Abstract
Epithelial cells of the small intestine are the site of zinc absorption. Intestinal uptake of zinc is inversely proportional to the dietary supply of this essential micronutrient. The mechanism responsible for this adaptive differential in apical zinc transport is not known. The zinc transporter Zip4 (Slc39a4) is essential for adequate enteric zinc uptake. In mice, Zip4 expression is upregulated at low zinc intakes with a concomitant ZIP4 localization to the apical enterocyte plasma membrane. With the present experiments, we show that the zinc finger transcription factor Krüppel-like factor 4 (KLF4), produced in high abundance in the intestine, is expressed at elevated levels in mice fed a low-zinc diet. In the murine intestinal epithelial cell (IEC) line MODE-K, zinc depletion of culture medium with cell-permeant and cell-impermeant chelators increased Zip4 and Klf4 mRNA and Zip4 heterogeneous nuclear RNA expression. Zinc depletion led to increased KLF4 in nuclear extracts. Knockdown of KLF4 using small interfering RNA transfection drastically limited ZIP4 induction upon zinc depletion and reduced 65Zn uptake by depleted IECs. EMSAs with nuclear extracts of IECs showed KLF4 binding to cis elements of the mouse Zip4 promoter, with increased binding under zinc-limited conditions. Reporter constructs with the Zip4 promoter and mutation studies further demonstrated that Zip4 is regulated through a KLF4 response element. These data from experiments with mice and murine IECs demonstrate that KLF4 is induced during zinc restriction and is a transcription factor involved in adaptive regulation of the zinc transporter ZIP4.
Keywords: nutrient zinc transport, absorption, transcription, gene regulation
several mutations of the Slc39a4 gene (Zip4) produce acrodermatitis enteropathica in humans (23, 33). Acrodermatitis enteropathica patients have low intestinal zinc absorption and severe zinc deficiency, including secondary cutaneous infections, as well as other manifestations of zinc deprivation (19, 22, 24, 31). The demonstration that pathological signs abate in acrodermatitis enteropathica patients upon zinc supplementation (22, 24, 31) suggests that ZIP4 is a major zinc transporter in the gastrointestinal tract responsible for adequate zinc balance and homeostasis in humans. During zinc deficiency in rodents, zinc absorption increases primarily through increased kinetics of uptake by the intestine (12, 20). Most likely, this adaptive capacity for absorption occurs through ZIP4 upregulation (7, 17); however, the mechanism for this response has not been elucidated. Since the upregulation of Zip4 occurs in the intestine, but not in other tissues such as liver, brain, and pancreas (unpublished data), we hypothesized that a zinc-responsive transcription factor with relatively high expression in intestinal cells would be involved in Zip4 upregulation by zinc deficiency.
The transcription factor Krüppel-like factor 4 (KLF4), originally named gut-enriched Krüppel-like factor, is highly expressed in some epithelial tissues, including the intestinal epithelium (29). KLF4 is 1 of the 17 members of the mammalian Krüppel-like transcription factor family (25). KLFs bind to GC-rich or CACCC consensus sequences through three COOH-terminal (C2H2) zinc fingers. KLF4 influences intestinal, skin, epithelial cell, and adipocyte differentiation (1, 3, 10). Furthermore, this transcription factor is involved in regulation of inducible nitric oxide synthase (8) and intestinal alkaline phosphatase (11). Two consensus DNA binding sequences for KLF4, both upstream of the murine Zip4 transcription start site, can be identified (27). In this report, we demonstrate the increased expression of KLF4 in response to zinc depletion in mouse intestine and the mouse intestinal epithelial cell (IEC) line MODE-K and regulation of the murine Zip4 gene by KLF4.
MATERIALS AND METHODS
Animals and diets.
Male 6-wk-old CD-1 mice (Charles River) were housed individually in stainless steel hanging wire cages and fed a zinc-adequate diet (30 mg Zn/kg; Research Diets) for 1 wk. Then the animals were randomly assigned to a zinc-deficient diet (<1 mg Zn/kg; ZnD) group or a zinc-adequate diet (ZnA) group for 3 wk as described elsewhere (17). Plasma zinc concentrations were measured by atomic absorption spectrophotometry. Mice were anesthetized with 2-bromo-2-chloro-1,1,1-trifluoroethane (Sigma) and killed by exsanguination via cardiac puncture. The protocol was approved by the University of Florida Institutional Animal Care and Use Committee.
Cell culture of mouse IEC MODE-K cells.
The IEC line MODE-K (kindly donated by Dr. Kenneth Croitoru, McMaster University, Hamilton, ON, Canada) was derived as previously described (32). Cells were cultured in DMEM (Mediatech) supplemented with 10% (vol/vol) FBS (Mediatech), 20 μM glutamine (Sigma), nonessential amino acids (Mediatech), and antibiotic-antimycotic solution (Sigma) (2). Cells were grown to confluence in 24-well plates before treatments were started. In some experiments, the culture medium contained diethylenetriamine pentaacetic acid (DTPA) at 50 μM alone or with 50 μM ZnSO4. In other experiments, the culture medium contained N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN) at 4 μM alone or with 4 μM ZnSO4. Cells were usually incubated for 24 h. For Klf4 small interfering RNA (siRNA) inhibition, cells were seeded at 1 × 105 per well in 24-well plates. After 12 h, the cells were treated with 80 nM nontargeting siRNA (siCONTROL #1) or 80 nM mouse KLF4 SMARTpool siRNA (Dharmacon). HyPerFect transfection reagent (Qiagen) was used for transfection of siRNA, and Zip4 expression was assessed 24 h after the cells were placed in control or DTPA-containing medium. In some experiments, confluent MODE-K cells were transfected with Klf4 siRNA or nontargeting siRNA, and 24 h later the cells were placed in fresh medium (2.5 μM Zn) containing 65Zn (57,000 cpm/ml) for measurement of zinc uptake. At specific times, the 65Zn-containing medium was removed and the cells were washed in EDTA-containing buffer. The cells were lysed with an SDS-NaOH solution for measurement of 65Zn by gamma spectrometry (18). Protein concentrations were measured spectrophotometrically using the RC DC assay (Bio-Rad) method here and in all other experiments.
mRNA quantitation by quantitative RT-PCR.
Total RNA was isolated by using TRIzol reagent (Invitrogen) and treated with Turbo DNase (Ambion). Quantitative PCR (qPCR) was performed using one- or two-step RT reactions (Applied Biosystems) as described previously (17). Primers used for qPCR are as follows: 5′-CAGCTGGCAAGCGCTACA-3′ (forward) and 5′-CCTTTCTCCTGATTATCCATTCACA-3′ (reverse) for Klf4 and 5′-TCATGGTCAAGTTCCCAGCAAGTCAGC-3′ for TaqMan probe, 5′-AACTCACCTCATGGGCCTCTT-3′ (forward) and 5′-GGGTTTCGGTTGGCATCATA-3′ (reverse) for alkaline phosphatase, and 5′-AGGGCGAGGCTTGGATTC-3′ (forward) and 5′-TAAGGCTCTTCCCCTTTCTAACC-3′(reverse) for Zip4 heterogeneous nuclear RNA (hnRNA). The latter two assays used SYBR Green (Applied Biosystems) for detection. The primer and probe sequences used for measurement of Zip4 mRNA were reported previously (17). Transcript abundances were normalized to levels of 18S rRNA and are expressed as relative values.
Western blot analysis and immunohistochemistry.
Total cell and tissue lysates were prepared by homogenization in 50 mM Tris buffer (pH 7.4) with 1% Triton X-100 and protease inhibitor cocktail (Pierce). Cytoplasmic and nuclear fractions were extracted using the NE-PER extraction system (Pierce) according to the manufacturer's protocol. Proteins were electrophoretically separated on 10% SDS-polyacrylamide gels and then transferred onto nitrocellulose membranes (Whatman). Blots were incubated with a polyclonal rabbit anti-KLF4 antibody (catalog no. H-180, Santa Cruz Biotechnology). Preparation of the affinity-purified polyclonal rabbit anti-Zip4 antibody was described previously (17). Horseradish peroxidase-conjugated anti-rabbit IgG antibody was used as the secondary antibody. Immunoreactive bands were visualized using enhanced chemiluminescence (Pierce) and X-ray film.
Sections of intestine were removed and fixed in 10% neutral buffered formalin. Histological slides were prepared as previously described (17). The anti-KLF4 antibody (catalog no. T-16, Santa Cruz Biotechnology) was used at 10 μg/ml with anti-rabbit IgG-Alexa 594 conjugate (Invitrogen) as the secondary antibody. Nuclei were stained with 4′,6-diamidino-2-phenylindole. Fluorescent images were captured with an Olympus confocal microscope or a Zeiss epifluoresence microscope and digitally processed.
EMSA.
Putative KLF4 binding sites in the Zip4 promoter were identified using the Genomatix suite of programs (http://www.genomatix.de, Genomatix Software, Munich, Germany) (27). EMSA was performed using nuclear extracts from small intestine and IEC cells according to protocols for the LightShift chemiluminescent system (Pierce). The gel concentration was 6% acrylamide. Double-stranded DNA probes (5′-ACGCGCTCAAACCTTTAATCCCCCTAGC-3′ and 5′-CGAGTTTGGAAATTAGGGGGATCGAAGA-3′) that corresponded to the KLF binding sites at positions −280 to −300 bp of the mouse Zip4 promoter were generated. Nucleotides were labeled using biotin 3′-end DNA labeling (Pierce). Detection of supershift mobility was assessed by reaction of KLF4 antibody (see above) with nuclear extracts for 30 min before addition of the labeled nucleotides for the EMSA.
Luciferase reporter gene assay.
Mouse genomic DNA was used as the template to generate a mouse Zip4 promoter (−1437 to +22) by PCR, and the promoter was cloned into the pGL3-Basic vector (Promega). Mouse Zip4 promoter constructs with mutated sequences for the −280- to −300-bp site were generated using site-directed mutagenesis (QuickChange II, Stratagene). The primers were designed using Stratagene's web-based QuickChange primer design program. The sequences were as follows: 5′-catgtttgacttgataacatctacgctttatatcctttaatccccctagcttctggtcc-3′ for KLF4E1mutF and 5′-ggaccagaagctagggggattaaaggatataaagcgtagatgttatcaagtcaaacatg-3′ for KLF4E1mutR. The Dual-Glo luciferase assay system (Promega) was used as recommended by the manufacturer to assay promoter activity. Luciferase activity was detected using a microplate reader (SpectraMax M5, Molecular Devices).
Statistical analysis.
Values are means ± SD and were analyzed for statistical significance by Student's t-test, ANOVA, and the Student-Newman post test as appropriate. The significance level was set at P < 0.05.
RESULTS
KLF4 and Zip4 are upregulated in zinc-depleted mice.
Intestinal metallothionein mRNA levels were significantly reduced in ZnD mice (Fig. 1A). The plasma zinc concentrations were 0.6 and 0.2 μM in ZnA and ZnD mice, respectively. These parameters are presented as evidence for zinc depletion of the mice, as we reported previously (17). We examined a number of transcription factors that had potential binding elements within 1 kb of the Zip4 promoter. These included SCL6, ZBPK1, RBPJκ, DREAM, ZBP/ZNF202, and KLF4. Expression of none of these transcription factors, except Klf4 (data not shown), showed consistent responsiveness to zinc status (data not shown). ZnD mice exhibited increased abundance of Zip4 and Klf4 transcripts in the intestine (Fig. 1, B and C). An increase in alkaline phosphatase mRNA was also found in the ZnD mice (Fig. 1D).
Fig. 1.
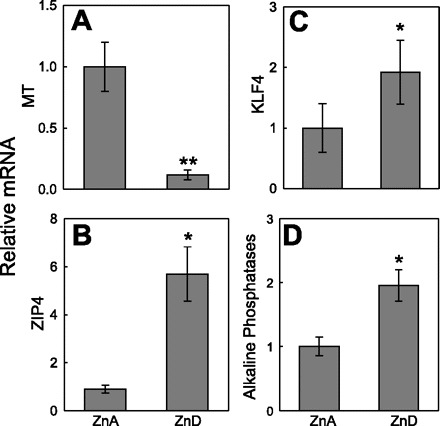
Characterization of Zip4 and Klf4 expression in intestines of control (ZnA) and zinc-depleted (ZnD) mice. mRNA levels in total RNA from intestines of the mice were determined by quantitative PCR (qPCR) and normalized to 18S rRNA levels. A: Mt mRNA, a positive control, demonstrates zinc depletion in ZnD mice. B–D: Zip4, Klf4, and alkaline phosphatase transcripts. Values are means ± SD (n = 3–4 mice per group). *P < 0.05; **P < 0.02 vs. ZnA.
Immunofluoresence confocal microscopy with anti-KLF4 antibody shows upregulation of this transcription factor in nuclei of enterocytes of the ZnD mice. The abundance of KLF4 in nuclei of enterocytes of the ZnA mice (Fig. 2A) is clearly less than the robust presence in nuclei of enterocytes of the ZnD mice (Fig. 2B). In particular, the KLF4 nuclear localization, as indicated by red/pink spots in nuclei of the ZnD mice (Fig. 2B), is nearly absent from nuclei of the ZnA mice (Fig. 2A). Western analysis of nuclear extracts from intestines (Fig. 3) confirmed the increased abundance of KLF4 in nuclei of the ZnD mice as observed with confocal microscopy (Fig. 2, A and B). Similarly, ZIP4 is strikingly more abundant at the apical plasma membrane of enterocytes of the ZnD mice (Fig. 2D) than the ZnA mice (Fig. 2C). The latter images were captured at high magnification to show the plasma membrane localization of ZIP4. In companion experiments, in mice fed a diet supplemented with additional zinc at six times the usual content (180 mg/kg) for 1 wk, intestinal Zip4 and Klf4 transcript and protein abundance remained the same in control mice (data not shown). Distribution of Zip4 and Klf4 transcripts in tissue, including skin, liver, lung, brain, and small intestine, was determined. Significant, differential expression in response to zinc depletion was limited to the small intestine (Fig. 4). The transcript abundances for Zip4 and Klf4 in the lung are shown for comparison.
Fig. 2.
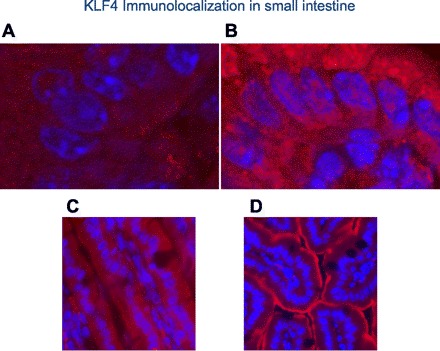
Immunofluoresence localization of Krüppel-like factor 4 (KLF4) and ZIP4 (both detected separately with Texas Red) in mouse intestine. A and C: KLF4 and ZIP4, respectively, in ZnA mice. B and D: KLF4 and ZIP4, respectively, in ZnD mice. Sections were stained with 4′,6-diamidino-2-phenylindole nuclear stain. KLF4 images were captured by confocal microscopy and ZIP4 images by epifluoresence microscopy. Images represent findings from numerous experiments. Magnification: ×63 + ×4 (A and B) ×630 (C and D).
Fig. 3.
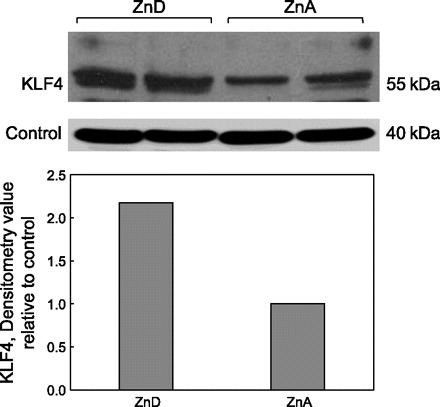
Western blot (top) and densitometric (bottom) analysis of KLF4 in nuclear extracts from intestines of ZnA and ZnD mice. Data represent results from replicated experiments.
Fig. 4.
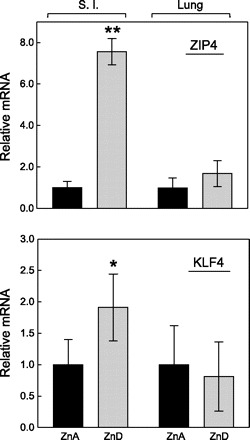
Expression of Zip4 and Klf4 in small intestine (SI) and lung of ZnA (solid bars) and ZnD (gray bars) mice. Transcript abundance is normalized to level of 18S rRNA. Values are means ± SD (n = 3 mice per group). *P < 0.05; **P < 0.02 vs. ZnA.
KLF4 and Zip4 expression in cultured IECs.
The metal chelators TPEN and DTPA were used to examine directly the influence of cellular zinc depletion on Zip4 and Klf4 expression in mouse IECs. Zip4 mRNA (Fig. 5A) and Klf4 mRNA (Fig. 5B) increased significantly, two- to threefold, in TPEN-treated cells. The effect of TPEN chelation on Zip4 and Klf4 mRNA levels was completely abolished by the equimolar addition of zinc to TPEN-containing medium. In addition, DTPA in the culture medium increased Zip4 (Fig. 5C) and Klf4 (Fig. 5D) transcript levels by twofold. Similarly, this effect was abolished by the equimolar addition of zinc to DTPA-containing medium. Although the specificity of Zn2+ chelation is greater for TPEN than DTPA, and DTPA also binds Fe3+, the finding that both chelators at equimolar concentrations with zinc fully blocked zinc depletion argues that the effect of KLF4 on Zip4 upregulation is specific for zinc. To examine the transcription rate for Zip4, we measured the Zip4 hnRNA level in control and DTPA-treated cells (Fig. 5E). Zinc depletion resulted in a 4.7-fold increase in Zip4 hnRNA after 24 h of incubation with DTPA. This suggests that zinc depletion increases the Zip4 transcription rate. A time-course study comparing the changes in Zip4 and Klf4 transcripts in cells treated with DTPA for up to 24 h showed that the increase in Klf4 mRNA occurred before the increase in Zip4 mRNA following zinc depletion (data not shown). This temporal finding supports our hypothesis that the induction of Zip4 by zinc depletion is KLF4 dependent.
Fig. 5.
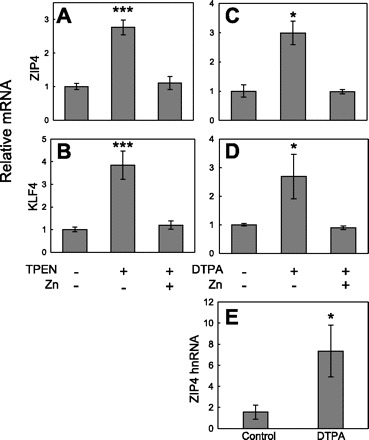
Response of Zip4 and Klf4 mRNA and Zip4 heterogeneous nuclear RNA (hnRNA) expression to zinc depletion in the intestinal epithelial cell line MODE-K. Cells were incubated for 24 h in medium alone (control), medium containing 4 μM N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN; with or without 4 μM zinc), or medium containing 50 μM diethylenetriamine pentaacetic acid (DTPA; with or without 50 μM zinc). Abundance of Zip4 and/or Klf4 mRNAs was measured by qPCR and normalized to level of 18S rRNA. A and B: TPEN comparison groups. C and D: DTPA comparison groups. E: relative abundance of Zip4 hnRNA after incubation for 24 h with or without 50 μM DTPA. Values are means ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001 vs. other groups.
KLF4 knockdown decreases Zip4 expression.
Targeting by siRNA was used to examine the role of KLF4 in Zip4 upregulation by zinc depletion. More than 90% knockdown of Klf4 mRNA was obtained by Klf4 siRNA treatment (Fig. 6A). A control (nontargeting) siRNA served as the transfection control. In cells transfected with the control siRNA, incubation with 50 μM DTPA produced a fourfold increase in Zip4 mRNA compared with cells not exposed to the zinc chelator (Fig. 6B). In contrast, in cells transfected with Klf4 siRNA and exposed to 50 μM DTPA, there was an 80% knockdown of the Zip4 mRNA induction. The successful reduction of ZIP4 protein by Klf4 siRNA is further evidence that KLF4 is involved in Zip4 upregulation by zinc depletion (Fig. 6C). Knockdown of KLF4 and reduced 65Zn uptake in DTPA-treated cells represent further functional evidence of the influence of this transcription factor on Zip4 transporter activity. As shown in Fig. 7, cells that had been pretreated with DTPA for 24 h to increase ZIP4 expression accumulated 15% more 65Zn during a 60-min uptake time course than cells also treated with Klf4 siRNA. Cells incubated for up to 60 min at 0°C accumulated negligible 65Zn.
Fig. 6.
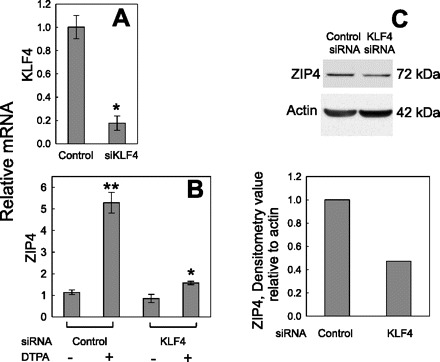
Knockdown of Klf4 with small interfering RNA (siRNA) in intestinal epithelial cells limits Zip4 induction by zinc depletion. Cells were transfected with control (nontargeting) siRNA or mouse Klf4 siRNA and then incubated for 24 h in medium alone or medium containing 50 μM DTPA. Abundance of Zip4 mRNA or Klf4 mRNA was measured by qPCR and normalized to level of 18S rRNA. A: Klf4 mRNA knockdown by Klf4 siRNA transfection. *P < 0.05 vs. control. B: knockdown of Zip4 mRNA by Klf4 siRNA transfection in response to DTPA. Values are means ± SD (n = 3). *P < 0.05; **P < 0.01 vs. non-DTPA-treated cells using nontargeting and Klf4 siRNA, respectively. C: densitometry of Zip4 expression. Western blot analysis demonstrates that upregulation of ZIP4 produced by DTPA is inhibited by Klf4 siRNA. Data represent results from numerous experiments.
Fig. 7.
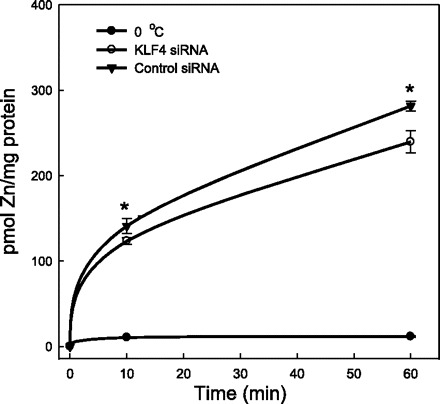
Knockdown of KLF4 with siRNA limits 65Zn transport activity of intestinal epithelial cells in response to zinc depletion. Nearly confluent cells were transfected with control (nontargeting) siRNA or mouse Klf4 siRNA and cultured in medium containing 50 μM DTPA for 24 h. Cells were then placed in fresh medium (2.5 μM Zn) containing 65Zn (57,000 cpm/ml) without DTPA and incubated for up to 60 min at 37°C. Cells were also incubated at 0°C as a control. Values are means ± SD (n = 3). *P < 0.05 vs. control.
EMSA and luciferase reporter assay of the mouse Zip4 promoter.
To determine whether the potential KLF4 binding sites on the mouse Zip4 promoter are capable of binding KLF4, we performed EMSAs with nuclear fractions from the IECs. Figure 8 shows the DNA-protein complexes formed using the KLF4 binding site at −280 to −300 bp of the Zip4 promoter. Zinc depletion using DTPA caused the binding of the DNA probes to KLF4 in the nuclear extract (Fig. 8). Specificity was verified by using unlabeled DNA probes. Furthermore, prior incubation of the nuclear extract with KLF4 antibody produced a supershift band (Fig. 8). These results showed that zinc depletion increased KLF4-DNA complex formation at the Zip4 promoter. Finally, a Zip4 promoter construct with luciferase as the reporter was generated and used to transfect the IECs. DTPA treatment of the transfected cells increased luciferase activity significantly (Fig. 9), indicating a direct effect of zinc depletion on Zip4 promoter activity. The effect of zinc chelation with DTPA was eliminated when this KLF4 binding site of the promoter was mutated.
Fig. 8.
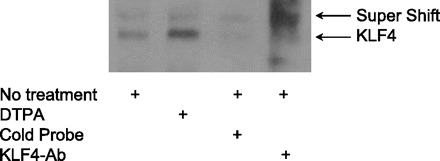
Representative chemiluminescence-based EMSA demonstrates KLF4 binding to cis elements in mouse Zip4 promoter. Cells were incubated for 48 h in medium alone or medium containing 50 μM DTPA, and nuclear extracts were prepared. In one reaction, unlabeled (cold) probe was added at an excess (100×). Another reaction included anti-KLF4 antibody to obtain evidence for a supershift of the complex.
Fig. 9.
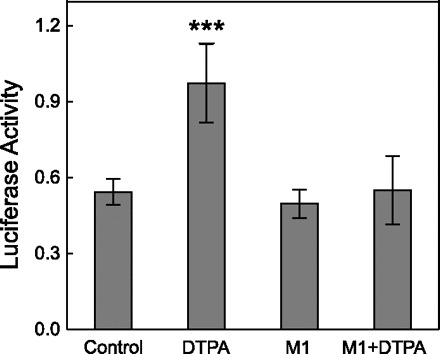
Effect of zinc depletion on activity of the Zip4 promoter. Confluent cells were cultured in medium containing 50 μM DTPA for 48 h and transiently transfected with native or mutated Zip4 promoter luciferase constructs. M1 indicates the mutated KLF4 binding element of the Zip4 promoter. Firefly luciferase activity of lysed cells was normalized to Renilla luciferase activity. Values are means ± SD (n = 4). ***P < 0.05 vs. control.
DISCUSSION
Substantial evidence suggests that ZIP4 is a transporter of zinc in the intestine that is necessary to maintain health (7, 17). An extensive qPCR analysis of mRNAs of zinc transporter genes expressed in mouse small intestine identified Zip4 as being particularly sensitive to dietary zinc restriction (17). The upregulation in expression of Zip4 under zinc depletion conditions is very high in the murine intestine relative to other tissues (7). Zip4 expression in response to a low-zinc diet changes rapidly, within 24 h; however, the mechanism of Zip4 regulation is unknown. An analysis of the murine Zip4 promoter region shows two binding sites for the transcription factor KLF4 within the first 800 bp upstream (27). In our experiments, mutation of the KLF4 binding site at −280 to −300 bp completely eliminated the increased Zip4 promoter activity produced by zinc depletion with DTPA (Fig. 9). Therefore, in the mouse, this proximal KLF4 binding site may be the most important in transcriptional control of Zip4. Human and rat Zip4 have one and two KLF4 binding regions, respectively. Collectively, the limited expression of KLF4 in tissue, principally the intestine (29), the responsiveness of Zip4 to zinc restriction, which also appears to be limited to the intestine, and the presence of KLF4 binding elements within Zip4 regulatory regions support a role for KLF in Zip4 expression.
The present results show that Klf4 is upregulated in mice by zinc dietary deficiency or in cells treated with zinc chelators and that zinc depletion increases KLF4 abundance in the nucleus. Particularly relevant to our present findings are the experiments reported by Kindermann et al. (13, 14) that demonstrated the marked upregulation of Klf4 by zinc chelation with TPEN in a human colon carcinoma cell line (HT-29). They suggested that the metal-responsive transcription factor MTF-1 was involved in regulation of Klf4 expression, since the human KLF4 promoter has MRE sequences upstream in the regulatory region (13). MTF-1 occupancy by zinc causes nuclear translocation and binding to MREs of some genes, including Mt and ZnT1 (37). Our genomic analysis revealed one MTF-1 binding site on either side of the transcription start site murine Klf4 and only one MTF-1 site in human Klf4 (27). This occurrence supports the notion that MTF-1 could be involved in upregulation of Klf4 in mouse intestine during zinc depletion. Conditional knockout of liver Mtf-1 produced upregulation of a number of genes, including the zinc transporter Zip10 (37). This suggests that MTF-1 may repress transcription of some MRE-containing genes under zinc-replete conditions. In zinc depletion, where nuclear MTF-1 translocation does not occur, repression is removed, allowing for transcription to occur for some genes with MREs. Such an MTF-1-related repression mechanism may explain the response of Klf4 to zinc depletion.
In contrast to our data with intact mice and mouse IECs, others have presented evidence interpreted to indicate that Zip4 is controlled through mRNA stability, rather than changes in transcription (35). Those interpretations may reflect the fact that the bulk of the experiments were conducted with cell lines that are nonintestinal in origin and, hence, do not express large amounts of Zip4. The one intestinal cell line tested in those experiments was an intestinal crypt-like cell line, and crypt cells do not express Klf4 (11) or appreciable amounts of Zip4 (17).
The reduction in 65Zn transport by KLF4 knockdown in DTPA-treated cells of only 15% was surprising considering the proposed critical role of Zip4 in enteric zinc absorption (7, 17). However, in the IEC culture system, 65Zn uptake (retention) may involve simultaneous transport activity by ZIP4, ZIP5, ZIP10, and ZIP14, as well as ZnT1, ZnT4, ZnT5, ZnT6, and ZnT7, all of which are expressed in these cells. Furthermore, there may also be some residual Zip4-mediated transport in the cells after KLF4 knockdown that does not have an efficiency of 100% and may not fully inhibit ZIP4 expression. A differential of 13% is evident after 10 min. This suggests that decreased ZIP4 produced via KLF4 knockdown influences the initial rate of zinc uptake. Furthermore, these uptake measurements are comparable to those we observed with HEK-293H cells transfected with Zip14 (18). A more comprehensive analysis of the magnitude and role for each of these specific transporters in polarized enterocytes is beyond the scope of the present experiments.
A broader role for KLF4 may exist with respect to zinc restriction. KLF4 has a regulatory function for inducible nitric oxide synthase and alkaline phosphatase (11, 19). Both genes appear to upregulate in the intestine during zinc deficiency (4, 5). These changes may occur via KLF4, but that connection has not been directly investigated. Similarly, since KLF4 may regulate the p53 gene and oxidative stress (6, 9, 34), the upregulation of this transcription factor by zinc depletion could influence these processes. In addition, KLF4 has three zinc fingers (25); hence, DNA binding activity by this transcription factor could be sensitive to intracellular zinc levels. However, in this regard, zinc chelation or dietary restriction would be expected to result in loss of function for these zinc motifs.
Most nutrients are absorbed from exogenous sources via the small intestine, and a transcription factor expressed in intestinal cells (e.g., KLF4) would be an ideal target for adaptation to specific nutrient intake levels. Such a role for KLF4 is supported by its action in regulating the sodium-dependent multivitamin transporter (SMVT) (28). KLF4 binding sites have been identified within the hSMVT promoter, and this transcription factor most likely is responsible for adaptive hSMVT upregulation in biotin deficiency.
The current focus of these studies has been on the mechanism of Zip4 regulation during zinc restriction. By contrast, we have found that excess dietary zinc has minimal effects on Zip4 mRNA (7). It has been demonstrated that elevated zinc in cells leads to posttranslational processing, endocytosis of plasma membrane-localized Zip4, and degradation or ubiquitination (14). Those findings agree with our results on ZIP4 protein induction and that such degradative pathways are negligible during zinc-limited conditions.
KLF4 is known to be involved in cell cycle regulation, differentiation, inflammation, and cancer (1, 3, 10, 29), particularly gastrointestinal cancer (36). In two prostate cancer cell lines, methylselenol, an active selenium metabolite, upregulates Klf4 (16). It was suggested that the growth-suppressing effect of selenium in prostate cancer cells occurs via KLF4 regulation of genes that control apoptosis and cell growth. A mechanism for this action was not established. The demonstration that induced pluripotent stem cells, produced from fibroblasts via transfection with four transcription factors including KLF4 (30), is of interest, inasmuch as zinc has undefined effects on growth and differentiation. Zip4 could also be a mediator of some of the biological effects reported for KLF4. For example, in pancreatic cancer, where KLF4 is usually increased (26), ZIP4 expression is highly upregulated and promotes tumor growth (15). Similarly, Zip4 is expressed in the colon, and a KLF4-colon cancer association has been documented (36). It is possible that other KLF transcription factors may be modulated by zinc status. These transcription factors exhibit tissue-specific expression; therefore, although KLF4 may be critical for zinc homeostasis for the intestine, other KLF zinc-responsive family members may control other genes in other tissues and cell types. Such a possibility has not been investigated.
GRANTS
This research is supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant DK-31127 and Boston Family Endowment Funds of the University of Florida to R. J. Cousins.
Acknowledgments
The authors acknowledge helpful discussions with Louis Lichten and Moon-Suhn Ryu.
Present address of J. P. Liuzzi: School of Public Health, Florida International University, Miami, FL 33199 (e-mail: jliuzzi@fiu.edu).
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Alder JK, Georgantas RW 3rd, Hildreth RL, Kaplan IM, Morisot S, Yu X, McDevitt M, Civin CI. Krüppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol 180: 5645–5652, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhardhani MS, Borojevic R, Basak S, Ho E, Zhou P, Croitoru K. IL-10 protects mouse intestinal epithelial cells from Fas-induced apoptosis via modulating Fas expression and altering caspase-8 and FLIP expression. Am J Physiol Gastrointest Liver Physiol 291: G820–G829, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Birsoy K, Chen Z, Friedman J. Transcriptional regulation of adipogenesis by KLF4. Cell Metab 4: 339–347, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchard RK, Cousins RJ. Regulation of intestinal gene expression by dietary zinc: induction of urogyanylin mRNA by zinc deficiency. J Nutr 130: 1393S–1398S, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Cui L, Blanchard RK, Cousins RJ. The permissive effect of zinc deficiency on urogyanylin and inducible nitric oxide synthase gene upregulation in rat intestine induced by interleukin 1α is rapidly reversed by zinc repletion. J Nutr 133: 51–56, 2003 [DOI] [PubMed] [Google Scholar]
- 6.Cullingford TE, Butler MJ, Marshall AK, Tham EL, Sugden PH, Clerk A. Differential regulation of Krüppel-like factor family transcription factor expression in neonatal rat cardiac myocytes: effects of endothelin-1, oxidative stress and cytokines. Biochim Biophys Acta 1783: 1229–1236, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dufner-Beattie J, Wang F, Kuo YM, Gitschier J, Eide D, Andrews GK. The acrodermatitis enteropathica gene Zip4 encodes a tissue-specific, zinc-regulated zinc transporter in mice. J Biol Chem 278: 33474–33481, 2003 [DOI] [PubMed] [Google Scholar]
- 8.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Krüppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem 280: 38247–38258, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Ghaleb AM, Katz JP, Kaestner KH, Du JX, Yang VW. Krüppel-like factor 4 exhibits antiapoptotic activity following gamma-radiation-induced DNA damage. Oncogene 16: 2365–2373, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Krüppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res 15: 92–96, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinnebusch BF, Siddique A, Henderson JW, Malo MS, Zhang W, Athaide CP, Abedrapo MA, Chen X, Yang VW, Holdin RA. Enterocyte differentiation marker intestinal alkaline phosphatase is a target gene of the gut-enriched Krüppel-like factor. Am J Physiol Gastrointest Liver Physiol 286: G23–G30, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Hoadley JE, Leinart AS, Cousins RJ. Kinetic analysis of zinc uptake and serosal transfer by vascularly perfused rat intestine. Am J Physiol Gastrointest Liver Physiol 252: G825–G831, 1987 [DOI] [PubMed] [Google Scholar]
- 13.Kindermann B, Döring F, Budczies J, Daniel H. Zinc-sensitive genes as potential new target genes of the metal transcription factor-1 (MTF-1). Biochem Cell Biol 83: 221–229, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Kindermann B, Döring F, Pfaffl M, Daniel H. Identification of genes responsive to intracellular zinc depletion in the human colon adenocarcinoma cell line HT-29. J Nutr 134: 57–62, 2004 [DOI] [PubMed] [Google Scholar]
- 15.Li M, Zhang Y, Liu Z, Bharadwaj U, Wang H, Wang X, Zhang S, Liuzzi JP, Chang SH, Cousins RJ, Fisher WE, Brunicardi FC, Logsdon CD, Chen C, Yao Q. Aberrant expression of zinc transporter Zip4 (SLC39A4) significantly contributes to human pancreatic cancer pathogenesis and progression. Proc Natl Acad Sci USA 104: 18636–18641, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Zhang H, Zhu L, Zhao L, Dong Y. Krüppel-like factor 4 is a novel mediator of selenium in growth inhibition. Mol Cancer Res 2: 306–313, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liuzzi JP, Bobo JA, Lichten LA, Samuelson DA, Cousins RJ. Responsive transporter genes within the murine intestinal-pancreatic axis form a basis of zinc homeostasis. Proc Natl Acad Sci USA 101: 14355–14360, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liuzzi JP, Aydemir F, Nam H, Knutson MD, Cousins RJ. ZIP14 (Slc39a14) mediates non-transferrin-bound iron uptake into cells. Proc Natl Acad Sci USA 103: 13612–13617, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lombeck I, Schnippering HC, Ritzi F, Fernendegen LE, Bremer HJ. Absorption of zinc in acrodermatitis enteropathica. Lancet 1: 855, 1975 [DOI] [PubMed] [Google Scholar]
- 20.Lönnerdal B. Intestinal absorption of zinc. In: Zinc in Human Biology. New York: Springer, 1989, chapt. 3, p. 33–55.
- 21.Mao X, Kim BE, Wang F, Eide DJ, Petris MJ. A histidine-rich cluster mediates the ubiquitization and degradation of the human zinc transporter, hZip4, and protects against zinc cytotoxicity. J Biol Chem 282: 6992–7000, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Maynahan EJ. Acrodermatitis enteropathica: a lethal inherited human zinc deficiency disorder. Lancet 2: 399–400, 1974 [DOI] [PubMed] [Google Scholar]
- 23.Nakano A, Nakano H, Nomura K, Toyomaki Y, Hanada K. Novel SLC39A4 mutations in acrodermatitis enteropathica. J Invest Dermatol 120: 963–966, 2003 [DOI] [PubMed] [Google Scholar]
- 24.Neldner KH, Hambidge KM. Zinc therapy of acrodermatitis enteropathica. N Engl J Med 292: 879–882, 1975 [DOI] [PubMed] [Google Scholar]
- 25.Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Krüppel-like transcription factors: a functional family. Int J Biochem Cell Biol 40: 1996–2001, 2008 [DOI] [PubMed] [Google Scholar]
- 26.Prasad NB, Biankin AV, Fukushima N, Maitra A, Dhara S, Elkahloun AG, Hruban RH, Goggins M, Leach SD. Gene expression profiles in pancreatic intraepithelial neoplasia reflect the effects of Hedgehog signaling on pancreatic ductal epithelial cells. Cancer Res 65: 1619–1626, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23: 4878–4884, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reidling JC, Nabokina SM, Said HM. Molecular mechanisms involved in the adaptive regulation of human intestinal biotin uptake: a study of the hSMVT system. Am J Physiol Gastrointest Liver Physiol 292: G275–G281, 2007 [DOI] [PubMed] [Google Scholar]
- 29.Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Krüppel-like factor expressed during growth arrest. J Biol Chem 271: 20009–20017, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 1–12, 2007 [DOI] [PubMed] [Google Scholar]
- 31.Thyresson N. Acrodermatitis enteropathica. Acta Dermatol 54: 383–385, 1974 [PubMed] [Google Scholar]
- 32.Vidal K, Grosjean I, evillard JP, Gespach C, Kaiserlian D. Immortalization of mouse intestinal epithelial cells by the SV40-large T gene. Phenotypic and immune characterization of the MODE-K cell line. J Immunol Methods 1: 63–73, 1993 [DOI] [PubMed] [Google Scholar]
- 33.Wang K, Zhou B, Kuo YM, Zemansky J, Gitschier J. A novel member of a zinc transporter family is defective in acrodermatitis enteropathica. Am J Hum Genet 71: 66–73, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wassmann S, Wassmann K, Jung A, Velten M, Knuefermann P, Petoumenos V, Becher U, Werner C, Mueller C, Nickenig G. Induction of p53 by GKLF is essential for inhibition of proliferation of vascular smooth muscle cells. J Mol Cell Cardiol 3: 301–307, 2007 [DOI] [PubMed] [Google Scholar]
- 35.Weaver B, Dufner-Beattie J, Taiho K, Andrews GK. Novel zinc-responsive post-transcriptional mechanisms reciprocally regulate expression of the mouse Slc39a4 and Slc39a5 zinc transporters (Zip4 and Zip5). Biol Chem 388: 1301–1312, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis 27: 23–31, 2006 [DOI] [PubMed] [Google Scholar]
- 37.Wimmer U, Wang Y, Georgiev O, Schaffner W. Two major branches of anti-cadmium defense in the mouse: MTF-1 metallothioneins and glutathione. Nucleic Acids Res 33: 5715–5727, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]


