). Less α-SMA immunostaining was observed in the livers of bile duct-ligated HIF-1α-deficient mice (Fig. 11C).
Fig. 11.
Control mice and HIF-1α-deficient mice were subjected to bile duct ligation or sham operation. Fourteen days later, α-smooth muscle actin levels were quantified by real-time PCR and immunohistochemistry. A: quantification of α-smooth muscle actin mRNA levels in the livers of mice. aSignificantly different from sham-operated mice (P < 0.05). bSignificantly different from control mice subjected to bile duct ligation (P < 0.05). Data are expressed as means ± SE; n = 6. Representative photomicrograph of a section of liver from a bile duct-ligated control mouse (B) and HIF-1α-deficient mouse (C) stained for α-smooth muscle actin. Positive staining for α-smooth muscle actin appears dark gray in the photomicrographs.
Effect of HIF-1α deletion on levels of profibrotic mediators in the liver after bile duct ligation.
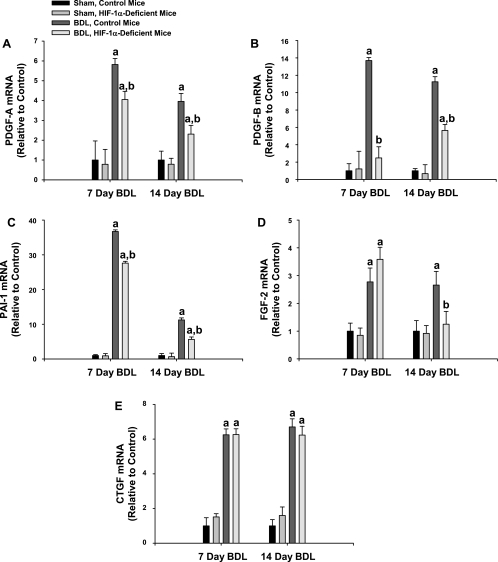
Levels of PDGF-A, PDGF-B, PAI-1, FGF-2, and CTGF mRNAs were significantly increased in the livers of control mice subjected to bile duct ligation 7 and 14 days earlier (Fig. 12). Levels of PDGF-A, PDGF-B, and PAI-1 mRNAs were significantly decreased in HIF-1α-deficient mice at both time points compared with control mice 14 days after bile duct ligation, whereas levels of FGF-2 mRNA were only significantly decreased at 14 days (Fig. 12). Levels of CTGF mRNA were not significantly different at either time point (Fig. 12). Another important growth factor regulated by HIFs is vascular endothelial growth factor (VEGF); therefore, levels of VEGF mRNA were measured. Surprisingly, VEGF mRNA levels were not increased in control mice or HIF-1α-deficient mice subjected to bile duct ligation at either time point compared with sham-operated mice (data not shown).
Fig. 12.
Control mice and HIF-1α-deficient mice were subjected to bile duct ligation or sham operation. Seven and 14 days later, platelet-derived growth factor (PDGF)-A (A), PDGF-B (B), plasminogen activator inhibitor-1 (PAI-1; C), fibroblast growth factor-2 (FGF-2; D), and connective tissue growth factor (CTGF; E) levels were quantified by real-time PCR. Values indicate fold change relative to control (i.e., control mice, sham operated). Data are expressed as means ± SE; n = 8 mice/group. aSignificantly different (p < 0.05) from sham-operated mice. bSignificantly different (p < 0.05) from control, bile duct-ligated mice.
DISCUSSION
During chronic injury in the liver, cells become stimulated to secrete growth factors, such PDGF, FGF-2, and CTGF. These proteins stimulate hepatic stellate cells and other cell types to differentiate into myofibroblasts and to produce collagen. Furthermore, proteins such as PAI-1 are produced that modulate protease activity, which may indirectly facilitate collagen deposition. Although it is clear that PDGF, FGF-2, CTGF, and PAI-1 contribute to the development of fibrosis, what remains largely unknown is the mechanism by which liver injury stimulates production of these proteins. Our studies are the first to indicate that hypoxia through activation of the transcription factor, HIF-1α, is critical for this process.
The present studies show that HIF-1α was activated in the livers of mice subjected to bile duct ligation. Activation occurred by 3 days after bile duct ligation, a time before which significant fibrosis is typically present in this model (Fig. 2). Furthermore, HIF-1α activation occurred primarily in hepatocytes and macrophages at the periphery of bile infarcts and within regions of proliferating bile ducts (Figs. 3 and 4). These are both regions where fibrosis occurs. Interestingly, although we detected HIF-1α activation within areas of bile duct proliferation, we did not observe hypoxia in this region by pimonidazole immunostaining (Fig. 1). It is possible that other stimuli activate HIF-1α in these cell types. Studies have shown that several mediators, including cytokines, growth factors, bacterial lipopolysaccharide, and reactive oxygen species, activate HIF-1α in normoxic cells (3, 13, 15, 26, 27). Levels of all of these mediators are increased after bile duct ligation, suggesting that one or more of them may activate HIF-1α in cells within periportal regions (2, 29, 32, 33).
To test the hypothesis that HIF-1α is critical for upregulation of profibrotic mediators, such as PDGF, FGF-2, CTGF, and PAI-1, we used an inducible Cre/lox system to delete HIF-1α in adult mice. As mentioned previously, deletion of HIF-1α in mice causes embryonic lethality (17). Therefore, to study the role of HIF-1α in the development of liver fibrosis, we crossed HIF-1αfl/fl mice with mice expressing Cre recombinase under control of the Mx interferon-inducible promoter. The advantage of this system is that it allows investigation of the function of HIF-1α in adult mice while bypassing the need for HIF-1α for normal embryonic development. Using this system, we were able to delete HIF-1α from 90–100% in the livers of HIF-1α-deficient mice and demonstrate for the first time that HIF-1α is important for the development of fibrosis. In these studies, measures of fibrosis, including type I collagen and α-SMA levels, were reduced in HIF-1α-deficient mice after bile duct ligation (Figs. 9–11). The mechanism by which liver fibrosis was reduced in HIF-1α-deficient mice appeared to be related to a reduction in the levels of PDGF, FGF-2, and PAI-1 (Fig. 12). All of these mediators have been shown to contribute to the development of liver fibrosis, and hypoxia-response elements are present in the promoters of all of these genes (4, 19, 34). Furthermore, studies have confirmed that the promoters of these genes are activated in hypoxic cells in a HIF-1α-dependent manner, suggesting that the hypoxia-response elements present in the promoters of these genes are functional (4, 19, 34). Therefore, during the development of fibrosis, HIF-1α may directly regulate production of these mediators by hypoxic cells in the liver. Interestingly, although CTGF is another HIF-1α target gene (16), CTGF levels were not affected in HIF-1α-deficient mice (Fig. 12). This suggests that other pathways regulate this growth factor during the development of fibrosis. Recent studies showed that transforming growth factor-β regulates CTGF in hepatocytes (12). Therefore, it is possible that this growth factor stimulates production of CTGF in the livers of bile duct-ligated mice.
Although the inducible Cre/lox system allowed investigation of the role of HIF-1α in the development of fibrosis, one disadvantage of this system is that HIF-1α deletion is not cell type specific. Therefore, although we can conclude that HIF-1α is an important regulator of profibrotic mediator production during the development of fibrosis, we cannot specifically determine in which cell type(s) in liver HIF-1α performs this important function. Our studies did, however, demonstrate that HIF-1α was primarily activated in hepatocytes and macrophages (Fig. 4). Furthermore, previous studies have demonstrated that hypoxia modulates expression of genes important for fibrosis in these cells. For instance, in vitro studies have shown that HIF-1α directly regulates PAI-1 in hypoxic hepatocytes (19). Furthermore, exposure of macrophages to hypoxia upregulates PAI-1, PDGF, and FGF-2 (24, 25). It is possible that these genes may be similarly upregulated in hypoxic Kupffer cells in a HIF-1α-dependent manner.
Overall, we propose the following model for the role of HIF-1α in the development of fibrosis. During chronic liver injury, regions of hypoxia develop in the liver possibly due to disrupted blood flow and/or other factors. Hepatocellular hypoxia stimulates activation of HIF-1α, which regulates production of growth factors and other mediators that are intended to facilitate repair and revascularization of the liver. As liver injury continues, however, liver hypoxia and HIF-1α activation become persistent, causing continuous production of growth promoting factors that ultimately stimulate overproduction of collagen and liver fibrosis. Overall, our studies suggest that HIF-1α regulates numerous profibrotic mediators and thus serves as the critical link between chronic liver injury and the initiation of fibrosis.
GRANTS
This study was supported by National Institutes of Health Grants DK073566 (B. L. Copple) and Center of Biomedical Research Excellence (COBRE) P20 RR021940 as well as the Molecular Biology Core and the Histology Core supported by the COBRE grant.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Arteel GE, Thurman RG, Raleigh JA. Reductive metabolism of the hypoxia marker pimonidazole is regulated by oxygen tension independent of the pyridine nucleotide redox state. Eur J Biochem 253: 743–750, 1998. [DOI] [PubMed] [Google Scholar]
- 2.Beierle EA, Vauthey JN, Moldawer LL, Copeland EM 3rd. Hepatic tumor necrosis factor-alpha production and distant organ dysfunction in a murine model of obstructive jaundice. Am J Surg 171: 202–206, 1996. [DOI] [PubMed] [Google Scholar]
- 3.Blouin CC, Page EL, Soucy GM, Richard DE. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1alpha. Blood 103: 1124–1130, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Calvani M, Rapisarda A, Uranchimeg B, Shoemaker RH, Melillo G. Hypoxic induction of an HIF-1alpha-dependent bFGF autocrine loop drives angiogenesis in human endothelial cells. Blood 107: 2705–2712, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cash TP, Pan Y, Simon MC. Reactive oxygen species and cellular oxygen sensing. Free Radic Biol Med 43: 1219–1225, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159, 1987. [DOI] [PubMed] [Google Scholar]
- 7.Coleman ML, Ratcliffe PJ. Oxygen sensing and hypoxia-induced responses. Essays Biochem 43: 1–15, 2007. [DOI] [PubMed] [Google Scholar]
- 8.Copple BL, Roth RA, Ganey PE. Anticoagulation and inhibition of nitric oxide synthase influence hepatic hypoxia after monocrotaline exposure. Toxicology 225: 128–137, 2006. [DOI] [PubMed] [Google Scholar]
- 9.Corpechot C, Barbu V, Wendum D, Kinnman N, Rey C, Poupon R, Housset C, Rosmorduc O. Hypoxia-induced VEGF and collagen I expressions are associated with angiogenesis and fibrogenesis in experimental cirrhosis. Hepatology 35: 1010–1021, 2002. [DOI] [PubMed] [Google Scholar]
- 10.Friedman SL Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev 88: 125–172, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaber T, Dziurla R, Tripmacher R, Burmester GR, Buttgereit F. Hypoxia inducible factor (HIF) in rheumatology: low O2! See what HIF can do! Ann Rheum Dis 64: 971–980, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gressner OA, Lahme B, Demirci I, Gressner AM, Weiskirchen R. Differential effects of TGF-beta on connective tissue growth factor (CTGF/CCN2) expression in hepatic stellate cells and hepatocytes. J Hepatol 47: 699–710, 2007. [DOI] [PubMed] [Google Scholar]
- 13.Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, Simon MC, Hammerling U, Schumacker PT. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab 1: 401–408, 2005. [DOI] [PubMed] [Google Scholar]
- 14.Han YP Matrix metalloproteinases, the pros and cons, in liver fibrosis. J Gastroenterol Hepatol 21, Suppl 3: S88–S91, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellwig-Burgel T, Rutkowski K, Metzen E, Fandrey J, Jelkmann W. Interleukin-1beta and tumor necrosis factor-alpha stimulate DNA binding of hypoxia-inducible factor-1. Blood 94: 1561–1567, 1999. [PubMed] [Google Scholar]
- 16.Higgins DF, Biju MP, Akai Y, Wutz A, Johnson RS, Haase VH. Hypoxic induction of Ctgf is directly mediated by Hif-1. Am J Physiol Renal Physiol 287: F1223–F1232, 2004. [DOI] [PubMed] [Google Scholar]
- 17.Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev 12: 149–162, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji S, Lemasters JJ, Christenson V, Thurman RG. Periportal and pericentral pyridine nucleotide fluorescence from the surface of the perfused liver: evaluation of the hypothesis that chronic treatment with ethanol produces pericentral hypoxia. Proc Natl Acad Sci USA 79: 5415–5419, 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kietzmann T, Roth U, Jungermann K. Induction of the plasminogen activator inhibitor-1 gene expression by mild hypoxia via a hypoxia response element binding the hypoxia-inducible factor-1 in rat hepatocytes. Blood 94: 4177–4185, 1999. [PubMed] [Google Scholar]
- 20.Kim ND, Moon JO, Slitt AL, Copple BL. Early growth response factor-1 is critical for cholestatic liver injury. Toxicol Sci 90: 586–595, 2006. [DOI] [PubMed] [Google Scholar]
- 21.Kinnman N, Francoz C, Barbu V, Wendum D, Rey C, Hultcrantz R, Poupon R, Housset C. The myofibroblastic conversion of peribiliary fibrogenic cells distinct from hepatic stellate cells is stimulated by platelet-derived growth factor during liver fibrogenesis. Lab Invest 83: 163–173, 2003. [DOI] [PubMed] [Google Scholar]
- 22.Kisseleva T, Brenner DA. Hepatic stellate cells and the reversal of fibrosis. J Gastroenterol Hepatol 21, Suppl 3: S84–S87, 2006. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science 269: 1427–1429, 1995. [DOI] [PubMed] [Google Scholar]
- 24.Kuwabara K, Ogawa S, Matsumoto M, Koga S, Clauss M, Pinsky DJ, Lyn P, Leavy J, Witte L, Joseph-Silverstein J, Furie MB, Torcia G, Cozzolino F, Kamada T, Stern DM. Hypoxia-mediated induction of acidic/basic fibroblast growth factor and platelet-derived growth factor in mononuclear phagocytes stimulates growth of hypoxic endothelial cells. Proc Natl Acad Sci USA 92: 4606–4610, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao H, Hyman MC, Lawrence DA, Pinsky DJ. Molecular regulation of the PAI-1 gene by hypoxia: contributions of Egr-1, HIF-1α, and C/EBPα. FASEB J 21: 935–949, 2007. [DOI] [PubMed] [Google Scholar]
- 26.Mansfield KD, Guzy RD, Pan Y, Young RM, Cash TP, Schumacker PT, Simon MC. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab 1: 393–399, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMahon S, Charbonneau M, Grandmont S, Richard DE, Dubois CM. Transforming growth factor beta1 induces hypoxia-inducible factor-1 stabilization through selective inhibition of PHD2 expression. J Biol Chem 281: 24171–24181, 2006. [DOI] [PubMed] [Google Scholar]
- 28.Rosmorduc O, Wendum D, Corpechot C, Galy B, Sebbagh N, Raleigh J, Housset C, Poupon R. Hepatocellular hypoxia-induced vascular endothelial growth factor expression and angiogenesis in experimental biliary cirrhosis. Am J Pathol 155: 1065–1073, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saperstein LA, Jirtle RL, Farouk M, Thompson HJ, Chung KS, Meyers WC. Transforming growth factor-beta 1 and mannose 6-phosphate/insulin-like growth factor-II receptor expression during intrahepatic bile duct hyperplasia and biliary fibrosis in the rat. Hepatology 19: 412–417, 1994. [PubMed] [Google Scholar]
- 30.Tomita S, Sinal CJ, Yim SH, Gonzalez FJ. Conditional disruption of the aryl hydrocarbon receptor nuclear translocator (Arnt) gene leads to loss of target gene induction by the aryl hydrocarbon receptor and hypoxia-inducible factor 1alpha. Mol Endocrinol 14: 1674–1681, 2000. [DOI] [PubMed] [Google Scholar]
- 31.Tomita S, Ueno M, Sakamoto M, Kitahama Y, Ueki M, Maekawa N, Sakamoto H, Gassmann M, Kageyama R, Ueda N, Gonzalez FJ, Takahama Y. Defective brain development in mice lacking the Hif-1alpha gene in neural cells. Mol Cell Biol 23: 6739–6749, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai LY, Lee KT, Liu TZ. Evidence for accelerated generation of hydroxyl radicals in experimental obstructive jaundice of rats. Free Radic Biol Med 24: 732–737, 1998. [DOI] [PubMed] [Google Scholar]
- 33.Van Bossuyt H, Desmaretz C, Gaeta GB, Wisse E. The role of bile acids in the development of endotoxemia during obstructive jaundice in the rat. J Hepatol 10: 274–279, 1990. [DOI] [PubMed] [Google Scholar]
- 34.Yoshida D, Kim K, Noha M, Teramoto A. Hypoxia inducible factor 1-alpha regulates of platelet derived growth factor-B in human glioblastoma cells. J Neurooncol 76: 13–21, 2006. [DOI] [PubMed] [Google Scholar]