Abstract
Toll-like receptors (TLRs) and members of their signalling pathway play an important role in the initiation of the innate immune response to a wide variety of pathogens1,2,3. The adaptor protein TIRAP mediates downstream signalling of TLR-2 and -44,5,6. We report a case-control genetic association study of 6106 individuals from Gambia, Kenya, United Kingdom, and Vietnam, with invasive pneumococcal disease, bacteraemia, malaria and tuberculosis. Thirty-three SNPs were genotyped, including TIRAP S180L. Heterozygous carriage of this variant was found to associate independently with all four infectious diseases in the different study populations (P=0.003, OR=0.59, 95%CI 0.42-0.83 for IPD; P=0.003, OR=0.40, 95%CI 0.21-0.77 for bacteraemia; P=0.002, OR=0.47, 95%CI 0.28-0.76 for malaria; P=0.008, OR=0.23 95%CI 0.07-0.73 for tuberculosis). Substantial support for a protective effect of S180L heterozygosity against infectious diseases was observed when the study groups were combined (N=6106, Overall P≤9.6×10-8). Tirap S180L was also shown to be functionally impaired in TLR2 signal transduction.
Toll-like receptors (TLRs) recognise a diverse array of pathogens and initiate intracellular signalling via their Toll/Interleukin-1 receptor (TIR)-domains, leading to differential effects on gene expression and the generation of an inflammatory host response. TLR2 has been shown to recognise, among other agonists, lipoteichoic acid, lipoarabinomannan and mycobacterial lipopeptides1, whilst TLR4 recognises lipopolysaccharide (LPS)7,8,9. Both TLRs 2 and 4 sense P. falciparum glycosylphosphatidylinositol (GPI) (Aucan et al. unpublished data)10. The adaptor protein TIRAP (TIR domain-containing adaptor protein), also known as MAL (MYD88 adapter-like), is essential for MYD88-dependent signalling downstream of TLR2 (with TLR1 and TLR6 as co-receptors) and TLR41,5. Following stimulation of TLR2 or TLR4, TIRAP triggers a signalling cascade which culminates in the activation of the transcription factor NF-κB and the subsequent activation of pro-inflammatory genes. Based on the central position of TIRAP in the TLR2 and TLR4 pathways, and knowledge of the microbial components which associate with these TLRs, we hypothesised that genetic variation at TIRAP might underlie susceptibility to common infectious diseases.
TIRAP spans 14500 base pairs on Chr11q24.2 and encodes a protein of 221 amino acids. Analyzing thirty-three SNPs in TIRAP and surrounding region in multiple populations found TIRAP S180L consistently associated with disease (Supplementary Note, Fig. 1, Table 1 and 2 online). TIRAP S180L genotyping was performed in a total of 6106 individuals with four different diseases: two United Kingdom (UK) populations of European ancestry with invasive pneumococcal disease (IPD); a Kenyan population with bacteraemia; three populations with malaria, (from The Gambia, Kenya, and Vietnam); and two populations with tuberculosis (from Algeria as well as the West African populations of The Gambia, Guinea-Bissau and the Republic of Guinea).
In the UK population, heterozygosity at TIRAP S180L was associated with protection from invasive pneumococcal disease (3×2 χ2=8.72, P=0.013, Table 1). An excess of mutant homozygotes amongst IPD cases (Table 1) was also observed in this UK population. TIRAP S180L was then examined in a separate group of UK individuals with thoracic empyema and a second control group. Although no association was observed between genotype and susceptibility to thoracic empyema overall (n=584, 3×2 χ2=0.63, P=0.73), analysis of the small subgroup of individuals with pneumococcal empyema revealed a non-significant trend towards association (3×2 χ2=5.05, P=0.080; Table 1). Interestingly, an excess of mutant homozygotes was again observed amongst this second group of IPD cases (Table 1).
Table 1.
TIRAP S180L genotype frequencies in individuals with infectious disease and controls
| Population | Status | Genotype distribution. No. individuals (%) |
P valuea [adjusted P valueb] |
Heterozygote advantage model OR (95%c.i.) | ||
|---|---|---|---|---|---|---|
| Ser/Ser | Ser/Leu | Leu/Leu | ||||
| UK | Control (1) | 527 (71.1%) | 199 (26.9%) | 15 (2.0%) |
0.013h [0.031] |
0.65 (0.44-0.97)c,d |
| IPD | 146 (76.4%) | 36 (18.8%) | 9 (4.7%) | |||
| Control (2) | 246 (68.1%) | 107 (29.6%) | 8 (2.2%) | 0.080i [0.080] |
0.74 (0.32-1.68)e | |
| Pneumococcal empyema | 25 (69.4%) | 8 (22.2%) | 3 (8.3%) | |||
| Kenyan | Control | 398 (94.1%) | 25 (5.9%) | 0 (0%) |
0.003j [0.003] |
0.34 (0.17-0.69) |
| Overall bacteraemia+ | 719 (97.6%) | 18 (2.4%) | 0 (0%) | |||
| Pneumococcal bacteraemiag | 161 (98.2%) | 3 (1.8%) | 0 (0%) |
0.024|| [0.056] |
0.30 (0.06-0.99) | |
| Gambian | Control | 172 (96.6%) | 6 (3.4%) | 0 (0%) |
0.025f [0.004] |
0.15 (0.043-0.55) |
| General malaria | 652 (99.1%) | 6 (0.9%) | 0 (0%) | |||
| Severe malaria | 413 (99.3%) | 3 (0.7%) | 0 (0%) |
0.024f [0.003] |
0.04 (0.004-0.34) | |
| Vietnamese | Control | 81 (95.3%) | 4 (4.7%) | 0 (0%) |
0.046|| [0.041] |
0.23 (0.056-0.94) |
| Severe malaria | 357 (98.9%) | 4 (1.1%) | 0 (0%) | |||
| Kenyan | Control | 398 (94.1%) | 25 (5.9%) | 0 (0%) | 0.035k
|| [0.035] |
0.58 (0.31-1.09) |
| Severe malaria | 599 (96.5%) | 22 (3.5%) | 0 (0%) | |||
| West African(The Gambia, Guinea-Bissau, Republic of Guinea) | Control | 593 (98.0%) | 12 (2.0%) | 0 (0%) |
0.041f [0.013] |
0.23 (0.07-0.73) |
| Tuberculosis | 671 (99.4%) | 4 (0.6%) | 0 (0%) | |||
2×2 or 3×2 Chi-squared comparisons of genotypes, depending on the presence or absence of mutant homozygotes in the different populations.
Following logistic regression for confounding factors in the different diseases (sickle cell trait, age, ethnicity, and homestead for malaria; HIV status, malnutrition, and age for bacteraemia; HIV status, ethnicity, age, and sex for tuberculosis; comorbid conditions and age for IPD). The Hosmer-Lemeshow goodness of fit tests was not significant (P>0.05) for any of the logistic regression models, indicating that the phenotypes predicted by the logistic regression models fitted with the observed phenotypes.
Logistic regression using the wild-type Ser/Ser homozygotes as reference.
Odds ratio of the Leu/Leu homozygotes = 2.39, 95%CI: 0.95-5.92.
Odds ratio of the Leu/Leu homozygotes = 4.01, 95%CI: 0.65-17.66.
Two-tailed Fisher’s exact test.
The Kenyan pneumococcal bacteraemia cases are a subset of the Kenyan bacteraemia case population, and the result here is analysed against the same Kenyan controls. The data relating to pneumococcal cases was not included separately in the combined analysis; only the overall Kenyan bacteraemia case-control data was included.
3×2 Chi-square=8.72.
3×2 Chi-square=5.05.
2×2 Chi-square=9.05.
2×2 Chi-square=3.28
One tailed P values were used for replication sets.
Protection afforded by the heterozygote variant was seen for individuals suffering from both Gram-positive bacteraemia (2×2 χ2 =5.18, P=0.023, OR=0.40, 95%CI: 0.16-0.95) and Gram-negative bacteraemia (2×2 χ2 =6.82, P=0.009, OR=0.38, 95%CI: 0.17-0.85). Correction for the effects of HIV status, malnutrition, and age using binary logistic regression did not affect the observed associations.
We then studied TIRAP S180L in a second population with invasive bacterial disease, comprising Kenyan children with well-defined bacteraemia. Although the mutant allele was found to be less common in the Kenyan population than in UK individuals, the same pattern of association was observed. The TIRAP S180L heterozygotes were significantly more common amongst community controls (5.9%), compared to individuals with bacteraemia (2.4%) (2×2 χ2=9.05, P=0.003; Table 1). The heterozygote protective effect of the S180L locus was also significant within the subgroup of 164 Kenyan children with pneumococcal bacteraemia (Fexact=0.024, Table 1), thus replicating the findings in the UK studies.
In the Gambian malaria case-control study, TIRAP S180L heterozygosity demonstrated a significant protective effect against both general malaria (Wald=8.35, P=0.004, Table 1) and severe malaria (Wald=8.706, P=0.003, Table 1). This result was replicated in a second malaria case-control study, this time in a Vietnamese population whose design included only cases of severe malaria: TIRAP S180L heterozygotes were again found to be more prevalent among the controls (4.7%) compared to the severe malaria cases (1.1%) (Fexact=0.046, Table 1). An attempt to replicate this in a third malaria case-control study (from Kenya) demonstrated similar results (2×2 χ2 =3.28, P=0.035; Table 1).
Finally, the possible effect of the TIRAP S180L polymorphism on susceptibility to tuberculosis was examined in 1280 individuals from The Gambia, Guinea-Bissau and the Republic of Guinea (collectively known as the EUTB West African study), again with ethnically matched controls. The TIRAP S180L heterozygous genotype was found to be protective against clinical tuberculosis (Wald=6.23, P=0.013). This finding was then replicated in two family-based studies on tuberculosis: from the EUTB West African study, as well as from Algeria. In both studies, the variant TIRAP S180L was found to be under-transmitted in subjects with clinical tuberculosis compared to subjects free from clinical tuberculosis (TRANSMIT P=0.075 and P=0.038 respectively; Table 2).
Table 2.
Transmission Disequilibrium Test (TDT) results of TIRAP S180L in West Africa and Algeria
| Algeria | ||||
| All sibs | One sib | |||
| Genotype | Observed | Expected | Observed | Expected |
| 180-SS | 159 | 150.92 | 74 | 70.346 |
| 180-SL | 15 | 23.079 | 10 | 13.654 |
| P || | 0.020 | 0.038 | ||
| West Africa | ||||
| All sibs | One sib | |||
| Genotype | Observed | Expected | Observed | Expected |
| 180-SS | 259 | 258.23 | 259 | 258.23 |
| 180-SL | 1 | 1.779 | 1 | 1.779 |
| P || | 0.075 | 0.075 | ||
| Combined Algerian and West African family study: P||=0.016 | ||||
| Combined data from all studies: P≤9.6×10-8. | ||||
One tailed P values were used.
When all the study groups are combined and stratified for the different populations, there is a clear association between heterozygosity at the TIRAP S180L locus and protection against multiple infectious diseases (N=6106, P≤9.6×10-8). To our knowledge, there are no other samples where we (or others) have tried and failed to replicate this finding. Studies on the functional effect of the S180L variant were consequently undertaken.
We first assayed the effect of the S180L polymorphism on TLR2 signalling using transfected murine embryonic fibroblasts (MEFs) from Tirap knockout mice (Fig. 1). Treatment of wild type MEFs with the TLR2 ligand Malp2 induced I-κBα degradation at 30 and 50 minutes (first panel). This effect was absent in Tirap-deficient MEFs (second panel). However, transfection of Tirap knock-out cells with the wild-type 180-Ser fully reconstituted TLR2 signalling, with degradation of IκB-α occurring at 30 and 50 minutes post-treatment (third panel). Transfection with the mutant 180-Leu failed to result in IκB-α degradation following TLR2 stimulation (fourth panel). Both wild-type 180-Ser and variant 180-Leu were expressed at similar levels in the cells (not shown).
Figure 1.
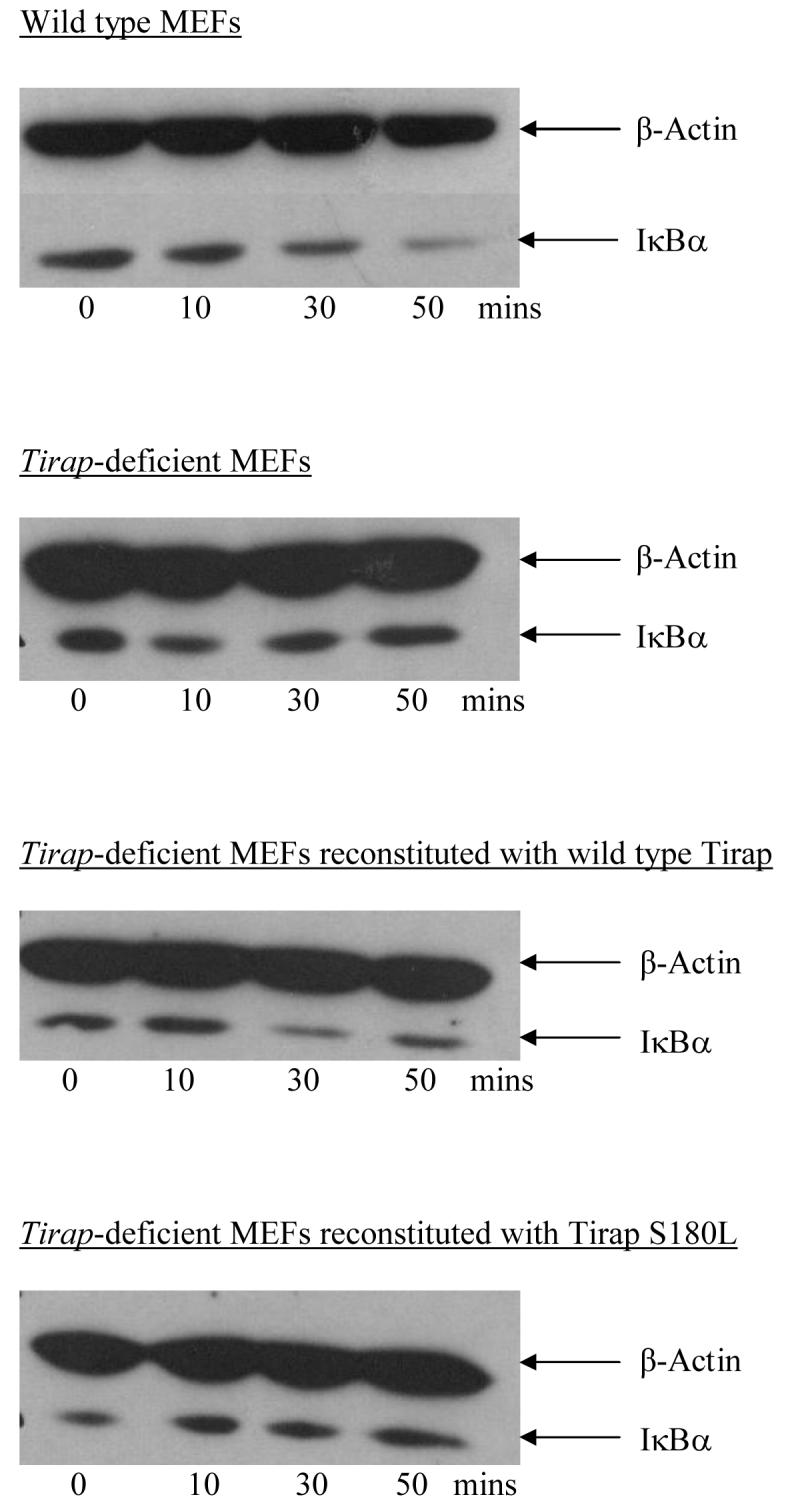
Macrophage-activating lipopeptide 2 (Malp2) induced, TLR2 signalling assay as measured by degradation of IκB-α at various time points. First panel: Malp2 (5nM) -stimulated wild-type murine embryonic fibroblast (MEF) cells. Second panel: Malp2-stimulated Tirap knockout cells. Third panel: Malp2-stimulated Tirap knockout cells transfected with Tirap 180-Ser. Fourth panel: Malp2-stimulated Tirap knockout cells transfected with Tirap 180-Leu. Samples were immunoblotted with an anti-I IκB-α antibody. Protein loading was controlled for using the β-actin housekeeping gene. Results shown are representative of 3 separate experiments.
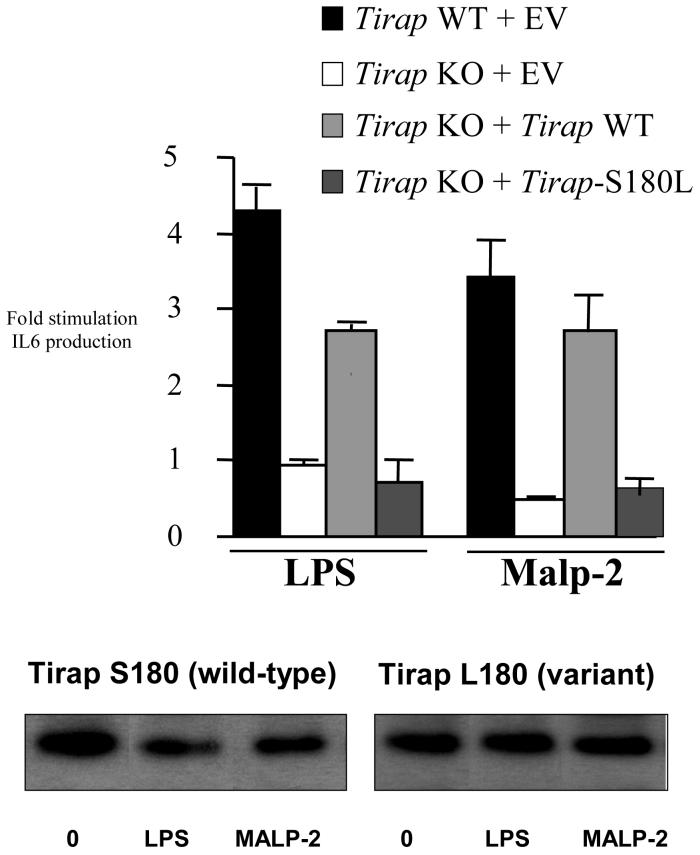
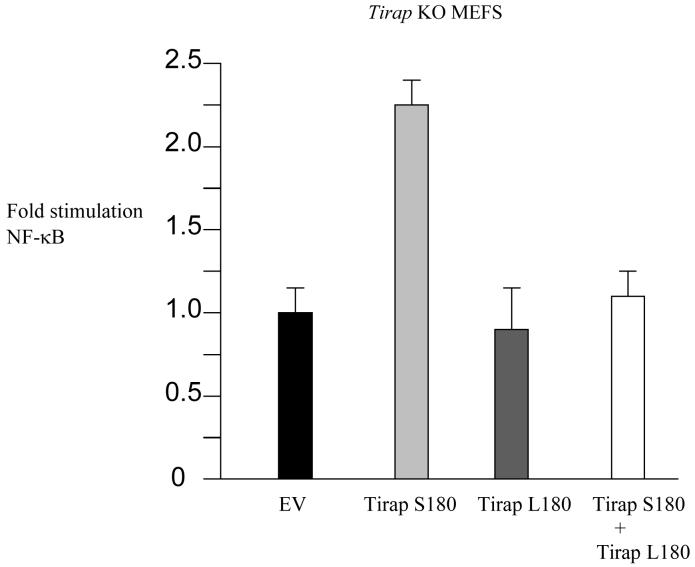
A similar lack of reconstitution was observed when IL-6 was measured, as shown in Fig. 2. Both LPS and Malp2 induced IL-6 in wild-type MEFs causing a 4-fold increase over untreated cells (first histobar in each set). The effect of both stimuli was however abolished in Tirap-deficient cells (second histobar in each set). Transfection of the Tirap-deficient cells with a plasmid encoding wild-type Tirap 180-Ser reconstituted the effect of both LPS and Malp2, leading to a 3-fold stimulation over untreated cells (third histobar in each set). The variant Tirap 180-Leu was however unable to reconstitute the effect of either stimulus (fourth histobar in each set). Both wild-type 180-Ser and variant 180-Leu were expressed at similar levels in the cells (lower panel). We also tested the functional consequences of co-expression of wild-type 180-Ser and variant 180-Leu in Tirap-deficient cells. As shown in Fig. 3, expression of wild-type 180-Ser Tirap activated an NF-κB-linked reporter gene (second histobar). The variant 180-Leu form was however inactive in cells lacking endogenous Tirap (third histobar), attesting further to the functional deficiency of this form of Tirap. Importantly, the variant form inhibited the ability of the wild-type form of Tirap to activate NF-κB (fourth histobar). This suggests that TIRAP signalling will be directly impaired in heterozygotes, since the variant might interfere with signalling by the wild-type form.
Figure 2.
Wild-type MEFs or Tirap knockout MEFs transfected with empty vector or plasmids encoding Tirap 180-Ser or Tirap 180-Leu as indicated, were treated with LPS (1μg/ml) or Malp2 (5nM). After 24h, supernatants were removed and assayed for IL-6 by ELISA. Expression levels of Tirap S180 and Tirap L180 in the cells analysed (untreated or treated with LPS or Malp2 for 24h) are shown in the lower panel. Results shown are mean ± s.e.m. of triplicate determinations, similar results being obtained in 2 further experiments.
Figure 3.
Tirap knockout MEFs were transfected with 100ng empty vector, or plasmids encoding Tirap 180-Ser or Tirap 180-Leu (50ng of each, supplemented to 100ng with 50ng of empty vector), or a combination of plasmids encoding Tirap 180-Ser and Tirap 180-Leu (50ng of each) as indicated, each in combination with luciferase reporter gene (80 ng per well 5 x NF-κB-luciferase) with Renilla-luciferase reporter gene (40 ng) as an internal control. Cell lysates were prepared as previously described4. Results shown are mean + S.D. from triplicate determinations.
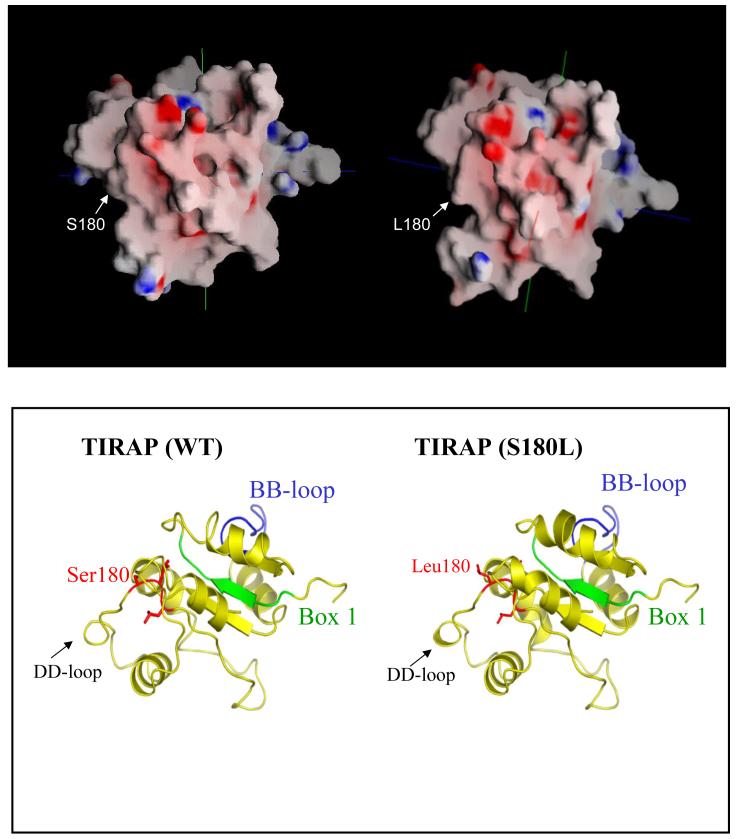
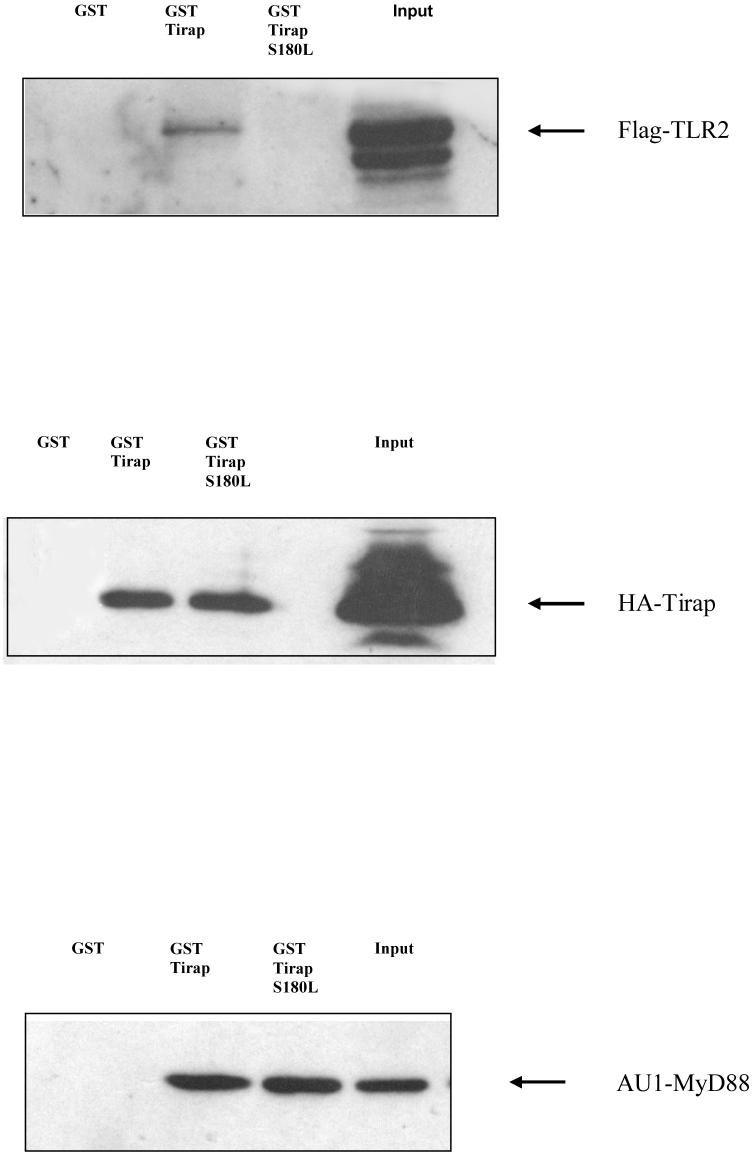
Wild-type TIRAP has been shown previously to interact with TLR211. We generated models of the wild-type and mutant protein in order to assess the effect of the S180L mutation on TIRAP structure. According to our model (Fig. 4), serine 180 is located on the edge of a surface-exposed loop opposite the BB-loop, the region that contains a critical proline residue that has been shown for other TIR containing proteins to form a point of contact with downstream signalling molecules. The model predicts that the electrostatic surface potential does not differ significantly between the wild-type and mutant protein. However the leucine residue appears to be more exposed (Fig. 4 upper panel). Previous modelling and interaction studies have suggested that TIRAP may bind to TLR2 via a region termed the DD-loop11. In our current model, Serine 180 is located in close proximity to this DD-loop (Fig. 4 lower panel). We therefore performed an in vitro binding assay in order to assess the effect of the mutation on the TIRAP: TLR2 interaction. As shown in Fig. 5 upper panel, a GST-tagged version of the TIRAP 180-Leu containing allotype, unlike wild-type TIRAP, failed to bind to TLR2 suggesting that the defect in reconstitution of signalling by TIRAP S180L occurs as a result of TLR2 not recruiting the variant. In addition to TLR2, TIRAP has also been shown to bind to both itself and MyD884. We therefore tested the effect of the S180L mutation on these interactions. As shown in Fig. 5, middle and lower panel, respectively, the S180L mutation had no effect on the interaction of TIRAP with both itself and MyD88.
Figure 4.
Molecular models of wild-type and mutant TIRAP. The computational utility Molecular Operating Environment (MOE2006.02) (www.chemcomp.com) was used to generate the models. Electrostatic surface potentials are highlighted in blue (positive) and red (negative) (upper panel). Structural features are highlighted in the lower panel. Box 1, the TIRAP BB loop and the position of Serine 180 are all indicated.
Figure 5.
HEK-293 cells (1×106) were transfected with 3 μg of Flag-tagged TLR2, HA-Tirap or AU1-tagged MyD88. Lysates were incubated with purified glutathione-coupled GST, GST-Tirap and GST-Tirap S180L for 2 hrs. After washing, the complex was analysed by SDS-PAGE and western blotting. Results shown are representative of 3 separate experiments.
Studies in Tirap-deficient mice have demonstrated impaired cytokine responses and NF-κB activation following stimulation with ligands at TLR4 and TLR2, as well as the TLR2 co-receptors TLR1 and TLR65,6. Unlike bacterial components1,12,13, the interaction of malaria parasites with TLRs is less well-described, although it has been demonstrated that P. falciparum-derived GPI is a ligand for both TLR2 and TLR410 (Aucan et al. unpublished data). The association between TIRAP variants and malaria in Gambian and Vietnamese populations presented here provides further evidence for a role of the TLR pathway in malaria pathogenesis. In addition, a role for the TLR2 system in host defence against malaria is also suggested from mutations in CD36, which associate with susceptibility to severe malaria14. Recently, CD36 has been shown to be a co-receptor for TLR215. Taken with the TIRAP variant presented here, the TLR2 system is clearly important for the host response in malaria.
On the basis of our co-transfection experiments (Fig. 3), the protective heterozygote state is likely to be associated with attenuated signaling and reduced NF-κB activation. This finding is consistent with increasing evidence that an excessive host inflammatory response may render individuals more vulnerable to developing severe forms of malaria and bacterial disease16,17,18. On the other hand, too little signaling would lead to an inadequate anti-pathogen response. This provides the opportunity for intermediate genotypes regulating signaling to be of optimal protective value. The mutant homozygotes are too rare to assess in African and Vietnamese populations, although in each of the UK studies there is a significant association with increased susceptibility to IPD amongst mutant homozygotes. This may have occurred by chance through analysis of the relatively small number of mutant homozygote individuals, or alternatively it may reflect a true heterozygote protective effect. Heterozygote protection against infectious disease is well recognized with examples such as sickle cell trait and malaria, prion protein gene variation and spongiform encephalopathy, and HLA and HIV/AIDS disease progression19,20,21. If the TIRAP 180-Leu homozygous state does indeed confer increased susceptibility to IPD, the mechanism by which this occurs is unclear, although we speculate that it may result in severely impaired or abolished signaling and increased susceptibility to severe infection, as has also been demonstrated in patients with rare functional mutations in IRAK4 and NEMO in the same pathway22,23. We suggest that heterozygosity at S180L may therefore confer a protective phenotype characterized by intermediate levels of pathway activation and an ‘optimal’, balanced inflammatory response. Additional host and microbial factors such as pathogen virulence and dose and prior immunity might be expected to interact with this model, as described in murine models of pneumococcal susceptibility24, and the optimal level of inflammatory response to a given exposure will likely vary depending on such factors.
The rarity of the mutant allele in African populations is also noteworthy. The burden of infectious disease mortality is very high in African populations, and this may strongly select against mutant homozygote individuals and act to reduce the 180-Leu allele frequency. The observed allele frequency may reflect a balance between protection afforded to heterozygotes and increased susceptibility to infectious disease in mutant homozygotes. It is also possible that the Leu allele may be subject to ongoing selective pressure and has not yet achieved fixation in the African population. Furthermore, it is conceivable that TIRAP S180L may influence susceptibility to other common infectious and inflammatory causes of death and so be subject to multiple selective pressures which may differ between human populations and ecological settings. Investigation of the role of this functional variant of TIRAP in susceptibility to other diseases may shed light on this area. We report here the first single nucleotide change that appears to impact on numerous major infectious diseases, roughly halving the risk in heterozygous individuals. This should represent a major evolutionary pressure as these diseases together account for over five million deaths each year in the developing world.
Supplementary Material
Acknowledgements
The authors would like to thank all the patients and the many investigators involved in the original case-control studies in Algeria, The Gambia, Guinea-Bissau, Republic of Guinea, Kenya, Vietnam and the UK for their contributions. This paper was published with the permission of the director of the Kenya Medical Research Institute (KEMRI). We thank Kate Fitzgerald, University of Massachusetts, Worcester, MA, USA, for the gift of the Mal-deficient fibroblasts. This work was funded by the Wellcome Trust, Science Foundation Ireland, Irish Health Research Board and the Agency for Science, Technology and Research (A-STAR), Singapore. CCK and RTG are scholars of A-STAR, and are members of the Bachelor of Medicine and Surgery (MBBS)-PhD programme, Faculty of Medicine, National University of Singapore. FOV is supported by the European Commission FP6 GenoSept grant and the UK ORS Scheme. SJC is a Wellcome Trust Clinical Research Fellow; AVSH is a Wellcome Trust Principal Fellow.
Appendix
METHODS
Sample collections
The UK IPD, thoracic empyema, Gambian malaria, West African tuberculosis and Kenyan bacteraemia study groups have previously been described in detail. Informed consent and written approval from the relevant ethical review boards has been granted for all sample collections. Overviews of the sample collections are presented in the online Supplementary Methods.
Sample genotyping
Direct sequencing of the TIRAP coding exons, a 1500 base pair region immediately 5′ to the transcription start site, and the 3′ untranslated region was performed in 48 Gambian individuals. The databases searched include dbSNP and Ensembl (www.ensembl.org). The TIRAP S180L polymorphism was genotyped using the Sequenom Mass-Array® MALDI-TOF primer extension assay for the Gambian malaria, Kenyan bacteraemia and UK IPD study groups25. Confirmation via direct sequencing with BigDye v3.1 terminator mix (ABI) was then performed on 12 randomly selected wild type and 12 heterozygous mutant samples from each of these populations. For the Vietnamese sample set, genotyping was performed using a site-directed mutagenesis assisted restriction fragment length polymorphism (RFLP) assay, and for the Gambian tuberculosis study group, genotyping was performed using the RFLP assay and confirmed by both the Sequenom Mass-Array® MALDI-TOF primer extension assay and direct sequencing. The rarity of the S180L polymorphism in the African populations studied increases the risk of bias due to genotyping error. To address this issue, all genotypes containing the mutant allele in these populations were confirmed by direct sequencing. The -80708, -68168, -52070, -29144, -23852, -18304, -12534, -5289, -4570, -1580, -386, -355, +177, +3977, +6032, +7685, R13W, S55N, Q101Q, S180L, A186A, V197I, +10122, +10610, +11347, +13466, +13955, +14472, +21162, +35464, +44402, +60339 and +71446 polymorphisms were genotyped via the Sequenom Mass-Array® assay. Details of the genotyping methodology for all markers genotyped above are available in the Supplementary Note and Methods, as well as appended in Supplementary Table 1 and 2 online. The individual assay details can be found in Supplementary Table 3 online. All control genotype frequencies were in Hardy-Weinberg equilibrium.
Statistical methods
Statistical analysis of genotype frequencies was performed with Chi-squared tests, two tailed Fisher’s exact tests if the Chi-squared test was inappropriate and forward, stepwise logistic regression, using the program SPSS. P<0.05 was considered significant. One tailed P values were computed if statistical testing was performed on a sample set replicating the same disease state. The overall, two-tailed P value for the combined analysis was computed using the method similar to that previously described26. Analysis of linkage disequilibrium was performed using the HaploView v3.2 program27. Analysis of transmission disequilibrium was performed using the program TRANSMIT28.
Functional analysis
Reporter Gene Assays
Tirap-deficient MEFs (2 × 104 cells/well) were seeded into 96-well plates, transfected as described4 on the following day with pcDNA3-Mal-HA or pcDNA3-MalS180L-HA as indicated (which was generated using the Quikchange™ kit (Stratagene)), in combination with luciferase reporter gene (80 ng well-1 κB-luciferase) and with Renilla-luciferase reporter gene (40 ng) as an internal control. Cell lysates were prepared as previously described4.
Reconstitution Assays
Murine embryonic fibroblasts (MEFs) were transfected with plasmids encoding Tirap, Tirap-S180L or empty vector using metafectene essentially as described by the manufacturers (Biontex). After 48 hrs, cells were treated with LPS (1μg/ml) or Malp2 (5 nM) for the indicated times. Immunoblot analysis was performed using a monoclonal anti-IκBα antibody, phosphor-p38 antibody or anti-β-actin as loading control (all from Santa Cruz, CA, USA) as indicated. IL-6 levels were measured by ELISA according to the manufacturer’s recommendation (R and D Systems).
GST-Pulldown Assays
The pGEX-Mal construct has been described previously11. The S180L point mutation was generated using the QuickChange® site-directed mutagenesis kit (Stratagene) according the protocol of the manufacturer. HEK293 cells were seeded (105 ml-1) onto 100 mm dishes 24 hours prior to transfection with 3 μg of Flag-tagged TLR2, HA-tagged Tirap or AU1-tagged MyD88 using Genejuice (Novagen) according to the manufacturer’s recommendations. 24 hours post-transfection cells were washed by the addition of 3 ml of ice-cold phosphate buffered saline (PBS). Cells were lysed at 4°C (1 hr) in buffer containing 150 mM NaCl, 2 mM EDTA, 10% glycerol, 1% NP-40, 0.2 mM phenylmethylsulfonyl fluoride, 0.2 mM Na3VO4 and 1μg of leupeptin ml-1. GST-pulldown assays were performed using the recombinant wild-type and mutant GST-Tirap proteins coupled to GSH-sepharose. Lysis extracts were incubated for 2 hours with the fusions indicated in the figure legends. The complexes were washed 3 times in lysis buffer and subjected to SDS-PAGE and western blotting. Monoclonal antibodies against the FLAG (12CA5) and AU-1 epitopes were obtained from Sigma and Berkeley Antibody Company (BAbCo), respectively. The polyclonal antibody against the HA-epitope tag (Y-11) was obtained from Santa Cruz Biotechnology.
Footnotes
Competing interests statement
The authors declare that they have no competing financial interests.
Accession Number
TIRAP is identified by MIM:606252 and GeneID:114609.
References
- 1.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunology. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 3.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–80. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald KA, et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001 Sep 6;413(6851):78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto M, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 2002;420:324–329. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
- 6.Horng T, Barton GM, Flavell RA, Medzhitov R. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. 2002;420:329–333. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- 7.Poltorak A, et al. Defective LPS signalling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 8.Hoshino K, et al. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–52. [PubMed] [Google Scholar]
- 9.Shimazu R, et al. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J Exp Med. 1999;189:1777–82. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnegowda G, Hajjar AM, Zhu J, Douglass EJ, Uematsu S, Akira S, Woods AS, Gowda DC. Induction of proinflammatory responses in macrophages by the glycosylphosphatidylinositols of Plasmodium falciparum: cell signalling receptors, glycosylphosphatidylinositol (GPI) structural requirement, and regulation of GPI activity. J Biol Chem. 2005 Mar 4;280(9):8606–16. doi: 10.1074/jbc.M413541200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunne A, Ejdeback M, Ludidi PL, O’Neill LA, Gay NJ. Structural complementarity of Toll/interleukin-1 receptor domains in Toll-like receptors and the adaptors TIRAP and MyD88. J.Biol.Chem. 2003;278(42):41443–51. doi: 10.1074/jbc.M301742200. [DOI] [PubMed] [Google Scholar]
- 12.Thoma-Uszynski S, et al. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science. 2001;291:1544–1547. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]
- 13.Malley R, et al. Recognition of pneumolysin by Toll-like receptor 4 confers resistance to pneumococcal infection. PNAS. 2003;100(4):1966–1971. doi: 10.1073/pnas.0435928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aitman TJ, et al. Malaria susceptibility and CD36 mutation. Nature. 2000 Jun 29;405(6790):1015–6. doi: 10.1038/35016636. [DOI] [PubMed] [Google Scholar]
- 15.Hoebe K, et al. CD36 is a sensor of diacylglycerides. Nature. 2005 Feb 3;433(7025):523–7. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 16.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 17.Annane D, Bellissant E, Cavaillon JM. Septic shock. Lancet. 2005;365(9453):63–78. doi: 10.1016/S0140-6736(04)17667-8. [DOI] [PubMed] [Google Scholar]
- 18.Cundell DR, Gerard NP, Gerard C, Idanpaan-Heikkila I, Tuomanen EI. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature. 1995;377:435–438. doi: 10.1038/377435a0. [DOI] [PubMed] [Google Scholar]
- 19.Dean M, Carrington M, O’Brien SJ. Balanced polymorphism selected by genetic versus infectious human disease. Annu Rev Genomics Hum Genet. 2002;3:263–92. doi: 10.1146/annurev.genom.3.022502.103149. [DOI] [PubMed] [Google Scholar]
- 20.Mead S, Stumpf MP, Whitfield J, Beck JA, Poulter M, Campbell T, Uphill JB, Goldstein D, Alpers M, Fisher EM, Collinge J. Balancing selection at the prion protein gene consistent with prehistoric kurulike epidemics. Science. 2003;300:640–3. doi: 10.1126/science.1083320. [DOI] [PubMed] [Google Scholar]
- 21.Carrington M, Nelson GW, Martin MP, Kissner T, Goedert JJ, Kaslow R, Buchbinder S, Hoots K, O’Brien SJ. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–52. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 22.Picard C, Puel A, Bonnet M, Ku CL, Bustamante J, Yang K, Soudais C, Dupuis S, Feinberg J, Fieschi C, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299:2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 23.Doffinger R, Smahi A, Bessia C, Geissmann F, Feinberg J, Durandy A, Bodemer C, Kenwrick S, Dupuis-Girod S, Blanche S, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat Genet. 2001;27:277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 24.Clatworthy MR, Smith KG. FcgammaRIIb balances efficient pathogen clearance and the cytokine-mediated consequences of sepsis. J Exp Med. 2004;199:717–23. doi: 10.1084/jem.20032197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jurinke C, van den Boom D, Cantor CR, Koster H. The use of MassARRAY technology for high throughput genotyping. Adv Biochem Eng Biotechnol. 2002;77:57–74. doi: 10.1007/3-540-45713-5_4. [DOI] [PubMed] [Google Scholar]
- 26.Altshuler D, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000 Sep 26;(1):76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Clayton D. Am J Hum Genet. 1999;65:1170. doi: 10.1086/302577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.