Abstract
Enteroaggregative Escherichia coli (EAEC) pathogenesis is thought to comprise intestinal colonization followed by the release of enterotoxins and cytotoxins. Here, we use PCR to determine the prevalence of ten genes encoding serine protease autotransporter toxins (SPATEs) in a collection of clinical EAEC isolates. Eighty-six percent of EAEC strains harbored genes encoding one or more Class I cytotoxic SPATE protein (Pet, Sat, EspP, or SigA). Two Class II, non-cytotoxic, SPATE genes were found among EAEC strains: pic and sepA, each originally described in Shigella flexneri 2a. Using a multiplex PCR for five SPATE genes (pet, sat, sigA, pic and sepA), we found that most of the Shigella isolates also harbored more than one SPATE, whereas members of most other E. coli pathotypes rarely harbored a cytotoxic SPATE gene. SPATEs may be relevant to the pathogenesis of both EAEC and Shigella spp..
Enteroaggregative Escherichia coli (EAEC) has been associated with several clinical scenarios, including travelers' diarrhea 1, 2, 3, endemic pediatric diarrhea among children in industrialized 4 and developing countries 5, as well as persistent diarrhea among HIV-infected patients 6-10. The current pathogenetic paradigm for EAEC includes colonization of the intestinal mucosa followed by the elaboration of one or more cytotoxins and enterotoxins, of which several have been described 11-18. Enterotoxins include the enteroaggregative heat-stable toxin (EAST1) and the Shigella enterotoxin-1 (ShET1). The roles of these toxins in EAEC pathogenesis and epidemiology are not yet known.
Infection of human intestinal explants suggests that most EAEC strains elicit frank mucosal damage, accompanied by rounding and exfoliation of colonocytes. Henderson et al. have reported that the serine protease autotransporter (SPATE) toxin called Pet (plasmid-encoded toxin) is required for strain 042 to elicit cytotoxic effects on human explants 14; however, only a small minority of EAEC strains carry the pet gene, though a much larger number of strains are toxic to explants. This paradox occurs in the context of substantial heterogeneity of EAEC adhesins and other putative virulence factors, presenting a confusing clinical and epidemiologic scenario 19.
The Pet protease is a member of the serine protease autotransporters of Enterobacteriaceae (SPATE) family of secreted proteases. SPATEs comprise a large group of trypsin-like serine proteases, which are secreted by Shigella spp., uropathogenic E. coli, and all of the diarrheagenic E. coli (DEC) pathotypes (reviewed in refs. 19 and 20). The toxins are translocated across the outer membrane by the autotransporter pathway, in which translocation requires a dedicated C-terminal beta barrel domain. The N-terminal, mature SPATE toxins are 104-110 kDa in size and feature a typical N-terminal serine protease catalytic domain, followed by a highly conserved beta-helix motif, which is present in nearly all autotransporters 20. The SPATE family has been organized phylogenetically into two classes. Members of the Class I SPATEs (which include Pet) are all cytotoxic to epithelial cells 21. In addition to Pet, the Class I SPATEs include, prominently, EspP from enterohemorrhagic E. coli (EHEC), EspC from enteropathogenic E. coli (EPEC), SigA from Shigella flexneri, and Sat, from uropathogenic and diffusely adhering E. coli (DAEC) 16. Class II SPATEs are more diverse with regard to phenotype, though several are known to cleave mucin. Many EAEC and Shigella strains encode Pic, a mucinase encoded on the bacterial chromosome 22. Pic may promote intestinal colonization via an unknown mechanism (Harrington and Nataro, unpublished observations). Class II includes, besides Pic, SepA from Shigella flexneri 23 and Tsh from avian pathogenic E. coli 24, and several others.
We sought to resolve the paradox between observed cytotoxic effects attributed to most EAEC strains and the low carriage rate of the Pet cytotoxin. Specifically, we hypothesized that other Class I SPATE cytotoxins would be commonly found among EAEC strains. We tested this hypothesis on 55 EAEC and 10 each of ETEC, EHEC, EPEC, EIEC, and DAEC strains, as well as 12 Shigella strains. All strains were isolated in the course of epidemiologic studies 27, 29 and were derived from the collections of the Statens Serum Institut (SSI) in Denmark or the Center for Vaccine Development (CVD) of the University of Maryland School of Medicine (Table 1 and Table 2) 25, 26, 27, 28. In addition, we selected 12 non-pathogenic E. coli strains: six strains were isolated from healthy humans in the course of the Danish antimicrobial resistance surveillance program (DANMAP), and six strains were isolated from minced meat as part of the Danish food surveillance program. Shigella flexneri 2a strain 2457T was used as a control30. Stock cultures were frozen at - 80 °C in Luria broth (LB) or SSI broth containing 10% (v/v) glycerol. All strains were grown at 37°C. Serotyping of the EAEC strains was performed at the SSI using standard methods 31.
Table 1.
Fifty-five EAEC strains used in this study including, origin, serotype and monoplex PCR results.
| SPATE proteins | Aggregative Adherence Fimbriae (AAF) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class I | Class II | AAF/I | AAF/II | AAF/III | AAF/IV | |||||||
| Species | Strain | Origin | Serotype | pet | sat | sigA | pic | sepA | aggA | aafA | agg3A | agg4A |
| EAEC | 144-1-1 | Thailand (H) | O36:H18 | − | + | − | + | − | − | − | − | + |
| 6-1-1 | Thailand (H) | ORough:H2 | − | + | − | − | − | − | − | − | + | |
| 199-1-4 | Thailand (H) | ORough:H1 | + | − | − | + | − | − | + | − | − | |
| 103-1-1 | Thailand (H) | O148:H28 | − | + | − | − | − | − | − | − | − | |
| 44-1-1 | Thailand (H) | O77:H− | − | + | − | − | − | − | − | − | − | |
| 435-1-1a | Thailand (H) | O33:H16 | − | − | − | + | + | − | + | − | − | |
| 501-1-1 | Thailand (H) | O148:H53 | − | + | − | − | + | − | − | − | + | |
| 253-1-1 | Thailand (H) | O3:H2 | − | + | − | − | − | + | − | − | − | |
| 309-1-1 | Thailand (H) | O130:H27 | − | + | − | + | − | + | − | − | + | |
| 133 | Peru (H) | ONT:H1 | − | + | − | + | − | − | − | − | + | |
| H46-2 | Peru (H) | O130:H26 | − | + | − | + | − | + | − | − | + | |
| H92-1 | Peru (H) | O33:H16 | + | + | − | + | − | − | − | − | + | |
| H32-1a | Peru (H) | O15:H− | − | + | − | + | + | − | − | − | + | |
| H223-1 b | Peru (H) | O101:H33 | − | − | + | + | + | − | − | − | + | |
| H232-1 | Peru (H) | O20:H1 | − | + | − | + | − | − | − | − | + | |
| H145-1 | Peru (H) | O130:H27 | − | + | − | + | − | + | − | − | + | |
| 191-1 | Peru (H) | O101:H9 | − | + | − | + | + | − | − | − | − | |
| H38-1 | Peru (H) | O15:H− | − | + | − | − | + | + | − | − | − | |
| H32-1b | Peru (H) | O73:H16 | − | + | + | − | + | + | − | − | − | |
| DS65-R3 | Philipines (H) | O103:H43 | − | + | + | − | + | + | − | − | − | |
| DS67-R2 | Philipines (H) | O25:H4 | − | + | − | − | + | − | − | − | + | |
| DS244- R3 | Philipines (H) | O103:H43 | + | − | − | + | − | − | + | − | + | |
| 60Ac | Mexico (H) | O20:H+ | − | + | − | + | − | − | − | − | − | |
| H194-2c | Peru (H) | O175:H27 | − | + | − | + | + | − | − | − | + | |
| H77-1 | Peru (H) | O92:H33 | − | + | − | + | + | − | − | − | − | |
| 239-1 | Thailand (H) | ORough:H21 | − | + | − | + | + | − | − | − | − | |
| 278-1-1 | Thailand (H) | O125ac:H21 | − | + | − | + | + | − | − | − | + | |
| 216-1 | Thailand (H) | ORough:H27 | + | − | − | + | − | − | + | − | ||
| 495-1 | Thailand (H) | O130:H27 | − | + | − | + | − | + | − | − | + | |
| 232-1-1 | Thailand (H) | O127:H21 | − | + | − | + | + | − | − | − | + | |
| 99-1 | Japan (H) | O5:H10 | − | + | − | + | + | − | − | − | − | |
| 96-1 | Japan (H) | O5:H10 | − | + | − | + | + | − | − | − | − | |
| 86-1 | Japan (H) | O5:H10 | − | + | − | − | − | − | − | − | + | |
| 101-1 | Japan (H) | O36:H10 | − | − | − | − | − | − | − | − | + | |
| C585-00* | Denmark (H) | O86:H− | − | + | − | − | − | + | − | − | − | |
| C790-00 | Denmark (H) | O+:H− | − | − | − | − | − | − | − | − | − | |
| C809-00 | Denmark (H) | O86:H1 | − | + | − | − | − | + | − | − | − | |
| C1010-00 d | Denmark (H) | ORough:H− | − | + | − | + | + | − | − | − | + | |
| C1046-00 | Denmark (H) | O44:H18 | − | + | − | − | − | + | − | − | − | |
| C1059-00 | Denmark (H) | O73:K5:H18 | − | − | − | + | + | − | − | − | + | |
| C1082-00 | Denmark (H) | O107, O117:H32 | − | − | − | − | − | − | − | − | + | |
| C246-01 | Denmark (H) | O34:H9 | − | − | − | − | + | − | − | − | + | |
| C247-01* | Denmark (H) | ONT:H− | − | − | − | − | − | − | − | − | − | |
| C252-01 | Denmark (H) | O111:H21 | − | + | − | + | + | − | − | − | − | |
| C254-01 | Denmark (H) | ONT:H10 | − | − | − | + | − | − | − | − | + | |
| C390-01 | Denmark (H) | O134:H27 | − | + | − | + | − | + | − | − | − | |
| C763-01 | Denmark (H) | O153:H2 | − | + | − | + | − | + | − | − | − | |
| C798-01 | Denmark (H) | O92:H33 | − | + | − | + | − | + | − | − | − | |
| C1239-01 | Denmark (H) | O86:H18 | − | + | − | + | − | + | − | − | − | |
| C1275-01 | Denmark (H) | O91:H1 | − | − | − | − | + | + | − | − | − | |
| C274-02 | Denmark (H) | O86:H2 | − | + | − | + | − | + | − | − | − | |
| JM221 | Mexico (H) | O92:K-:H33 | − | + | − | + | − | + | − | − | − | |
| 17-2 | Serbia (H) | O3:H2 | − | + | − | − | − | + | − | − | − | |
| 042 | Peru (H) | O44:H18 | + | − | − | + | − | − | + | − | − | |
| 55989 | CAR (H) | O104:H4 | − | + | + | + | − | − | − | + | − | |
ONT, O not typeable; H+, motile but not typeable; H−, non motile; (H), human origin; (A)
Strains positive for EspP
EAEC strain 435-1 was found probe positive for Pet in a previous study 27
EAEC strain H223-1 was found probe negative for SigA in a previous study 27
EAEC strains 60A and H194-2 were found probe positive for AggA in a previous study 27
Reported in a previous study as ORough:H1 41
Table 2.
Shigella, ETEC and non-pathogenic E. coli strains used in this study.
| SPATE proteins |
||||||||
|---|---|---|---|---|---|---|---|---|
| Class I | Class II | |||||||
| Species | Strain | Origin | Serogroup/type | pet | sat | sigA | pic | sepA |
| Shigella | 389 | Chile (H) | S. flexneri 6 | − | − | + | − | − |
| 691 | Chile (H) | S. flexneri 1b | − | + | − | − | − | |
| 718 | Chile (H) | S. flexneri 2a | − | + | + | + | − | |
| 970 | Chile (H) | S. flexneri 2b | − | + | − | − | − | |
| D570 | Chile (H) | S. sonnei | − | − | + | − | − | |
| D405 | Chile (H) | S. flexneri 3a | − | + | − | − | + | |
| D445 | Chile (H) | S. flexneri 2a | − | + | + | + | + | |
| D427 | Chile (H) | S. flexneri 2a | − | + | + | + | + | |
| 1427 | Chile (H) | S. flexneri 2b | − | + | − | − | + | |
| D274 | Chile (H) | S. flexneri 6 | − | − | + | − | − | |
| 1303 | Chile (H) | S. flexneri 1b | − | + | − | − | − | |
| 1534 | Chile (H) | S. flexneri 2a | − | + | + | + | + | |
| ETEC | 24337/A | (H) | ND | − | − | − | − | − |
| 1392/752A | (H) | ND | − | − | − | − | − | |
| B7A-7318 | (H) | ND | − | − | − | − | − | |
| H10407 | (H) | ND | − | − | − | − | − | |
| E11881D | (H) | ND | − | − | − | − | − | |
| E11881A | (H) | ND | − | − | − | − | − | |
| E11881L | (H) | ND | − | − | − | − | − | |
| 214-4 | (H) | ND | − | − | − | − | − | |
| 60R75 | (H) | ND | − | − | − | − | − | |
| TD225 | (H) | ND | − | − | − | − | − | |
| EHEC* | 84-067 | (H) | O126:K7:H8 | − | − | − | − | − |
| 83-377 | (H) | O2:K1:H1 | − | − | − | − | − | |
| 83-378 | (H) | O1:K1:H− | − | + | + | − | − | |
| 83-408 | (H) | O145:H− | − | − | − | − | − | |
| 86-420 | (H) | O6:H1 | − | + | − | + | − | |
| 10433-88 | (H) | O157:H7 | − | − | − | − | − | |
| 8350-86 | (H) | O157:H7 | − | − | − | − | − | |
| 8504-86 | (H) | O157:H7 | − | − | − | − | − | |
| 8868-86 | (H) | O157:H7 | − | − | − | − | − | |
| 23379-85 | (H) | O157:H7 | − | − | − | − | − | |
| EPEC | E2348/6942 | England (H) | O127:K63:H6 | − | − | − | − | − |
| 50869 | (H) | O114 | − | − | − | − | − | |
| 1 | (H) | ND | − | − | − | − | − | |
| 6 | (H) | O55 | − | − | − | + | − | |
| 7 | (H) | O111 | − | − | − | − | − | |
| 9 | (H) | O55 | − | − | − | − | − | |
| 10 | (H) | O119 | − | − | − | − | − | |
| 11 | (H) | O55 | − | − | − | − | − | |
| 13 | (H) | O111 | − | − | − | − | − | |
| 18 | (H) | O119 | − | − | − | − | − | |
| EIEC | E134 | (H) | O28ac:H− | − | − | + | − | − |
| 439-1 | Thailand (H) | O114:H25 | − | − | − | − | − | |
| 705-1 | Thailand (H) | O114:H25 | − | − | − | − | − | |
| 949-1 | Thailand (H) | O114:H25 | − | − | − | − | − | |
| 1109-1 | Thailand (H) | O170:H− | − | − | − | − | − | |
| 205-1 | Thailand (H) | O170:H− | − | − | + | − | − | |
| 145-1 | Thailand (H) | O164:H− | − | − | + | − | − | |
| 905-2 | Thailand (H) | O28ac:H− | − | − | + | − | − | |
| 164-3 | Thailand (H) | O28ac:H− | − | − | + | − | − | |
| 183-4 | Thailand (H) | O28ac:H− | − | − | + | − | − | |
| DAEC | C184543 | (H) | O75:H− | − | + | − | − | − |
| DS55R3 | (H) | ND | − | − | − | − | − | |
| DS112R3 | (H) | ND | − | − | − | − | − | |
| DS126R1 | (H) | ND | − | − | − | − | − | |
| 1 | Mexico (H) | ND | − | + | − | − | − | |
| 4 | Mexico (H) | ND | − | − | + | − | − | |
| 5 | Mexico (H) | ND | − | − | − | − | − | |
| 17 | Mexico (H) | ND | − | − | − | − | − | |
| 18 | Mexico (H) | ND | − | + | − | − | − | |
| 20 | Mexico (H) | ND | − | − | − | − | − | |
|
Non-pathogenic E. coli |
C 123-01 a | Denmark (H) | O73:H18 | − | + | − | − | − |
| C 129-01 a | Denmark (H) | O8:H9 | − | − | − | − | − | |
| C 130-01 a | Denmark (H) | O81:H27 | − | − | + | − | − | |
| C 131-01 a | Denmark (H) | O9:H19 | − | − | − | − | − | |
| C 133-01a | Denmark (H) | O125ab:H4 | − | − | − | − | − | |
| C 134-01a | Denmark (H) | O40:H− | − | − | − | − | − | |
| C 107-01b | Denmark (MM) | O129:H31 | − | − | − | − | − | |
| C 108-01b | Denmark (MM) | O35:H29 | − | − | − | − | − | |
| C 109-01Ab | Denmark (MM) | O150:H8 | − | − | − | − | − | |
| C 109-01Bb | Denmark (MM) | O140:H21 | − | − | − | − | − | |
| C 110-01b | Denmark (MM) | O25:H28 | − | − | − | − | − | |
| C 111-01b | Denmark (MM) | O32:H37 | − | − | − | − | − | |
ONT, O not typeable; H+, motile but not typeable; H−, non motile; ND, not determined; (H), human origin
All EHEC strains positive for Shiga toxin (Stx); (MM) Minced meat
Non-pathogenic E. coli from healthy humans isolated as part of the Danish antimicrobial resistance surveillance program DANMAP
Non-pathogenic E. coli from minced meat isolated as part of the Danish food surveillance program.
PCR was used to detect the presence of genes corresponding to known SPATE sequences. Primers used are listed in Table 3. DNA template was obtained as previously described 25. The accuracy of a subset of PCR reactions was verified by nucleotide sequencing in the Biopolymer Laboratory Core Facility, University of Maryland School of Medicine. A multiplex PCR was designed to detect the most common SPATE genes; the Multiplex PCR kit was used according to manufacturer's instructions (Qiagen inc., Valencia, Ca, USA). Products were amplified by using the Eppendorf Mastercycler Gradient thermal cycler (Eppendorf North America Inc, Westbury, NY, USA). The specific temperatures and primers are listed in Table 3. Monoplex PCR reactions were performed as previously described 25. Multiplex PCR cycles comprised (a) 15 min. denaturation at 95°C, (b) 30 sec. denaturation (depending on the size of the product, with a 30 sec. increase for each 500 bp) at 94°C, (c) annealing for 1.5 min, and (d) extension 1.5 min. at 72°C with 35 cycles from step (b). The final extension was 10 min. at 72°C.
Table 3.
Primers used for monoplex PCR and multiplex PCR
| Factor class |
Gene | Primer (pmol/μL) |
Primer sequence (5′-3′) | PCR product in bp a |
Accession no.b |
Annealing temp |
Reference |
|---|---|---|---|---|---|---|---|
| SPATE- gene |
sat | 25 | TCAGAAGCTCAGCGAATCATTG CCATTATCACCAGTAAAACGCACC |
930 | AE014075 | 59 °C | This study |
| sigA | 25 | CCGACTTCTCACTTTCTCCCG CCATCCAGCTGCATAGTGTTTG |
430 | NC_004337 | 58 °C | This study | |
| pet | 25 | GGCACAGAATAAAGGGGTGTTT CCTCTTGTTTCCACGACATAC |
302 | AF056581 | 58 °C | 44 | |
| pic | 25 | ACTGGATCTTAAGGCTCAGGAT GACTTAATGTCACTGTTCAGCG |
572 | AF097644 | 58 °C | 44 | |
| sepA | 25 | GCAGTGGAAATATGATGCGGC TTGTTCAGATCGGAGAAGAACG |
794 | Z48219 | 58 °C | 44 | |
| tsh | 25 | CCGTACACAAATACGACGG GGATGCCCCTGCAGCGT |
900 | AF218073 | 59 °C | 44 | |
| vat | 25 | AACGGTTGGTGGCAACAATCC AGCCCTGTAGAATGGCGAGTA |
420 | AY151282 | 58 °C | 44 | |
| eatA | 25 | CAGGAGTGGGAACATTAAGTCA CGTACGCCTTTGATTTCAGGAT |
743 | AY163491 | 60 °C | 44 | |
| espP | 25 | GTCCATGCAGGGACATGCCA TCACATCAGCACCGTTCTCTAT |
547 | NC_002128 | 60 °C | 44 | |
| espC | 25 | TAGTGCAGTGCAGAAAGCAGTT AGTTTTCCTGTTGCTGTATGCC |
839 | AF297061 | 59 °C | 44 | |
| AAF gene |
aggA | 25 | AAATATGAGAAGAAAGAA AAAAATTAATTCCGGTATGG |
500 | ECU12894 | 50 °C | 41 |
| aafA | 25 | CAGAATGTTTGCGATTGCTAC TTTGTCACAAGCTCAGCATT |
468 | AF012835 | 50 °C | 41 | |
| agg3A | 25 | GTATCATTGCGAGTCTGGTATTCAG GGGCTGTTATAGAGTAACTTCCAG |
462 | AF411067 | 50 °C | 45 | |
| agg4A | 25 | TCCATTATGTCAGGCTGCAA GGCGTTAACGTCTGATTTCC |
411 | EU637023 | 59 °C | 41 | |
bp, Base pair
GenBank database accession number for the SPATE and fimbrial genes
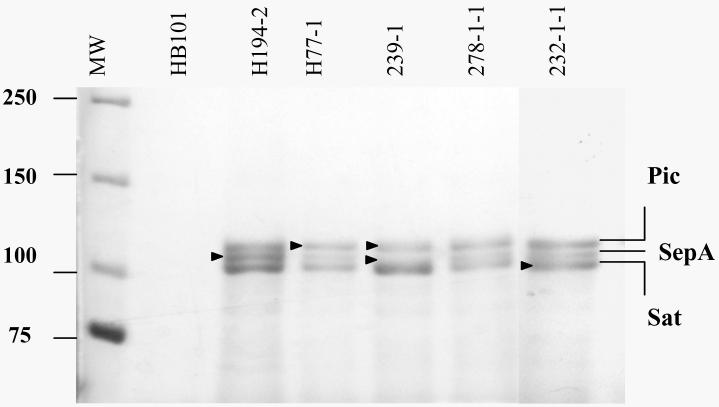
Results of monoplex PCR for ten SPATE-encoding genes (pet, sigA, espC, espP, sat, vat, pic, sepA, tsh and eatA) are presented in Table 1. In order to corroborate results of the PCR assays, the supernatants of several strains which were positive for SPATE sequences by PCR were separated by 17.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE); strains positive for up to three SPATEs by PCR also showed the expected number of supernatant proteins at the predicted molecular mass (104-130 kDa). Several protein bands were excised and subjected to tryptic digest and mass spectrometry in a Finnigan LCQ Advantage Ion Trap Mass Spectrometer in the Protein Analysis Core of the Center for Vascular and Inflammatory Diseases, University of Maryland School of Medicine. These analyses yielded protein identifications that correlated completely with PCR assays for pic, sat and sepA (Figure 1).
Figure 1. Secretion of SPATE proteins from EAEC strains.
Supernatants from 5 representative EAEC strains are shown. Supernatant proteins were precipitated with 10% TCA and separated by SDS-PAGE followed by Coomassie Staining. SPATE proteins represented by bands directly identified by mass spectrometry were SepA from strains H194-2 and 239-1; Pic from strains H77-1 and 239-1; Sat from strain 232-1-1 (indicated by arrows). MW, Molecular weight; HB101, negative control strain.
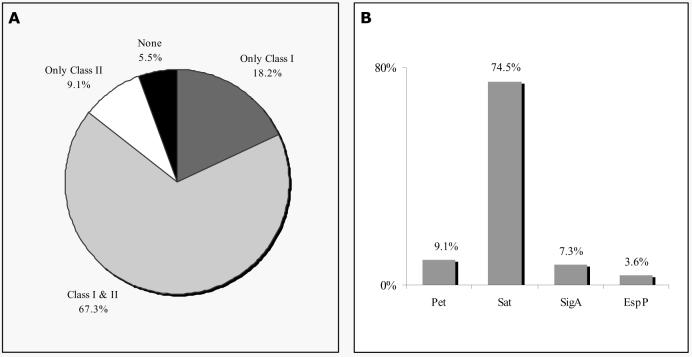
Out of the 55 EAEC strains tested, 94.5% harbored genes for one or more of the SPATE proteins, including members of Class I (Pet, Sat, SigA, EspP) and/or Class II (Pic, SepA, Tsh). No EAEC strain yielded a product corresponding to vat, espC, eatA, or tsh. 18.2% of the strains had only a Class I SPATE and no Class II sequence, 9.1% of the strains had only a Class II, and 67.3% were positive for both a Class I and II SPATE (Fig. 2A). Interestingly, the single most common SPATE among the EAEC strains was Sat (74.5% of strains), which was first described in uropathogenic E. coli strains, but which has more recently been reported among DAEC. Interestingly, we found pet in only 9.1% of our strains, and one strain (H92-1, Fig. 2A, Table 1) also harbored the sat gene. The mature Sat and Pet toxins are 52% identical at the amino acid level and thus they may be allelic toxins which fulfill the same roles in pathogenesis 32. Sat is cytotoxic to urinary epithelial cells in vitro 32, and Guignot et al. have suggested that Sat induces cytoskeletal perturbation in intestinal epithelium accompanied by rearrangement of tight junction proteins 33. Though the fundamental mode of action of Sat is unknown, Maroncle et al. have suggested that the protein enters epithelial cells and directly cleaves spectrin34, an effect also attributed to its closest homolog, Pet 35. Further experiments are required to determine whether the two toxins induce identical effects.
Figure 2. Distribution of Class I and Class II SPATEs among EAEC strains.
The distribution of the SPATE genes among 55 EAEC strains was determined by PCR. (A) Distribution of Class-I and Class-II SPATEs among the EAEC collection. (B) Frequency of Pet, Sat, SigA, and EspP.
Some EAEC strains carried neither pet nor sat, but interestingly, the majority of these strains harbored another Class I SPATE (Fig. 2B). 7.3 % of the EAEC strains harbored sigA, a cytotoxin originally described in S. flexneri, and 3.6% harbored espP, reported as a cytotoxin in Shiga toxin-producing E. coli strains. Overall, nearly 85% of EAEC strains harbored a gene encoding a Class I cytotoxic SPATE protein.
The Class II SPATEs were also common among the EAEC collections: 63.6 % of the strains harbored pic and 38.2 % carried sepA. Pic, originally described in strains of S. flexneri 2a, has been shown to cleave submaxillary mucin 22, and our data suggest that Pic promotes intestinal colonization in a mouse model (S. Harrington and J. Nataro, unpublished). SepA, also originally reported in S. flexneri 2a, may promote intestinal inflammation induced by Shigella strains, but its mode of action has not been described 23.
We hypothesized that the distribution of SPATE proteases would correlate with that of the AAF adhesins and/or with EAEC phylogeny as previously published 27. All five strains positive for the AAF/II pilin also carried the gene encoding Pet; none of these strains carried genes encoding Sat or SigA. Interestingly, these five Pet-encoding strains were distributed into three distantly related clusters on the EAEC phylogram, suggesting either horizontal co-transmission of AAF/II with Pet-encoding genes, or functional linkage of these factors. In contrast, strains harboring aggA (encoding the pilin of AAF/I) or aag4A (the pilin of AAF/IV, also called Hda) were commonly found to encode sat or sigA but not pet.
SigA was first reported to be encoded on a large chromosomal pathogenicity island in S. flexneri, which also encoded the Pic protease. In our collection, four EAEC strains harbored sigA; two of the four also carried pic. In contrast, 33 strains carried pic but not sigA. These data suggest that the linkage of pic and sigA as originally described is uncommon. No other correlations among toxins or adhesins were observed and no correlation of toxin genes with the phylogenetic clusters was apparent.
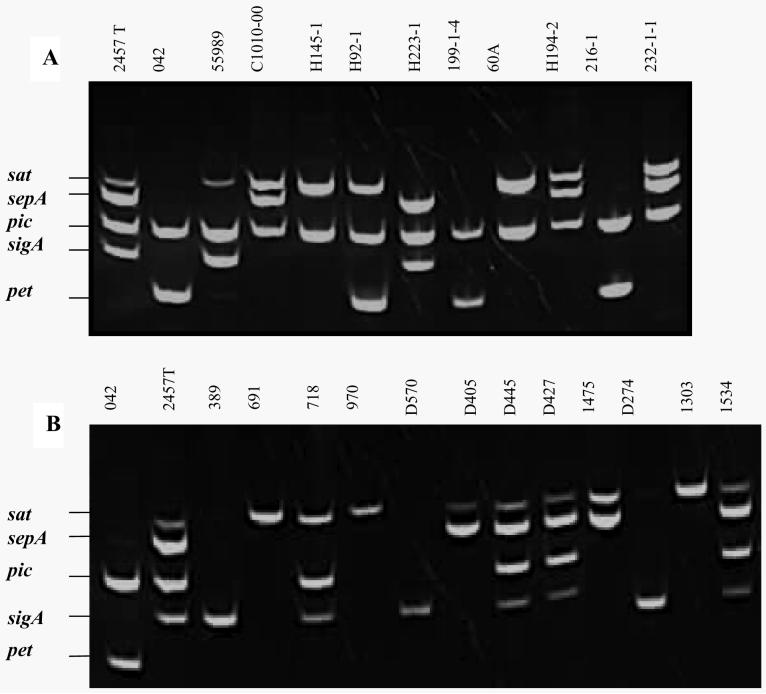
To facilitate later analysis for the five SPATEs commonly found in EAEC strains (pet, sat, sigA, pic and sepA), we developed a multiplex PCR. The multiplex assay yielded clear products at the appropriate mass in all relevant controls, with no erroneous bands detected. All EAEC strains positive for these genes by monoplex were re-tested with the multiplex, revealing perfect correlation between the multiplex and monoplex PCR assays. A representative image of multiplex PCR products derived from a subset of the EAEC and Shigella strains is shown in Fig. 3. Use of this assay as a tool for prospective epidemiologic analysis to address the potential role of SPATEs in human disease will be reported elsewhere.
Figure 3. Identification of SPATE sequences from EAEC and Shigella isolates by multiplex PCR.
PCR products representing genes for Sat, SepA, Pic, SigA and Pet were separated in 1.2% agarose gels. (A) PCR products from 11 EAEC strains (B) PCR products from 12 Shigella strains. EAEC strain 042 is the positive control for pet and pic; S. flexneri 2a strain 2457T is the positive control for sat, sepA, pic and sigA.
We performed our multiplex PCR analysis on sets of ETEC, EHEC, EPEC, EIEC, DAEC, Shigella and non-pathogenic E. coli strains (Table 2). The prevalence of the genes varied markedly by pathotype. In contrast to EAEC, we did not find Pic, SepA, Sat, SigA or Pet-encoding genes among any of the 10 ETEC strains (Table 2). Similarly, only two of 12 non-pathogenic E. coli strains were positive for any of these five SPATEs (one for sat and one for sigA; Table 2). One EPEC strain was positive for Pic, two EHEC strains were positive for Sat and either Pic or SigA, three DAEC strains harbored Sat and one Pic (Table 2). Previous studies have reported the presence of sat in the diarrheagenic E. coli pathotypes DAEC, EHEC and EPEC 32, 36. Since several of the SPATEs have previously been reported among Shigella strains 22, 23, 37, 38, we subjected 12 clinical Shigella isolates (11 S. flexneri and one S. sonnei) to our multiplex PCR assay. As we observed in EAEC strains, most of the Shigella strains harbored more than one SPATE protein (Fig. 3). Whereas none of the Shigella strains harbored the pet gene, all were positive for sat and/or sigA, with four strains harboring both genes. Interestingly six out of the ten EIEC strains harbored SigA as the only SPATE protein. EIEC is distinguished from Shigella by biochemical tests, but EIEC strains and Shigella spp. share essential virulence factors 39, 40 Thus it is plausible that SigA plays an important role in the pathogenesis of both EIEC and Shigella.
In our hands, the cytotoxic SPATE proteins were found almost exclusively among microbial pathogens that have been shown to induce mucosal damage and inflammation. Clinically, Shigella are more virulent in this regard than EAEC, and they commonly carried more than one cytoxic SPATE toxin. Whether the SPATE toxins exhibit primary or secondary roles in pathogenesis by Shigella and EAEC is under further investigation in our laboratory.
Acknowledgement
We would like to thank Dr. Søren Persson for his helpful advice regarding primer design, Statens Serum Institut. Also, Erik Juncker Boll for verifying PCR results. In Table 2 the six strains from the Danish antimicrobial resistance surveillance program DANMAP were kindly provided by Thomas Lund Sørensen and the six strains from the Danish food surveillance program were provided by Jeppe Boel. This project was funded by US. PHS grant, AI33096 to JPN and the Danish Counsil for Strategic Research grant 2101-07-0023 to KAK. NBO was supported in part by Statens Serum Institut, Copenhagen.
Reference list
- 1.Glandt M, Adachi JA, Mathewson JJ, Jiang ZD, DiCesare D, Ashley D, Ericsson CD, DuPont HL. Enteroaggregative Escherichia coli as a cause of traveler's diarrhea: clinical response to ciprofloxacin. Clin Infect Dis. 1999;29:335–8. doi: 10.1086/520211. [DOI] [PubMed] [Google Scholar]
- 2.Adachi JA, Jiang ZD, Mathewson JJ, Verenkar MP, Thompson S, Martinez-Sandoval F, Steffen R, Ericsson CD, DuPont HL. Enteroaggregative Escherichia coli as a major etiologic agent in traveler's diarrhea in 3 regions of the world. Clin Infect Dis. 2001;32:1706–9. doi: 10.1086/320756. [DOI] [PubMed] [Google Scholar]
- 3.Adachi JA, Ericsson CD, Jiang ZD, DuPont MW, Pallegar SR, DuPont HL. Natural history of enteroaggregative and enterotoxigenic Escherichia coli infection among US travelers to Guadalajara, Mexico. J Infect Dis. 2002;185:1681–3. doi: 10.1086/340419. [DOI] [PubMed] [Google Scholar]
- 4.Tompkins DS, Hudson MJ, Smith HR, Eglin RP, Wheeler JG, Brett MM, Owen RJ, Brazier JS, Cumberland P, King V, Cook PE. A study of infectious intestinal disease in England: microbiological findings in cases and controls [see comments] Commun Dis Public Health. 1999;2:108–13. [PubMed] [Google Scholar]
- 5.Okeke IN, Lamikanra A, Czeczulin J, Dubovsky F, Kaper JB, Nataro JP. Heterogeneous virulence of enteroaggregative Escherichia coli strains isolated from children in Southwest Nigeria. J Infect Dis. 2000;181:252–60. doi: 10.1086/315204. [DOI] [PubMed] [Google Scholar]
- 6.Wanke CA, Gerrior J, Blais V, Mayer H, Acheson D. Successful treatment of diarrheal disease associated with enteroaggregative Escherichia coli in adults infected with human immunodeficiency virus. J Infect Dis. 1998;178:1369–72. doi: 10.1086/314443. [DOI] [PubMed] [Google Scholar]
- 7.Wanke CA, Mayer H, Weber R, Zbinden R, Watson DA, Acheson D. Enteroaggregative Escherichia coli as a potential cause of diarrheal disease in adults infected with human immunodeficiency virus. J Infect Dis. 1998;178:185–90. doi: 10.1086/515595. [DOI] [PubMed] [Google Scholar]
- 8.Durrer P, Zbinden R, Fleisch F, Altwegg M, Ledergerber B, Karch H, Weber R. Intestinal infection due to enteroaggregative escherichia coli among human immunodeficiency virus-infected persons [In Process Citation] J Infect Dis. 2000;182:1540–4. doi: 10.1086/315885. [DOI] [PubMed] [Google Scholar]
- 9.Mossoro C, Glaziou P, Yassibanda S, Lan NT, Bekondi C, Minssart P, Bernier C, Le Bouguenec C, Germani Y. Chronic diarrhea, hemorrhagic colitis, and hemolyticuremic syndrome associated with HEp-2 adherent Escherichia coli in adults infected with human immunodeficiency virus in Bangui, Central African Republic. J Clin Microbiol. 2002;40:3086–8. doi: 10.1128/JCM.40.8.3086-3088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gassama-Sow A, Sow PS, Gueye M, Gueye-N'diaye A, Perret JL, M'Boup S, Aidara-Kane A. Characterization of pathogenic Escherichia coli in human immunodeficiency virus-related diarrhea in Senegal. J Infect Dis. 2004;189:75–8. doi: 10.1086/380489. [DOI] [PubMed] [Google Scholar]
- 11.Menard LP, Dubreuil JD. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1): a new toxin with an old twist. Crit Rev Microbiol. 2002;28:43–60. doi: 10.1080/1040-840291046687. [DOI] [PubMed] [Google Scholar]
- 12.Paiva de Sousa C, Dubreuil JD. Distribution and expression of the astA gene (EAST1 toxin) in Escherichia coli and Salmonella. Int J Med Microbiol. 2001;291:15–20. doi: 10.1078/1438-4221-00097. [DOI] [PubMed] [Google Scholar]
- 13.Savarino SJ, Fasano A, Robertson DC, Levine MM. Enteroaggregative Escherichia coli elaborate a heat-stable enterotoxin demonstrable in an in vitro rabbit intestinal model. J Clin Invest. 1991;87:1450–5. doi: 10.1172/JCI115151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eslava C, Navarro-Garcia F, Czeczulin JR, Henderson IR, Cravioto A, Nataro JP. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect Immun. 1998;66:3155–63. doi: 10.1128/iai.66.7.3155-3163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vila J, Vargas M, Henderson IR, Gascon J, Nataro JP. Enteroaggregative escherichia coli virulence factors in traveler's diarrhea strains. J Infect Dis. 2000;182:1780–3. doi: 10.1086/317617. [DOI] [PubMed] [Google Scholar]
- 16.Henderson IR, Nataro JP. Virulence functions of autotransporter proteins. Infect Immun. 2001;69:1231–43. doi: 10.1128/IAI.69.3.1231-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson IR, Hicks S, Navarro-Garcia F, Elias WP, Philips AD, Nataro JP. Involvement of the enteroaggregative Escherichia coli plasmid-encoded toxin in causing human intestinal damage. Infect Immun. 1999;67:5338–44. doi: 10.1128/iai.67.10.5338-5344.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fasano A, Noriega FR, Liao FM, Wang W, Levine MM. Effect of shigella enterotoxin 1 (ShET1) on rabbit intestine in vitro and in vivo. Gut. 1997;40:505–11. doi: 10.1136/gut.40.4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang DB, Okhuysen PC, Jiang ZD, DuPont HL. Enteroaggregative Escherichia coli: an emerging enteric pathogen. Am J Gastroenterol. 2004;99:383–9. doi: 10.1111/j.1572-0241.2004.04041.x. [DOI] [PubMed] [Google Scholar]
- 20.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala'Aldeen D. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev. 2004;68:692–744. doi: 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dutta PR, Cappello R, Navarro-Garcia F, Nataro JP. Functional comparison of serine protease autotransporters of enterobacteriaceae. Infect Immun. 2002;70:7105–13. doi: 10.1128/IAI.70.12.7105-7113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson IR, Czeczulin J, Eslava C, Noriega F, Nataro JP. Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect Immun. 1999;67:5587–96. doi: 10.1128/iai.67.11.5587-5596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjelloun-Touimi Z, Sansonetti PJ, Parsot C. SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol Microbiol. 1995;17:123–35. doi: 10.1111/j.1365-2958.1995.mmi_17010123.x. [DOI] [PubMed] [Google Scholar]
- 24.Provence DL, Curtiss R., 3rd Isolation and characterization of a gene involved in hemagglutination by an avian pathogenic Escherichia coli strain. Infect Immun. 1994;62:1369–80. doi: 10.1128/iai.62.4.1369-1380.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boisen N, Struve C, Scheutz F, Krogfelt KA, Nataro JP. New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect Immun. 2008;76:3281–92. doi: 10.1128/IAI.01646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol. 2006;61:1267–82. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 27.Czeczulin JR, Whittam TS, Henderson IR, Navarro-Garcia F, Nataro JP. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect Immun. 1999;67:2692–9. doi: 10.1128/iai.67.6.2692-2699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheikh J, Hicks S, Dall'Agnol M, Phillips AD, Nataro JP. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol Microbiol. 2001;41:983–97. doi: 10.1046/j.1365-2958.2001.02512.x. [DOI] [PubMed] [Google Scholar]
- 29.Olesen B, Neimann J, Bottiger B, Ethelberg S, Schiellerup P, Jensen C, Helms M, Scheutz F, Olsen KE, Krogfelt K, Petersen E, Molbak K, Gerner-Smidt P. Etiology of diarrhea in young children in Denmark: a case-control study. J Clin Microbiol. 2005;43:3636–41. doi: 10.1128/JCM.43.8.3636-3641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei J, Goldberg MB, Burland V, Venkatesan MM, Deng W, Fournier G, Mayhew GF, Plunkett G, 3rd, Rose DJ, Darling A, Mau B, Perna NT, Payne SM, Runyen-Janecky LJ, Zhou S, Schwartz DC, Blattner FR. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect Immun. 2003;71:2775–86. doi: 10.1128/IAI.71.5.2775-2786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orskov F, Orskov I. Escherichia coli serotyping and disease in man and animals. Can J Microbiol. 1992;38:699–704. [PubMed] [Google Scholar]
- 32.Guyer DM, Henderson IR, Nataro JP, Mobley HL. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol Microbiol. 2000;38:53–66. doi: 10.1046/j.1365-2958.2000.02110.x. [DOI] [PubMed] [Google Scholar]
- 33.Guignot J, Chaplais C, Coconnier-Polter MH, Servin AL. The secreted autotransporter toxin, Sat, functions as a virulence factor in Afa/Dr diffusely adhering Escherichia coli by promoting lesions in tight junction of polarized epithelial cells. Cell Microbiol. 2007;9:204–21. doi: 10.1111/j.1462-5822.2006.00782.x. [DOI] [PubMed] [Google Scholar]
- 34.Maroncle NM, Sivick KE, Brady R, Stokes FE, Mobley HL. Protease activity, secretion, cell entry, cytotoxicity, and cellular targets of secreted autotransporter toxin of uropathogenic Escherichia coli. Infect Immun. 2006;74:6124–34. doi: 10.1128/IAI.01086-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canizalez-Roman A, Navarro-Garcia F. Fodrin CaM-binding domain cleavage by Pet from enteroaggregative Escherichia coli leads to actin cytoskeletal disruption. Mol Microbiol. 2003;48:947–58. doi: 10.1046/j.1365-2958.2003.03492.x. [DOI] [PubMed] [Google Scholar]
- 36.Taddei CR, Moreno AC, Fernandes Filho A, Montemor LP, Martinez MB. Prevalence of secreted autotransporter toxin gene among diffusely adhering Escherichia coli isolated from stools of children. FEMS Microbiol Lett. 2003;227:249–53. doi: 10.1016/S0378-1097(03)00688-8. [DOI] [PubMed] [Google Scholar]
- 37.Al-Hasani K, Henderson IR, Sakellaris H, Rajakumar K, Grant T, Nataro JP, Robins-Browne R, Adler B. The sigA gene which is borne on the she pathogenicity island of Shigella flexneri 2a encodes an exported cytopathic protease involved in intestinal fluid accumulation. Infect Immun. 2000;68:2457–63. doi: 10.1128/iai.68.5.2457-2463.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy S, Thanasekaran K, Dutta Roy AR, Sehgal SC. Distribution of Shigella enterotoxin genes and secreted autotransporter toxin gene among diverse species and serotypes of shigella isolated from Andaman Islands, India. Trop Med Int Health. 2006;11:1694–8. doi: 10.1111/j.1365-3156.2006.01723.x. [DOI] [PubMed] [Google Scholar]
- 39.Parsot C. Shigella spp. and enteroinvasive Escherichia coli pathogenicity factors. FEMS Microbiol Lett. 2005;252:11–8. doi: 10.1016/j.femsle.2005.08.046. [DOI] [PubMed] [Google Scholar]
- 40.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–40. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 41.Boisen N, Struve C, Sheutz F, Krogfelt KA, Nataro JP. A new adhesin of Enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect Immun. 2008 doi: 10.1128/IAI.01646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine MM, Bergquist EJ, Nalin DR, Waterman DH, Hornick RB, Young CR, Sotman S. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet. 1978;1:1119–22. doi: 10.1016/s0140-6736(78)90299-4. [DOI] [PubMed] [Google Scholar]
- 43.Bilge SS, Clausen CR, Lau W, Moseley SL. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J Bacteriol. 1989;171:4281–9. doi: 10.1128/jb.171.8.4281-4289.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Restieri C, Garriss G, Locas MC, Dozois CM. Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl Environ Microbiol. 2007;73:1553–62. doi: 10.1128/AEM.01542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernier C, Gounon P, Le Bouguenec C. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect Immun. 2002;70:4302–11. doi: 10.1128/IAI.70.8.4302-4311.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]